Published online Dec 28, 2017. doi: 10.3748/wjg.v23.i48.8626
Peer-review started: July 30, 2017
First decision: August 30, 2017
Revised: October 9, 2017
Accepted: November 8, 2017
Article in press: November 8, 2017
Published online: December 28, 2017
Processing time: 151 Days and 17.1 Hours
To critically evaluate previous scientific evidence on Fusobacterium’s role in colorectal neoplasia development.
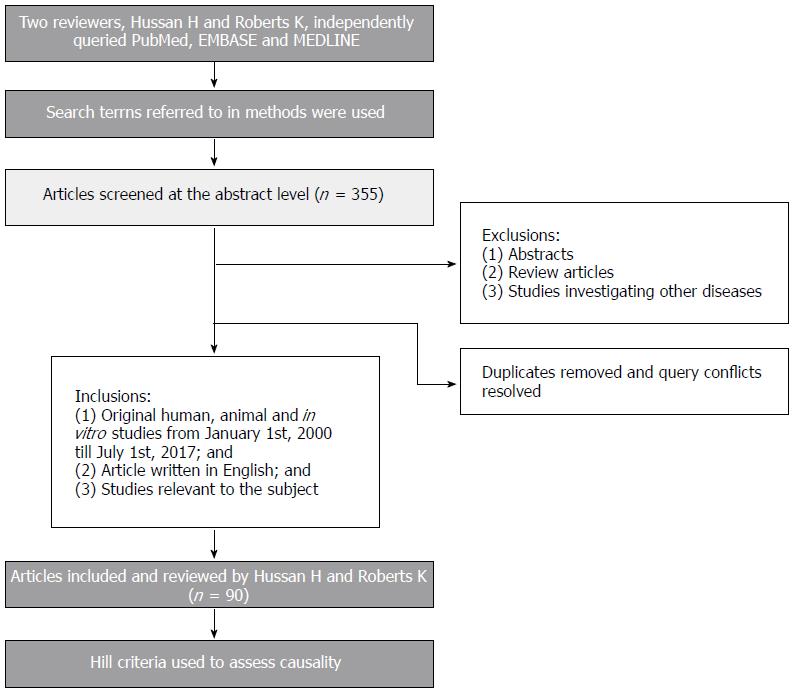
Two independent investigators systematically reviewed all original scientific articles published between January, 2000, and July, 2017, using PubMed, EMBASE, and MEDLINE. A total of 355 articles were screened at the abstract level. Of these, only original scientific human, animal, and in vitro studies investigating Fusobacterium and its relationship with colorectal cancer (CRC) were included in the analysis. Abstracts, review articles, studies investigating other colonic diseases, and studies written in other languages than English were excluded from our analysis. Ninety articles were included after removing duplicates, resolving disagreements between the two reviewers, and applying the above criteria.
Studies have consistently identified positive associations between Fusobacterium, especially Fusobacterium nucleatum (F. nucleatum), and CRC. Stronger associations were seen in CRCs proximal to the splenic flexure and CpG island methylator phenotype (CIMP)-high CRCs. There was evidence of temporality and a biological gradient, with increased F. nucleatum DNA detection and quantity along the traditional adenoma-carcinoma sequence and in CIMP-high CRC precursors. Diet may have a differential impact on colonic F. nucleatum enrichment; evidence suggests that high fiber diet may reduce the risk of a subset of CRCs that are F. nucleatum DNA-positive. Data also suggest shorter CRC and disease-specific survival with increased amount of F. nucleatum DNA in CRC tissue. The pathophysiology of enrichment of F. nucleatum and other Fusobacterium species in colonic tissue is unclear; however, the virulence factors and changes to the local colonic environment with disruption of the protective mucus layer may contribute. The presence of a host lectin (Gal-GalNAc) in the colonic epithelium may also mediate F. nucleatum attachment to CRC and precursors through interaction with an F. nucleatum protein, fibroblast activation protein 2 (FAP2). The clinical significance of detection or enrichment of Fusobacterium in colorectal neoplasia is ambiguous, but data suggest a procarcinogenic effect of F. nucleatum, likely due to activation of oncogenic and inflammatory pathways and modulation of the tumor immune environment. This is hypothesized to be mediated by certain F. nucleatum strains carrying invasive properties and virulence factors such as FadA and FAP.
Evidence suggests a potential active role of Fusobacterium, specifically F. nucleatum, in CRC. Future prospective and experimental human studies would fill an important gap in this literature.
Core tip: This is, to our knowledge, the first review to systematically examine the heterogeneous literature linking Fusobacterium to colorectal neoplasia. Accumulating evidence suggests that Fusobacterium, specifically Fusobacterium nucleatum (F. nucleatum), is more frequently detected in colorectal neoplasia, especially the pathway involving microsatellite instability. Multiple observational and animal experimental studies also suggest a procarcinogenic effect of F. nucleatum, likely due to activation of oncogenic and inflammatory pathways and modulation of the tumor immune environment. Virulence factors of F. nucleatum may contribute to its procarcinogenic effect. This information may be used to create novel strategies targeting colorectal cancer detection and chemoprevention.
- Citation: Hussan H, Clinton SK, Roberts K, Bailey MT. Fusobacterium’s link to colorectal neoplasia sequenced: A systematic review and future insights. World J Gastroenterol 2017; 23(48): 8626-8650
- URL: https://www.wjgnet.com/1007-9327/full/v23/i48/8626.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i48.8626
Colorectal cancer (CRC) is the most common gastrointestinal cancer, as well as the third leading cancer by incidence and mortality, in the United States[1]. The majority of CRC cases are sporadic, where a complex interaction among genetic and environmental factors impacts the carcinogenesis process. The underlying molecular changes follow at least two distinctive pathways of genomic dysfunction: the chromosomal instability (CIN) and CpG island methylator phenotype (CIMP)-high pathways. CIN is observed in 70%-85% of sporadic CRCs and describes aneuploidy due to gains or losses of whole or large portions of chromosomes[2,3]. CIN is likely due to defects in chromosome segregation pathways and is likely initiated by the adenomatous polyposis coli (APC) mutation with subsequent β-catenin/Wnt-signaling pathway activation[4]. APC mutation is followed by a cascade of molecular changes in a multistep fashion, as the flat mucosa evolves into a progressively larger adenoma that ultimately turns into cancer (adenoma-carcinoma sequence). CIMP-high CRCs include microsatellite instability (MSI)-high CRCs, with serrated polyps representing the main precursors[3]. These account for 15% of all CRCs and are characterized by inactivation of mismatch repair enzymes and other tumor suppressor genes via mutations or hypermethylation[5-8].
The human large intestine is a complex bacterial ecosystem that plays a significant role in health and disease. Increasing evidence suggests that a healthy symbiotic relationship between the host and microflora may be disrupted, leading to chronic metabolic and inflammatory changes promoting colorectal carcinogenesis[9,10]. Although the technology to define the microbiome continues to evolve, the prevalence of some bacteria appears to be elevated in CRC. These include Fusobacteria, Alistipes, Porphyromonadaceae, Coriobacteriaceae, Staphylococcaceae, Akkermansia, and Methanobacteriales. Conversely, other bacteria exhibit reduced prevalence in CRC, including Bifidobacterium, Lactobacillus, Ruminococcus, Faecalibacterium spp., Roseburia, and Treponema[11]. Although more research is warranted to establish firm causative links between CRC and flora diversity, patterns, specific microbial populations, and microbial functions, we are particularly intrigued by current data regarding Fusobacterium, a genus of the strictly anaerobic Fusobacteria phylum. Oral Fusobacterium consists mainly of the species Fusobacterium nucleatum (F. nucleatum), an adherent[12], invasive[13], and proinflammatory[14,15] bacterium that is linked to periodontal disease[16]. F. nucleatum is also the first anaerobic species to colonize the mouths of infants, indicating a potential prolonged exposure to F. nucleatum in adults who harbor it[17-19]. F. nucleatum is classified into subspecies animalis, fusiforme, nucleatum, polymorphum, and vincentii[20]. F. varium is another Fusobacterium species and has been associated with ulcerative colitis[21,22]. Other Fusobacterium species, such as F. naviforme, are mainly oral commensals and are associated with periodontal health[19,23]. The presence of Fusobacterium in the colon, specifically F. nucleatum, is increasingly linked to CRC through a variety of recent studies, albeit with significant heterogeneity in study methods and findings. Thus, a critical evaluation of the scientific literature regarding the link between Fusobacterium/F. nucleatum and CRC may contribute to the development of more comprehensive and novel studies to better define this relationship.
Two independent reviewers (HH and KR) systematically queried PubMed, Embase, and Medline using the following search terms: (“Fusobacterium ” {All fields} OR “Fusobacteria” {All fields}) AND (“colon” {All fields}), “rectum” {All fields}, “colorectal” {All fields}, “colorectal cancer” {All fields}, “polyps” {All fields}, “adenomas” {All fields}), “serrated” {All fields}, “SSA” {All fields}, “SSP” {All fields}, “CIMP” {All fields}, “MSI” {All fields}, OR “microsatellite” {All fields}). On the basis of this search, 355 articles were screened at the abstract level. The following inclusion criteria were used: (1) Original human, animal, and in vitro studies investigating Fusobacterium and colorectal neoplasia that were published between January 1, 2000, and July 1st, 2017; (2) articles written in English; and (3) studies relevant to colorectal neoplasia. We excluded: (1) abstracts; (2) review articles; and (3) studies investigating other colonic diseases, such as ulcerative colitis. Ninety original articles were included after removing duplicates, resolving disagreements between the two reviewers, and applying the above criteria. The resulting 90 articles were then independently reviewed at the manuscript level by HH and KR. We used the Hill criteria to assess causality in the current evidence linking F. nucleatum and CRC[24]. A brief illustration of our methods is shown in Figure 1.
Associations between Fusobacterium and CRC: Consistent case-control studies using various samples including both stool and fresh and formalin-fixed paraffin-embedded (FFPE) colonic tissue demonstrated increased detection, quantity, and/or relative percentage of Fusobacterium rDNA copies in CRC tissue compared with matched adjacent noncancerous tissue and compared with healthy controls without colorectal neoplasia, as summarized in Table 1[25-40]. The histopathology of these findings is ambiguous, but some data suggest that Fusobacteria have been observed within the colonic bacterial biofilms, in the colonic mucus layer, within colonic crypts, and invading the colonic epithelium[33,41-43]. F. nucleatum was the detected species of Fusobacterium in CRC tissue in 13 out of the 15 studies that presented species-level analysis[41,44-55]. In two out of three studies that presented subspecies-level analysis, F. nucleatum subspecies animalis was the most frequent subspecies of F. nucleatum in CRC tissue[40,54,56]. Other Fusobacterium species, such as F. periodonticum, F. varium, F. ulcerans F. necrophorum, and F. gonidiaformans, were also identified in CRC tissue in the five remaining studies[51,54,56-58]. F. nucleatum, F. periodonticum, F. varium, and F. ulcerans species can actively invade host cells, independently of mucosal compromise or presence of coinfection with other bacteria[59,60]. Conversely, F. necrophorum and F. gonidiaformans are termed passive invaders, and their presence in CRC could be due to the disruption of the mucus layer seen with CRC or to coinfection with other invasive bacteria. In the largest study comparing genes of Fusobacterium species, active invaders such as F. nucleatum were found to harbor larger genomes encode adhesions, and contain twice as many genes encoding membrane-related proteins compared with other Fusobacterial species termed passive invaders[59]. Thus, the presence of multiple Fusobacterial species could be due to their virulence and/or to early changes in the colonic environment that facilitate their presence in CRC tissue. Further studies are warranted to answer this question.
| Authors | Year | Cohort information | Specimen type | Detection method | Relation to CRC location | CRC features | Fusobacteria sequencing depth and associations with colorectal neoplasia |
| Ahn et al[25] | 2013 | United States cohort: 47 CRCs and 94 healthy controls. Matched for age, sex, BMI and hospital | Stool from cases and controls | 16S rRNA sequencing | - | - | Genus level: Fusobacterium rDNA was significantly more detected in of CRCs (31.9%) vs of controls (16%) |
| Vogtmann et al[26] | 2016 | United States cohort: 52 CRCs and 52 healthy controls, recruited between 1985-1987, matched by sex and BMI. Controls did not have a colonoscopy evaluation to rule out large polyps French validation cohort: 53 CRCs and 83 controls (61 healthy colons and 27 small adenomas) recruited from 2004-2006 | Stool from cases and controls | Whole-genome shotgun sequencing Compared to16S rRNA sequencing from Ahn et al[25] study | - | - | Genus level: Whole genome analysis: Fusobacterium rDNA was significantly more detected in CRCs (76.9%) vs controls (48.1%). 16S rRNA sequencing: Fusobacterium rDNA was significantly more detected in CRCs (31.9%) vs controls (16%) Genus level: Fusobacterium rDNA was significantly more detected in CRCs (10.08%) vs controls (0.01%) |
| Gao et al[27] | 2015 | Chinese cohort: 31 CRCs and 30 healthy controls | Fresh-frozen tissue from cancer, adjacent non-cancerous tissue and normal mucosa samples at the time of surgery/colonoscopy and after colonoscopy bowel preparation | 16S rRNA sequencing | Genus level: Fusobacterium rDNA was more detected in distal CRC compared to proximal CRC | - | Genus level: Higher relative percentage of Fusobacterium rDNA copies (relative to other bacterial rDNA) in CRCs (9.58%) vs adjacent non-cancerous tissues (0.57%) |
| Park et al[28] | 2016 | Korean cohort: 8 TAs, 10 SSA/Ps and 8 CRCs | Fresh-frozen tissue after colonoscopy bowel preparation | 16S rRNA sequencing | All CRCs were distal | - | Phylum level: Fusobacterium rDNA was detected in 37.5% of TAs, 50% of SSA/Ps and 100% of CRCs. Higher relative percentage of Fusobacterium rDNA copies in CRC tissue (33.8%) vs TA (4.3%) and SSA (1.9%). No difference in relative concentration of Fusobacterium rDNA copies between TAs and SSA/Ps |
| Feng et al[29] | 2015 | Austrian cohort: 46 CRCs, 44 advanced adenomas and 57 healthy controls. Ages between 45-86 yr, both genders and White race | Stool from cases and controls | Metagenomics Whole-genome shotgun sequencing | - | - | Genus level: Higher relative percentage of Fusobacterium rDNA copies in CRCs vs Carcinoma in situ. Higher relative percentage of Fusobacterium rDNA copies in CRCs vs advanced adenomas vs controls Genus level: No association between relative percentage of Fusobacterium rDNA copies and anthropometric measures or diet (meat, fiber, vegetables and fruit intake) |
| Burns et al[30] | 2015 | United States cohort: 44 CRCs | Fresh-frozen tissue from cancer and adjacent non-cancerous tissue at the time of surgery and after bowel preparation | 16S rRNA sequencing | - | - | Species level: No specific species identification. Higher relative concentration of Fusobacterium rDNA copies in CRCs vs adjacent non-cancerous tissues. Correlated enrichment with Providencia Species level: No specific species identification. Tumor microbiome was enriched with genes encoding virulence and toxin proteins that were associated with and dependent on the presence of Fusobacterium and Providencia |
| Viljoen et al[31] | 2015 | South African cohort: 55 CRCs. 70.4% mixed ancestry, 14.8% White, 11.1% African, equal gender, 7 MSI-high (4 CRCs with Lynch syndrome), 3 MSI-low. 41.5% received chemo-radiation prior to sample collection. 18 FFPE CRCs that are MSI high (2 CRCs with Lynch syndrome) | Fresh-frozen tissue from cancer and adjacent non-cancerous tissue at the time of surgery and after bowel preparation. FFPE samples after bowel preparation | 16S rRNA sequencing Metagenomics | No association between number of Fusobacterium rDNA copies in CRC tissue and colon vs rectum location | Association between higher number of Fusobacterium rDNA copies in CRC tissue and MSI-H status | Species level: No specific species identification. Fusobacterium rDNA was detected in 82% of CRCs and 81% adjacent non-cancerous tissues with concurrent presence in 80% of paired CRC and adjacent tissue. Higher number of Fusobacterium rDNA copies in CRCs vs adjacent non-cancerous tissues.Species level: No specific species identification. Higher number of Fusobacterium rDNA copies in CRC tissue was associated with African race and age < 60. Species level: No specific species identification. Higher number of Fusobacterium rDNA copies in CRC tissue correlated with pks-positve E. coli in normal tissue; but no correlation with Enteropathogenic Escherichia coli, Streptococcus gallolyticus, Enterococcus faecalis, Enterotoxigenic Bacteroides fragili or afaC-positive E. coli |
| Zhou et al[32] | 2016 | Chinese cohort: 97 CRCs and 48 healthy controls. Age and sex matched | Fresh-frozen tissue from cancer, adjacent non-cancerous tissue and normal mucosa samples at the time of surgery/colonoscopy and after bowel preparation | 16S rRNA sequencing | No association between relative percentage of Fusobacterium rDNA copies in CRC tissue and colon vs rectum location | No association between relative percentage of Fusobacterium rDNA copies in CRC tissue and CEA, P53, EGFR, Ki67, CA199 or CRP | Species level: No specific species identification. Higher number of Fusobacterium rDNA copies correlated positively with presence of chronic inflammation in CRC tissue Species level: No specific species identification. Fusobacterium rDNA was detected in 72.16% in CRCs vs 67.01% of adjacent non-cancerous tissues, both higher than controls. Species level: Higher number of Fusobacterium rDNA copies correlated positively with that of Enterotoxigenic Bacteroides fragilis, E. faecalis in CRC tissue when compared to adjacent non-cancerous tissue |
| Dejea et al[33] | 2014 | United States cohort: 30 CRCs, 6 adenomas and 22 healthy controls Malaysian cohort: 21 CRCs and 1 adenoma. | Fresh-frozen, formalin fixed tissue from tumors (adenomas and cancers), adjacent normal tissue and normal mucosa samples at the time of surgery/colonoscopy after bowel preparation | 16S rRNA sequencing | Similar relative percentage of Fusobacterium rDNA copies (≥ 25%) in CRC tissue proximal to hepatic flexure vs more distal CRC | - | Phylum level: Fusobacterium rDNA was detected in 32% of CRC, none of the matched normal tissue or healthy controls |
| Marchesi et al[34] | 2011 | Netherlands cohort: 6 CRCs | Fresh-frozen tissue from cancer and adjacent non-cancerous tissue at the time of surgery and after bowel preparation | 16S rRNA sequencing | - | - | Genus level: Fusobacterium rDNA was more detected with also higher relative percentage of Fusobacterium rDNA copies in CRC compared to adjacent non-cancerous tissue |
| Thomas et al[35] | 2016 | Brazilian cohort: 18 rectal cancers (no prior neoadjuvant therapy) and 18 healthy controls | Fresh-frozen tissue from cancer and normal mucosa samples at the time of surgery/colonoscopy and after bowel preparation | 16S rRNA sequencing | Rectal cancers only | - | Species level: No specific species identification. Higher number of Fusobacterium rDNA copies in rectal cancers compared to normal controls |
| Wang et al[36] | 2012 | Chinese cohort: 46 CRCs and 56 healthy controls | Stool, prior to bowel preparation | 16S rRNA sequencing | - | - | Genus level: Higher relative percentage of Fusobacterium rDNA copies in CRC tissue compared to controls |
| Gao et al[37] | 2017 | Chinese cohort: 65 CRC patients | Fresh-frozen tissue from cancer and matched adjacent non-cancerous tissue at the time of surgery after bowel preparation | 16S rRNA sequencing | Genus level: Fusobacterium rDNA was more detected in distal CRC compared to proximal CRC | No association between relative percentage of F. nucleatum rDNA copies in CRC tissue and presence of K-ras mutation | Phylum level: Fusobacterium rDNA was significantly more detected in CRC (8.5%) compared to matched normal tissue (4.13%) |
| Allali et al[38] | 2015 | United States and Spanish cohorts: 90 CRC patients | Fresh-frozen tissue from cancer and adjacent non-cancerous tissue at the time of surgery after bowel preparation | 16S rRNA | - | United States cohort: Higher relative percentage of Fusobacterium rDNA copies in both CRC and matched normal tissue in the right colon and splenic flexure vs left colon and sigmoid colon Spanish cohort: Higher relative percentage of Fusobacterium rDNA copies in CRC tissue in the left colon | Phylum level: Higher relative percentage of Fusobacterium rDNA copies in CRC vs matched normal tissue in Spanish cohort but not United States cohort. Higher relative percentage of Fusobacterium rDNA copies in matched adjacent non-cancerous tissue of the United States cohort compared to matched adjacent non-cancerous tissue of the Spanish cohort |
| Zackular et al[39] | 2014 | United States and Canadian cohort: 30 CRC, 30 TA and 30 healthy controls | Frozen fecal samples prior to colonoscopy and bowel preparation. | 16S rRNA | No relation between relative percentage of Fusobacterium rDNA copies in CRC tissue to CRC location | - | Genus level: Higher relative percentage of Fusobacterium rDNA copies in CRC compared to both adenoma and healthy controls |
| Zeller et al[40] | 2014 | French cohort (population F): 53 CRC, 42 TAs, and 61 healthy control patients. German cohort (Population G): 38 CRC patients. German, Danish and Spanish cohorts (Population H): 297 IBD and healthy controls. German cohort (48 CRC patients at the time of CRC surgery) | Populations F and G: Stool prior to colonoscopy bowel preparation Population H: Stool German cohort: CRC and matched normal tissue | 16S rRNA | - | - | Species level: Higher relative percentage of Fusobacterium rDNA copies in CRC compared to controls Subspecies level: F. nucleatum ssp. vincentii and F. nucleatum ssp. animalis are predominant in CRC tissue |
| Castellarin et al[41] | 2012 | Canadian cohort: 99 CRCs | Frozen tissue from cancer and adjacent non-cancerous tissue at the time of surgery after bowel preparation | Metagenomics F. nucleatum quantitative PCR | No association with proximal vs distal CRC location | Association between higher relative percentage of F. nucleatum rDNA copies in CRC and tumor involvement of more than 50% of the colon circumference | Subspecies level: Higher relative percentage of F. nucleatum subsp. Nucleatum rDNA copies in CRCs compared to matched normal tissues Species level: Confirmed in vitro invasion of F. nucleatum into human epithelial colonic cells Species level: No association between F. nucleatum and history of treatment or patient age |
| Chen et al[42] | 2017 | Chinese cohort: 14 CRCs 98 FFPE CRC | Frozen tissue at the time of surgery after bowel preparation FFPE CRC tissue after bowel preparation | 16S rRNA FISH F. nucleatum-targeted probe | Higher frequency of F. nucleatum rDNA detection in proximal CRC than distal location | - | 16s rRNA: Phylum level: Fusobacterium was a dominant phylum in CRC. FISH analysis. Species level: F. nucleatum rDNA was detected in 62.2% of FFPE CRC tissues No. difference in patients gender or age between CRCs that are F. nucleatum positive (detected) or absent |
| McCoy et al[43] | 2013 | United States Cohort: 10 CRCs, 48 adenomas and 67 healthy controls | Fresh-frozen normal rectal biopsies from adenoma and controls after bowel preparation Frozen tissue from CRC and adjacent non-cancerous tissue at the time of surgery after bowel preparation | F. nucleatum quantitative PCR 16S rRNA | - | Association between high number F. nucleatum rDNA copies in tissue and IL-6, IL-10, IL-17 and TNF-α in adenoma cases but no similar associations were seen in controls | Species level: Higher number of F. nucleatum rDNA copies seen in adenoma cases vs controls Species level: No association between F. nucleatum rDNA copy numbers and adenomas size/number Phylum level: Increased number of Fusobacterium rDNA copies in CRCs compared to matched normal tissues |
| Fukugaiti et al[44] | 2015 | Brazilian cohort: 7 CRCs and 10 healthy controls | Stool, prior to bowel preparation | 16S rRNA sequencing | - | - | Species level: Both F. nucleatum and Clostridium difficile had higher number of rDNA copies in stool of CRC patients when compared to controls |
| Warren et al[45] | 2013 | Canadian cohort: 65 CRCs | Frozen tissue from cancer and adjacent non-cancerous tissue at the time of surgery after bowel preparation | Metatran-scriptomics | - | - | Species level: Higher relative percentage of F. nucleatum rDNA copies in CRC compared to matched normal tissue Genus level: Co-occurrence of F. nucleatum with Campylobacter (in vitro co-aggregation with C. showae) and Leptotrichia in CRC tissue Genus level: Higher relative percentage of Fusobacterium rDNA copies in tumor tissue was correlated with host immune response genes and oncogenes |
| Ito et al[46] | 2015 | Japanese cohort: 138 Microvesicular HPs, 129 SSAs, 102 TSAs, 131 adenomas and 544 CRCs with matched adjacent non-cancerous tissue as well as 20 healthy controls | FFPE CRC tissue after bowel preparation | F. nucleatum quantitative PCR | No relation between F. nucleatum detection or higher number of rDNA copies in CRC and CRC location (Rectum to splenic flexure vs Transverse colon to cecum) Gradual increase in percentage of SSAs that are F. nucleatum positive from sigmoid colon to cecum; no similar finding seen for TA, TSA and HP | High number of F. nucleatum rDNA copies in CRC was associated with MLH1 methylation, CMP-high status and MSI-high status. No association between detection or number of F. nucleatum rDNA copies in CRC to KRAS mutation, PIK3A mutation or miRNA 31 expression | Species level: F. nucleatum rDNA was detected in 56% of CRCs. Higher number of F. nucleatum rDNA copies in CRC tissue compared to matched normal tissue. F. nucleatum rDNA was not detected in 17/20 of healthy controls; no significant difference in number of F. nucleatum rDNA copies between matched normal tissue and healthy controls Species level: F. nucleatum rDNA detected in 24% of HPs, 35% of SSAs, 30% of TSAs and 33% of TAs. No difference in frequency of F. nucleatum rDNA detection between these groups Species level: F. nucleatum rDNA more frequently detected in CRCs compared to polyps after adjustment for confounders Species level: High number of F. nucleatum rDNA copies in CRC was positively associated with large tumor size |
| Nosho et al[47] | 2016 | Japanese cohort: 511 CRCs | FFPE CRC tissue after bowel preparation | F. nucleatum quantitative PCR | - | F. nucleatum rDNA presence in CRC was associated with MSI high status | Species level: F. nucleatum rDNA was present in 8.6% of CRCs |
| Mima et al[48] | 2015 | United States cohort: 598 CRCs from the Nurses’ Health Study and Health Professionals Follow-up Study | FFPE CRC tissue after bowel preparation | F. nucleatum quantitative PCR | - | High number F. nucleatum rDNA copies in CRC tissue was associated with lower CD3+ T-cells density. No association with CD8+, CD45RO+, or FOXP3+ T-cells density in CRC. No significant association with Crohn’s-like histology, or tumor infiltrating lymphocytes | Species level: F. nucleatum rDNA was more frequently detected in CRCs (13%) compared to matched normal tissue (3.4%) |
| Kostic et al[49] | 2012 | Spanish, United States and Vietnamese cohort: 95 CRCs | Frozen tissue from cancer and adjacent non-cancerous tissue at the time of surgery after bowel preparation | Whole genome sequencing 16S rRNA F. nucleatum quantitative, PCR | No association with CRC location | No association with CRC purity, inflammation, necrosis, and vascularization | Species level: Higher relative percentage of Fusobacterium rDNA copies in CRC compared to matched normal tissue Detected species: F. nucleatum (most dominant species), F. necrophorum, F. mortiferum, and F. perfoeten. Species level: Higher relative percentage of Fusobacterium rDNA copies in Spanish vs US/Vietnamese cohorts Species level: No association with age, gender, ethnicity |
| Flanagan et al[50] | 2014 | Czech cohort: 49 CRCs German cohort: 45 CRCs. Irish cohort: | Frozen tissue from cancer, adjacent non-cancerous tissue at the time of surgery after bowel preparation. Stool 28 CRCs and 52 TAs Stool from 7 CRCs, 24 TAs patients (10 adenoma with HGDs, 12 TVAs and 2 adenomas) and 25 healthy controls | F. nucleatum quantitative PCR | No association between relative percentage of F. nucleatum rDNA copies in CRC and colon vs rectum location | Association between higher relative percentage of F. nucleatum rDNA copies in CRC tissue and TP53 mutation in the Irish cohort (Small sample size) Association between higher relative percentage of F. nucleatum rDNA copies in CRC tissue with KRAS mutation No association between F. nucleatum and BRAF mutation. No association between F. nucleatum and CRC grade | Species level: Higher relative percentage of F. nucleatum rDNA copies in CRC and HGD compared to matched normal tissue. Similar relative percentage of F. nucleatum rDNA copies in TA, TVA compared to their respective matched normal tissue Species level: Increased relative percentage of F. nucleatum rDNA copies during the adenoma-carcinoma progression in cancerous and matched normal tissue (TA to TVA to HGD to CRC) Species level: Increased relative percentage of F. nucleatum rDNA copies in stool of CRC patients compared to adenomas. Stool relative percentage of F. nucleatum rDNA copies is similar between adenoma and healthy controls patients. No significant correlation in relative percentage of F. nucleatum rDNA copies between disease tissue (CRC and adenomas) and stool samples from same patient |
| Wu et al[51] | 2013 | Chinese cohort: 19 CRCs and 20 healthy controls. Matched for age, sex and body mass index | Stool | 16S rRNA sequencing | Species level: Higher relative percentage of Fusobacterium rDNA copies compared to controls. Several species involved: F. nucleatum, F. periodonticum, F. necrophorum, F. ulcerans, F. varium, and F. gonidiaformans | ||
| Li et al[52] | 2016 | Chinese cohort: 101 CRC patients | Fresh-frozen tissue from cancer and adjacent non-cancerous tissue at the time of surgery after bowel preparation | F. nucleatum quantitative PCR | - | - | Species level: Increased relative percentage of F. nucleatum rDNA copies in 87.13% of CRCs compared to controls Higher relative percentage of F. nucleatum rDNA copies in CRC compared to controls |
| Mira-Pascual et al[53] | 2015 | Spanish cohort: 7 CRC, 8 TA, 7 healthy controls | Frozen fecal samples prior to colonoscopy and bowel preparation Biopsies from normal rectal mucosa of controls and neoplasm of cases after bowel preparation | F. nucleatum quantitative PCR | - | - | Species level: F. nucleatum rDNA more frequently present in fecal and tissue samples of tumor group (CRCs and polyps) compared to controls |
| Amitay et al[54] | 2017 | German cohort of patients aged 50 years old and above: 46 CRC, 113 advanced adenomas (TA > 1 cm in size, TVA, or with HGD), 110 adenomas, and 231 healthy controls | Frozen fecal samples prior to colonoscopy and bowel preparation. Median time between collection and storage was 7 d | F. nucleatum quantitative PCR | - | - | Subspecies level: Fusobacterium rDNA was more frequently present in CRC (54.3%) than advanced adenoma (23.9%), TA (23.6%) and healthy controls (25.1%) (P < 0.001) rDNA sequence of F. periodonticum was more detected in CRC compared to controls (P = 0.003). No difference in detection of rDNA of Fusobacterium simiae, F. nucleatum ssp. nucleatum, F. nucleatum ssp. animalis, F. nucleatum ssp. vincentii and F. nucleatum ssp. polymorphum between CRC and controls No significant difference between relative concentration of F. nucleatum rDNA copies in advanced adenomas/TAs vs Controls |
| Yu et al[55] | 2015 | Chinese cohort: 42 CRCs, 47 TAs and 52 healthy controls | Stool Left colonic biopsies | F. nucleatum quantitative PCR16S rRNA | - | - | Species level: Relative percentage of F. nucleatum rDNA copies per sample gradually increased from healthy control to TAs and to CRC. Results seen in both stool and tissue samples |
| Ye et al[56] | 2017 | United States cohort: 25 CRC patients | Fresh-frozen tissue from CRC and adjacent non-cancerous tissue at the time of surgery after bowel preparation | F. nucleatum quantitative PCR16S rRNA | - | Increased Chemokine (C-C motif) ligand 20 (CCL20) chemokine expression in all stages of CRC suggesting it is an early event in carcinogenesis. F. nucleatum ssp. Animalis induced CCL20 cytokine expression in CRC cell lines and monocytes. Monocytes are activated and migrate in the presence of F. nucleatum ssp. Animalis, this effect is inhibited by blocking CCL20 No control bacteria used in this experiment | Genus level: Increased relative percentage of Fusobacterium rDNA copies in CRC tissue vs normal matched tissue Species level: CRC samples contained F. periodonticum, F. canifelinum, F. varium, F. simiae, and F. nucleatum. F. nucleatum was the most frequently detected among Fusobacterium species Subspecies level: F. nucleatum ssp. Animalis most dominant among F. nucleatum subspecies in CRC samples |
| Chen et al[57] | 2012 | Chinese cohort: 46 CRCs and 56 healthy controls. BMI range 20-24, matched by sex | Stool and fecal swabs from cases and controls prior to bowel preparation Fresh-frozen tissue from cancer and adjacent non-cancerous tissue from cases at the time of surgery after bowel preparation | 16S rRNA sequencing | - | - | Species level: Increased relative concentration of F. varium rDNA copies in fecal swabs of CRCs compared to controls Genus level: Increased relative concentration of Fusobacterium rDNA copies in CRC tissues vs stool specimens (4.97% vs 0.47%, P < 0.001) Genus level: Unifrac PCA analysis found no difference in microbial composition of cancers and adjacent non-cancerous tissues |
| Kasai et al[58] | 2016 | Japanese cohort: 9 CRCs (3 invasive and 6 carcinoma in adenoma), 50 TAs and 49 healthy controls | Stool prior to colonoscopy bowel preparation | 16S rRNA sequencing | - | - | Species level: Increased relative percentage of F. varium rDNA copies in carcinoma in adenomas vs not detected in controls |
| Tahara et al[61] | 2014 | United States cohort: 149 CRCs and 89 adjacent tissues | Fresh-frozen CRC and adjacent non-cancerous tissue | Pan Fusobacterium and F. nucleatum quantitative PCR | - | Association with CIMP-high CRC, wild type TP53, MLH1 methylation, MSI-high and CHD7/8 mutation positivity | Genus and species level: Fusobacterium and F. nucleatum rDNA were more frequently detected in CRCs (74.3% and 52.3% respectively) compared to adjacent normal appearing mucosa (52.8% and 30.3% respectively). Higher relative percentage of Fusobacterium and F. nucleatum rDNA in CRC compared to normal appearing mucosa |
| Yazici et al[62] | 2017 | United States cohort: CRC (97 African Americans and 56 Whites). Healthy controls (100 African Americans and 76 Whites) | Fresh-frozen CRC, adjacent non-cancerous tissue and normal mucosa samples at the time of surgery/colonoscopy and after bowel preparation | F. nucleatum quantitative PCR 16S rRNA | - | - | Genera level: Fusobacterium was most abundant sulfidogenic bacteria identified in the study. No difference in relative concentrations of Fusobacterium rDNA copies between normal mucosa of cases and controls. Increased sulfidogenic bacteria in African Americans compared to non-Hispanic whites |
| Park et al[69] | 2017 | South Korean cohort: 160 MSI-high CRC. Excluded rectal carcinoma post neoadjuvant chemotherapy | FFPE CRC tissue after bowel preparation | F. nucleatum quantitative PCR | No association between relative concentrations of F. nucleatum rDNA copies in CRC and proximal versus distal/rectal CRC location | Association between high relative concentrations of F. nucleatum rDNA in CRC and BRAF mutation, CDKN2A (P16) promoter hypermethylation, tumor-infiltrating pan-macrophage density and CD68+ macrophages in tissue when compared with F. nucleatum-low/negative CRCs No association between high relative concentration of F. nucleatum rDNA in CRC tissue and CRC infiltrating CD3+ lymphocyte; PD-L1 expression status; Kras mutation; expression of MLH1, MSH2, MSH6 or PMS; CIMP status; or MLH 1 methylation when compared to CRCs that were F. nucleatum-low/negative | Species level: Of the MSI-high CRCs, 9% had high F. nucleatum rDNA relative concentrations, 58% had low F. nucleatum rDNA relative concentrations, and 33% of CRCs had no detected F. nucleatum in the tissue |
| Wei et al[70] | 2016 | Chinese cohort: 180 CRCs, all stages, median follow up 47 months | Fresh-frozen tissue from cancer and adjacent non-cancerous tissue at the time of surgery after bowel preparation | F. nucleatum quantitative PCR | No association between relative concentration of F. nucleatum rDNA copies in CRC and CRC location in colon vs rectum | High relative concentration of F. nucleatum rDNA copies in CRC was associated with high TNF-α, MMP9, NF-κB, β-catenin, CTNNB, KRAS and BRAF expression as well as lower MLH1 expression No association between relative concentration of F. nucleatum rDNA copies in CRC and COX1 or COX2 protein levels | Species level |
| Mima et al[72] | 2016 | United States cohort: 1,102 CRCs from the Nurses’ Health Study and Health Professionals Follow-up Study. Median follow up of 10.7 years | FFPE CRC tissue after bowel preparation | F. nucleatum quantitative PCR | Percentage of CRCs with high number of F. nucleatum rDNA copies increased gradually from rectum (2.5%) to cecum (11%) with a linear trend (P < 0.0001) | - | Species level |
| Mima et al[73] | 2015 | United States cohort: 1069 CRCs from the Nurses’ Health Study and Health Professionals Follow-up Study. Median follow up of 10.7 years | FFPE CRC tissue after bowel preparation | F. nucleatum quantitative PCR | Association with proximal CRCs location | Association between high number of F. nucleatum rDNA copies in CRCs and poor tumor differentiation Association between high number of F. nucleatum rDNA copies in CRCs and MSI-high CRC independent of CIMP and BRAF status Association between high number of F. nucleatum rDNA copies in CRC and MLH1methylation | Species level: |
| Yoon et al[78] | 2017 | North Korean cohort: 6 CRCs, 6 TAs, 6 SSAs and 6 healthy controls. Equal male female distribution | Normal rectal mucosa after bowel preparation | 16S rRNA | - | - | Species level: F. nucleatum found in only one SSA patient rectal biopsy |
| Kostic et al[79] | 2013 | United States and United Kingdom cohorts: 27 CRCs, 28 TAs and 31 healthy controls | Fresh-frozen tissue from adenomas and adjacent normal tissue at the time of colonoscopy after bowel preparation Stool | Fusobacterium quantitative PCR | - | - | Species level: No specific species identified Fusobacterium detected in 48% of adenomas. Increased number of Fusobacterium rDNA copies in adenomas compared to matched normal tissue Species level: No specific species identified Higher Fusobacterium detection and number of copies in stool from CRC and adenoma compared to controls |
No major associations were found between F. nucleatum and characteristics of CRC patients such as age, gender, ethnicity, body mass index (BMI), smoking, or alcohol consumption, except in one South African study, where researchers found an association between Fusobacterium and both African race and age less than 60[29,31,42,49,50,54,61,62]. In all studies, the prevalence of F. nucleatum rDNA in CRC tissue varied between 8.6% and 87.1%. This wide variability could be explained by heterogeneity in study design, sampling, analysis methodology, population, geographic location, or diet; these variations are summarized in Table 1[38,46-48,52,63,64].For instance, higher F. nucleatum detection is seen when using CRC tissue samples and whole-genome shotgun metagenomics sequencing methods, as compared with fecal samples or bacterial 16s sequencing[10,26,50]. Furthermore, caution should be taken when interpreting studies of FFPE samples, because FFPE samples provide a less accurate assessment of the microbiome when compared with fresh-frozen samples[65]. Finally, patients typically undergo bowel preparation when tissue samples are collected, which may affect Fusobacterium detection or abundance in tissue samples[66]. We conclude that consistent associations are seen between Fusobacterium, mainly F. nucleatum, and CRCs, with variable prevalence of F. nucleatum in CRC subjects, which is likely due to heterogeneous methodologies. It would be of value for future studies to utilize unprepared fresh-frozen colonic samples (for instance, unprepped flexible sigmoidoscopy with biopsies) when possible, combined with whole-genome shotgun metagenomic sequencing, in order to potentially yield more accurate detection and quantification of Fusobacterium and F. nucleatum.
Relation between Fusobacterium and dietary characteristics of CRC patients: Low-fiber, high-fat Western diet administration over 2 wk to 20 Native Africans was associated with altered colonic microbiome and increased number of F. nucleatum rDNA copies in colonic tissue, in association with increases in early colonic biomarkers of CRC[63]. It is interesting to note that colonic biopsies quantity of F. nucleatum rDNA copies did not decrease in 20 African Americans switched from a Western diet to a high-fiber, low-fat diet for 2 wk. This could be due to the small sample size, or it could take longer than 2 wk for dietary changes to reverse F. nucleatum rDNA abundance in colonic tissue[63]. Recently, vegetable consumption was also inversely associated with relative concentration of Fusobacterium rDNA in stool of patients with advanced adenomas[54]. However, the same study did not find associations between relative concentration of Fusobacterium rDNA in stools of 46 CRC patients and dietary habits such as consumption of red meat, processed meat, any meat, vegetables, or whole grains. This finding may be due to the cross-sectional nature of the study or to the researchers’ superficial assessment of dietary habits and use of fecal samples as opposed to colonic tissue in their study[54]. Mehta et al[64] prospectively investigated long-term dietary patterns in a cohort of 137217 patients using validated food frequency questionnaires. There were 1019 incidences of CRCs, which were classified in into F. nucleatum-positive or F. nucleatum-negative CRCs based on presence or absence of F. nucleatum rDNA in CRC tissue respectively. They identified that, when compared with a Western diet, a diet rich in whole grains and dietary fiber (prudent diet) was associated with a lower risk of F. nucleatum-positive CRCs, with a hazard ratio (HR) of 0.43 (95%CI: 0.25-0.72; P = 0.003). No associations between prudent diet and F. nucleatum-negative CRC risk was identified, indicating a differential impact of prudent diet on CRC risk that are F. nucleatum-positive specifically[64]. These inverse associations between prudent diet and F. nucleatum-positive CRCs were more pronounced when comparing high fiber intake (> 26 g/d for men and > 19 g/d for women) with the lowest fiber intake quartile (< 18 g/d for men and < 13 g/d for women; P = 0.04). Cereal-derived fiber had the strongest inverse association with F. nucleatum-positive CRCs (HR = 0.58; 95%CI: 0.34-0.99; P = 0.03)[64]. Fruit consumption was also shown to reduce the risk of both F. nucleatum-positive and F. nucleatum-negative CRCs, with no specific relation to F. nucleatum status of the CRC[64]. The researchers observed no impact of prudent diet subgroups (vegetables, legumes, or whole grains), on F. nucleatum-positive CRC risk, as also previously demonstrated[29,54,64]. Limitations to the Mehta et al[64] study include the use of FFPE as opposed to fresh colonic samples and the study’s observational design.
One explanation for the relationship between diet and colonic F. nucleatum is the potential impact of diet on oral Fusobacterium abundance. However, in a population-based case-control study, no associations were found between fiber intake and presence of oral Fusobacteria, and only modest positive correlations were found between consumption of saturated fatty acids, vitamin C, B vitamins, and vitamin E, on the one hand, and oral Fusobacteria abundance, on the other (P < 0.01)[67]. Furthermore, no associations were observed between oral Fusobacterium presence or abundance and CRC[68]. However, that could be due to the study design, whereby patients’ oral microbiomes were sampled after CRC resection and treatment; the study also lacks oral hygiene data[68].
In summary, diet may have a differential impact on colonic F. nucleatum enrichment, with increased abundance of F. nucleatum in the colons of patients consuming a Western diet. Long-term consumption of a high fiber diet may reduce the risk of a subset of CRCs that are F. nucleatum-positive. Future studies investigating the mechanisms of the impact of diet on colonic Fusobacterium and, subsequently, CRC risk are needed in order to determine dietary patterns targeting CRC chemoprevention and treatment. Furthermore, these data underline the importance of considering the impact of diet when investigating the links among Fusobacterium, other bacteria, and CRC.
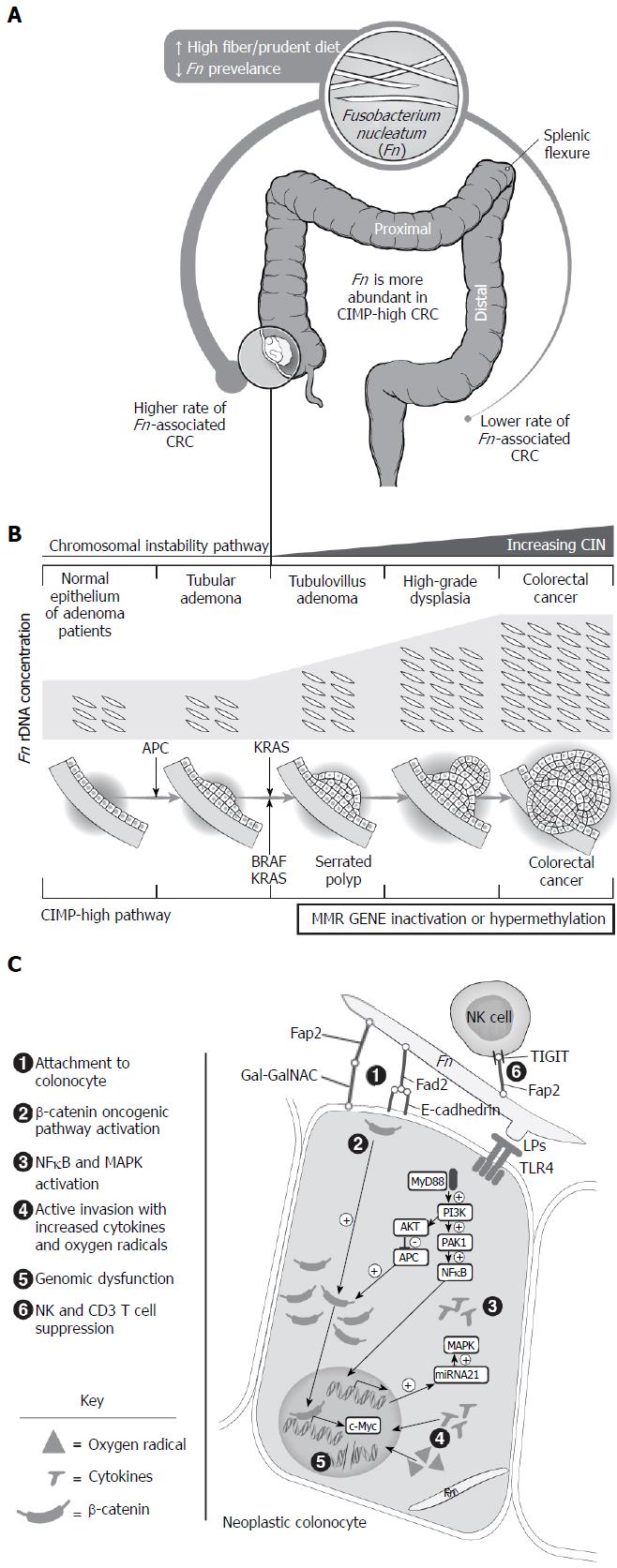
Fusobacterium associations with CRC anatomic location: Many previous publications have reported no difference in presence or relative percentage of Fusobacterium/F. nucleatum rDNA copies in tissue or stool with respect to CRC location, as illustrated in Table 1[31-33,39,43,46-48,69,70]. This could be due to varying definitions of high versus low F. nucleatum enrichment, unmeasured dietary confounders, and comparisons of colon versus rectum cancers, as opposed to proximal versus distal location. However, a few research teams have observed differences by CRC location. Yu et al[71] identified an increased F. nucleatum prevalence and relative concentrations in CRCs proximal to the splenic flexure as opposed to more distal CRCs[42]. A recent report by Mima et al[72,73] looked primarily at F. nucleatum enrichment in relation to CRC location and found significant relationships between F. nucleatum-high CRC and location. The study used FFPE samples from a large United States CRC cohort and found a gradual linear increment in CRCs that had high number of F. nucleatum rDNA copies from rectum to cecum (2.5% vs 11%, respectively; P < 0.0001)[72,73]. Contradictory findings were reported in two studies involving Chinese and Spanish cohorts with an increased detection and relative concentrations of Fusobacterium in CRCs distal to splenic flexure. These results could be due to small sample sizes, sampling bias, different geographic location and associated dietary patterns, or looking at Fusobacterium as opposed to F. nucleatum specifically[27,37,38]. The increased prevalence of F. nucleatum in proximal CRC coincides with the presence of invasive bacterial biofilms in 89% of right-sided colonic cancers and their surrounding normal mucosa, which may suggest a more active bacterial role in right CRC carcinogenesis[33]. Thus, current evidence is conflicting, but F. nucleatum may be more prevalent in CRC proximal to the splenic flexure, with a gradual increase in F. nucleatum-high CRCs from rectum to cecum. The increased F. nucleatum in proximal CRCs maybe due to F. nucleatum favoring anaerobic conditions, the presence of bacterial biofilms that facilitate its presence or to the differential impact of colonic lumen content on F. nucleatum abundance[63,64]. These associations are summarized in Figure 2A.
Fusobacterium associations with CRC molecular features: Associations have been observed between Fusobacterium/F. nucleatum and certain subsets of CRC, such as MSI-high and CIMP-high phenotypes (Table 1)[31,46,47,61,73]. F. nucleatum has also been associated with higher expression of BRAF and decreased MLH1 expression, both of which are seen in MSI-high sporadic CRCs[46,61,69,70,73]. KRAS mutations are usually associated with lower CRC methylation (CIMP negative), and conflicting results were seen when the relation of F. nucleatum to KRAS mutation was evaluated[37,46,50,69,70,74]. However, a large study by Ito et al[46] found no association between KRAS mutations and detection or number of F. nucleatum rDNA copies in CRC, which is consistent with an F. nucleatum predilection to CIMP-high CRCs. A recent, more in-depth investigation showed that presence of high number of F. nucleatum rDNA copies in CRC was associated with a 5-fold increased risk of having MSI-high CRCs, irrespective of CIMP-high status or BRAF mutation status[73]. This suggests that CRCs with MSI-high status are linked to F. nucleatum, whether owing to inherited, somatic, or epigenetic inactivation of MLH-1[5,6]. Further testing that was restricted to MSI-high CRCs showed that high relative concentrations of F. nucleatum rDNA was associated with CDKN2A (P16) promoter hypermethylation, a tumor suppressor gene associated with CIMP-high CRC[69].Thus, there is increasing evidence linking MSI-high CRC to Fusobacterium, but ambiguity exists regarding whether the increased detection of Fusobacterium is a cause or consequence of MSI-high status and associated molecular findings in colorectal neoplasia.
Fusobacterium associations with CRC stage and prognosis: Previous investigation has assessed Fusobacterium in relation to CRC staging and patient survival with variable results, as summarized in Table 2. High percentage of Fusobacterium rDNA copies in CRC tissue was associated with worse depth of invasion in two large studies that looked specifically at CRC prognosis[70,73]. Heterogeneity was seen when F. nucleatum abundance was evaluated in relation to lymph node metastasis[32,41,42,70,71,73]. None of the aforementioned studies found any associations between F. nucleatum and distal metastatic disease. Lastly, higher Fusobacterium rDNA was associated with more advanced CRC stage in 2 out of 11 studies, suggesting a lack of correlation between Fusobacterium abundance and the Duke’s or tumor, node, metastasis (TNM) staging classifications[31,32,41,46,50,72,73]. Conflicting observations were made when F. nucleatum was investigated as a predictor of CRC-specific survival. However, in two large studies by Wei et al[70] and Mima et al[73] with 10-year follow up, high quantity of F. nucleatum rDNA copies in CRC samples was associated with shorter CRC-specific survival after adjustment for multiple confounders. CRC-specific survival was assessed only secondarily in the other studies, with negative results[47,48,70,73]. Heterogeneous observations were made when Fusobacterium enrichment was assessed in relation to overall survival in CRC patients[41,46,50,70,73]. A comprehensive evaluation by Mima et al[73] adjusted for many confounders and found no association between high, low, or negative F. nucleatum rDNA copies presence in CRC tissue and CRC patients’ overall survival. The other two studies showing worsened overall survival had a shorter follow-up period and adjusted for fewer covariates than did Mima et al[50,70]. In one study, Fusobacterium subspecies were predominantly present in the goblet-like transcriptional CRC subtype[74-76]. This CRC subtype confers good prognosis in chemotherapy-untreated patients, but it has a detrimental effect on prognosis when adjuvant chemotherapy or chemoradiation are used[77]. Thus, CRC treatments, as well as CRC molecular subtypes, need to be investigated when looking at the impact of F. nucleatum presence on CRC survival. The above evidence is conflicting but suggests a more aggressive CRC biology with shorter CRC disease-specific survival periods in the presence of F. nucleatum; however, there are no relations between F. nucleatum and CRC staging. Thus, clarification is warranted of whether F. nucleatum modulation in CRC tissue is associated with better disease-free and CRC-specific survival rates after accounting for CRC molecular features and treatments.
| Authors | Sample size | CRC depth of invasion | CRC lymph nodes metastasis | CRC metastatic disease | CRC stage | CRC prognosis |
| Viljoen et al[31] | South African cohort: 55 CRCs. | - | - | - | Association between higher number of F. nucleatum rDNA copies and late stage CRC (stage III and IV compared to stage I and II) | - |
| Zhou et al[32] | Chinese cohort: 97 CRCs | No association with relative percentage of Fusobacterium rDNA copies | No association with relative percentage of Fusobacterium rDNA copies | No association with relative percentage of Fusobacterium rDNA copies | No association with relative percentage of Fusobacterium rDNA copies | - |
| Zackular et al[39] | United States and Canadian cohort: 30 CRC, 30 TA, 30 healthy controls | - | - | - | No association with relative percentage of F. nucleatum rDNA copies in CRC | - |
| Castellarin et al[41] | Canadian cohort: 99 CRCs | - | Association between relative percentage of F. nucleatum rDNA copies in CRC and regional lymph nodes metastasis | - | No association with relative percentage of F. nucleatum rDNA copies in CRC | No association to between relative percentage of F. nucleatum rDNA copies in CRC and CRC overall survival |
| Chen et al[42] | Chinese cohort: 98 CRCs | - | No association with presence of F. nucleatum rDNA in CRC | - | - | - |
| Ito et al[46] | Japanese cohort: 544 CRCs | - | - | - | No association with detection or number of F. nucleatum rDNA copies | No association between detection or number of F. nucleatum rDNA copies and CRC overall survival-unknown follow up period |
| Nosho et al[47] | Japanese cohort: 511 CRCs | - | - | - | No association with detection of F. nucleatum rDNA in CRC | No association between F. nucleatum rDNA presence in CRC and CRC-specific survival-unknown follow up period |
| Mima et al[48] | United States cohort: 598 CRCs. | - | - | - | - | No relation between F. nucleatum rDNA copies in CRC and CRC-specific survival or CRC overall survival- unknown follow up period |
| Flanagan et al[50] | Czech, German and Irish cohorts: 122 CRCs | - | - | - | No association with relative percentage of F. nucleatum rDNA copies | Higher relative percentage of F. nucleatum rDNA copies was associated with shorter CRC overall survival within 3-5 years follow up (HR = 19.96, 95%CI: 1.42-281.42) (no adjustment for other confounders) |
| Li et al[52] | Chinese cohort: 101 CRC | No association | Association between relative percentage of F. nucleatum rDNA copies in CRC and lymph nodes metastasis | - | No association with relative percentage of F. nucleatum rDNA copies in CRC | - |
| Amitay et al[54] | German cohort: 46 CRC | - | - | - | Relative percentage of F. nucleatum rDNA copies in CRC was associated with advanced stage [stage I vs II (P = 0.012) and stage I vs III (P = 0.042)] | - |
| Park et al[69] | South Korean cohort: 160 MSI-high CRC. | - | No association with F. nucleatum rDNA detection or number of copies | - | No association with F. nucleatum rDNA detection or number of copies | No association between F. nucleatum rDNA detection or number of copies and disease-free survival |
| Wei et al[70] | Chinese cohort: 180 CRCs | Association between high relative percentage of F. nucleatum rDNA copies in CRC and depth of invasion | Association between high relative percentage of F. nucleatum rDNA copies in CRC and lymph nodes metastasis | - | - | High relative percentage of F. nucleatum rDNA copies in CRC was associated shorter CRC overall survival within 3 years follow up [HR = 1.993 (1.024 to 3.879)] High relative percentage of F. nucleatum rDNA copies in CRC was associated with shorter CRC disease-free survival within 3 years follow up [HR = 1.829 (1.000 to 3.345)] |
| Yu et al[71] | Chinese cohort: 88 CRCs | - | F. nucleatum rDNA was more frequently detected in metastatic lymph nodes of proximal vs distal CRC F. nucleatum detected in 100% of metastatic lymph nodes compared to 40% of lymph nodes without metastasis (P < 0.001) | - | - | - |
| Mima et al[73] | United States cohort: 1069 CRCs | Association between number of F. nucleatum rDNA copies in CRC and higher pT of the TNM staging | No association with number of F. nucleatum rDNA copies in CRC | No association with number of F. nucleatum rDNA copies in CRC | No association with number of F. nucleatum rDNA copies in CRC | High number of F. nucleatum rDNA copies in CRC was associated with shorter CRC-specific survival within 10.7 years follow up [HR = 1.58 (1.04 to 2.39)] for F. nucleatum-high vs F. nucleatum-negative CRCs]. (Multivariable models included CRC stage, age, sex, year of diagnosis, family history of CRC, CRC location, MSI status, CIMP status, KRAS status, BRAF, PIK3CA and CRC LINE-1 methylation.) No association between F. nucleatum rDNA copies in CRC and CRC overall mortality |
F. nucleatum prevalence and enrichment was evaluated in CRC precursors in order to assess temporality and an earlier role in CRC carcinogenesis (Figure 2B and Table 1). The number of rDNA copies of F. nucleatum was higher in normal rectal/left colonic biopsies of patients with tubular adenomas (TAs) compared with controls[43,55]. The exception is a study that found no evidence of F. nucleatum in rectal biopsies of controls, TA patients, or CRC patients; this result is likely due to a small sample size[78]. Higher presence and relative percentage of Fusobacterium[29] and F. nucleatum[55,79] rDNA copies was seen in stools of patients with TAs compared with those of controls. On the contrary, two studies found no significant difference in relative percentage of Fusobacterium[39] and F. nucleatum[54] rDNA copies in fecal samples of patients with TAs, advanced TAs, and controls. Reasons for this discrepancy could include the use of fecal samples, which may represent transit from oral microbiome and may not necessarily correlate with true Fusobacterium abundance in colonic tissue, as well as absence of information on prior antibiotic use[10,26,50]. Detection and relative percentage of Fusobacterium[28] and specifically F. nucleatum[46,50,55] rDNA copies were found to increase in colonic tissue as it progressed through the CIN pathway (healthy control vs TA vs tubulovillous adenoma [TVA] vs high grade dysplasia vs CRC). Interestingly, no difference in relative percentage of F. nucleatum DNA copies was observed between TA and TVA tissue when compared with surrounding normal tissues[28,46,50]. This finding maybe be due to presence of bacterial biofilms or precancerous molecular changes in the surrounding normal mucosa, despite normal histological appearance, which may favor the attachment or invasion of Fusobacterium[33,80]. Data are limited but suggest no relation between adenoma size or burden and F. nucleatum rDNA copy numbers in rectal tissue[43]. Finally, F. nucleatum was associated with CIMP-high and right-sided sessile serrated adenomas/polyps (SSA/Ps) when SSA/Ps were compared with TAs[28,46,71]. Limitations to the above studies include an absence of information on concomitant preneoplastic tissue in patients who had hyperplastic polyps and the simultaneous use of FFPE and colonic preparation in specimens collected, which could reduce F. nucleatum detection[66,81]. All these data suggest that there may be a stepwise increase in F. nucleatum rDNA quantity and detection as colorectal neoplasms progress through the CIN pathway. Furthermore, F. nucleatum may play an earlier role in the CIMP-high CRC pathway. These results suggest temporality and a biological gradient of F. nucleatum in CRC development.
Despite the accumulating associations between Fusobacterium/F. nucleatum and colorectal neoplasia, establishing direct causality is challenging with the absence of prospective human studies supported by correlative laboratory science. In brief, multiple observational and animal experimental studies suggest plausible mechanisms by which F. nucleatum may contribute to CRC development, and these warrant additional investigation (Figure 2C).
F. nucleatum transmission to colorectal neoplastic tissue: Rats treated with 1,2-dimethylhydrazine (DMH) had increased Fusobacterium detection in tumors, whereas it was absent in nontreated controls, indicating a predilection of Fusobacterium to tumor tissue[82]. Oral administration of F. nucleatum into APCmin/+ and DMH-treated mice led to colorectal colonization and promoted colorectal neoplasia development, suggesting an active role of F. nucleatum in CRC neoplasia[55,79]. Similar findings were not seen in wild-type mice, suggesting that F. nucleatum can be contracted through oral ingestion if individuals are already predisposed to CRC. The mechanism by which F. nucleatum reaches the colonic epithelium are unclear. However, some F. nucleatum strains display the potential to disrupt the colonic mucosal barrier, suggesting that it can be transmitted from the colonic lumen to the epithelium, potentially causing colorectal disease[60]. Other Fusobacteria may take advantage of coinfection with other invasive bacteria or of disruption of the mucosal layer, seen with CRC. Another mechanism by which Fusobacteria home and localize to dysplastic colorectal epithelium is the blood-borne route[83]. In a novel study, a host lectin (Gal-GalNAc) was shown to mediate F. nucleatum attachment to CRC and precursor cells through interaction with an F. nucleatum protein, fibroblast activation protein 2 (FAP2)[83]. The expression of Gal-GalNAc is increased in a stepwise fashion in colorectal adenoma and matched surrounding normal tissue to villous adenomas with highest levels seen in CRC[83,84]. In a prior study, Gal-GalNAc was also more abundant in visually normal colonic epithelium of patients with CRC and its precursors, when compared to healthy controls[85]. Gal-GalNAc is mainly expressed in embryonic colonic goblet cells, and, in parallel, Fusobacterium was predominantly present in the goblet-like transcriptional CRC subtype[76,85-87]. The above data suggest that F. nucleatum can be localized through the lumen, or it can be blood borne. F. nucleatum’s preferential adherence to colorectal neoplasia maybe due to increased colonic epithelial Gal-GalNAc expression potentially due to goblet-like transformation of colorectal dysplastic epithelium. This increased Gal-GalNAc expression may explain F. nucleatum’s prevalence in visually normal colonic tissue of predisposed individuals, as well as F. nucleatum’s stepwise abundance through the adenoma-carcinoma sequence. Further evaluations confirming the goblet-like transformation of visually normal appearing colonic tissue of CRC patients in relation to Fusobacterium and bacterial biofilm formation are warranted.
F. nucleatum leads to increased expression of oncogenic and inflammatory factors early in CRC development: Stool metabolomics and CRC tissue inflammosome analysis supported associations between Fusobacterium[31,88], specifically F. nucleatum[45,89], and inflammatory metabolites as well as pathways implicated in colon carcinogenesis: Interleukin (IL) 6, IL8, IL10, IL17F, IL21, IL22, the Regenerating gene family, tumor necrosis factor (TNF), Matrix metallopeptidase 9 (MMP9) and Nuclear Factor kappa B (NF-κB)[70,76,90-92]. Quantity of F. nucleatum rDNA copies and inflammatory markers were both higher in visually normal rectal mucosa of adenoma patients compared with healthy controls[43,89]. Fluorescence in situ Hybridization (FISH) further confirmed the presence of F. nucleatum in the mucus layer and within colonic crypts of normal appearing colonic mucosa[43]. Bacterial biofilms were also found to cover normal appearing colorectal mucosa adjacent to CRC; and this was associated with an increase in colonic epithelial proliferation, IL6 and STAT3 activity as well as decreased E-cadherin in the normal appearing colonic epithelium[33]. All this suggests that F. nucleatum is associated with increased colorectal inflammation in CRC tissue. There is also an association between of presence of F. nucleatum rDNA and inflammation in visually normal appearing colorectal epithelium. The presence of inflammation in normal appearing colonic epithelium could potentially be due to presence of bacterial biofilms. These findings are interesting since inflammation is considered to be a marker of carcinogenesis which suggest a potential early role for F. nucleatum in carcinogenesis even prior to adenoma formation[93].
Indeed, data showed that incubation of F. nucleatum with CRC cell lines promotes proliferation and invasion of CRC cell in vitro and mice xenograft modules[91]. Experimental mouse data using APCMin/+ and DMH models are supportive showing that F. nucleatum administration increases the number and size of aberrant crypt foci and colorectal tumors, with activation of JAK/STAT and MAPK/ERK pathways critical for CRC development[55,79,91]. The mechanism for MAPK activation is thought to be due to recognition of F. nucleatum lipopolysaccharide by toll-like receptor 4 (TLR4) surface protein present on CRC cells leading to initiation of the TLR4/MYD88/NF-κB pathway, with subsequent binding of NF-κB to the micro RNA (miRNA)-21 promoter site[42,91]. This leads to increased expression of miRNA 21 which regulate RASA1 gene with subsequent activation of the MAPK pathway[91]. Similarly, F. nucleatum lipopolysaccharide possibly activates the TLR4/p21-activated kinase 1 (PAK1) cascade with subsequent increased β-catenin expression[42]. In parallel, it is proposed that F. nucleatum’s adhesion molecule, FadA, mediates induction of E-cadherin/β-catenin with subsequent abundance of target genes, such as C-myc and CCND1[89,91]. These proposed mechanisms are described in Figure 2C. In a recent study, F. nucleatum 2 equipped with FadA and FAP2 proteins did not increase inflammation or promote CRC in APCMin/+ nor in IL10-knockout mice, suggesting that FadA and FAP2 are necessary but not sufficient to promote CRC[94]. This could be due to some F. nucleatum strains having distinct virulence factors and/ or distinct lipopolysaccharides that are associated with more invasive and inflammatory behavior[60]. Functional pathway analysis supports this hypothesis, with increased bacterial virulence and motility protein pathways in F. nucleatum-invading CRC tumors[30]. Finally, F. nucleatum invasion and survival inside colorectal cells may cause increased production of reactive oxygen species[92,95]. The resultant activation of inflammatory cascades is hypothesized to induce DNA damage and epigenetic silencing of key targets, such as the mismatch repair gene MLH1, potentially leading to MSI seen frequently with F. nucleatum[46,61,89,90-92]. Additional investigation is warranted focusing upon the relationship between virulent Fusobacterium strains, specifically F. nucleatum, and induction of an inflammatory microenvironment that facilitates epigenetic and genetic alterations involved in early colorectal carcinogenesis.
F. nucleatum modulates the tumor immune microenvironment favorably towards carcinogenesis: Mounting evidence suggests that F. nucleatum modulates the microenvironment at the interface between the developing cancer and the host immune response. For instance, F. nucleatum rDNA abundance in tumor tissue was correlated with host immune response genes and oncogenes[45]. F. nucleatum can impact tumor T-cell abundance by inducing T-cell apoptosis, as well as by reducing T-cell proliferation, activation and response to certain mitogens and antigens[79,96-102]. This effect could be due to the FAP2 protein of F. nucleatum directly interacting with T-cell immunoreceptor with immunoglobulin (Ig) and ITIM domains (TIGIT), leading to the inhibition of natural killer (NK) cell-induced tumor cytotoxicity. Other tumor-infiltrating CD3+ T cells (CD4+ and CD8+) also have TIGIT and are possibly inhibited by FAP2[103]. This is consistent with the observation that Fusobacterium-high CRC cases are inversely associated with the density of CD3+ T cells, a type of T cell that is usually associated with better patient survival[48]. In parallel, Forkhead box P3 (FOXP3)-low T cells do not possess tumor suppressive activity and can secrete proinflammatory cytokines. FOXP3-low T-cell-infiltrated CRCs show increased expression of inflammation and immune-mediated genes such as IL12A, IL12B, transforming growth factor (TGF)-beta 1, and TNF, and they are associated with F. nucleatum abundance, paradoxically conferring better CRC-free survival[104]. F. nucleatum also recruits CD11b myeloid-derived immune cells, which are precursors to macrophages, consistent with the finding of increased tumor macrophages in the presence of F. nucleatum[69,79,105]. Furthermore, F. nucleatum induces activation of the CCL20/CCR6 axis in monocytes and CRC cells, potentially promoting monocyte migration and CRC development[56]. Thus, F. nucleatum abundance is associated with increased CD68 tumor-infiltrating macrophages, monocytes, and FOXP3-low T cells, but lower infiltration of CD3 lymphocytes. These findings support the hypothesis that F. nucleatum may exert an immunosuppressive effect in the cancer microenvironment that promotes the sustained survival of CRC cells. It may also explain the mystery of why the high load of MSI-induced antigens does not lead to immune eradication of MSI-high CRCs; this could be due to infiltration by F. nucleatum and associated immunosuppression. The relation between the immune microenvironment and prognosis is still controversial, and future studies linking bacteria such as Fusobacterium to survival through peripheral immune modulation are warranted.
The accumulating literature linking F. nucleatum to CRC led to efforts investigating the utility of F. nucleatum in CRC detection. Fecal-based F. nucleatum polymerase chain reaction (PCR) can serve as a noninvasive tool for CRC detection, with even better results when using digital PCR based on water-oil emulsion droplet technology[39,54,106-109]. Compared with PCR, loop-mediated isothermal amplification (LAMP) is a simple, noncostly and accurate method for bacterial testing that was shown to be more sensitive than PCR for F. nucleatum detection[110]. Two drawbacks of LAMP are the potential for false positivity and the complex design primer used. Metagenomic analysis of fecal microbiome across European and Chinese cohorts also showed that butyryl-CoA dehydrogenasegene F. nucleatum gene markers accurately distinguished CRC cases from controls, with area under the curve (AUC) = 0.84 and an odds ratio of 23[111]. Finally, Wang et al[112] demonstrated that F. nucleatum can also induce a serological anti-F. nucleatum-IgA immune response that is higher in CRC patients compared with patients with benign colonic polyps, those with inflammatory bowel disease, and healthy controls. In that study, the combination of anti-F. nucleatum-IgA and carcinoembryonic antigen (CEA) was found to better for diagnosing CRC compared with either one alone (sensitivity: 53.10%; specificity: 96.41%; AUC = 0.848).
The finding that diet can alter the microbiome and associated colonic carcinogenesis led to efforts investigating F. nucleatum modulation in CRC chemoprevention and therapeutics through the use of probiotics and herbals[63]. Probiotics including Bifidobacterium longum, Lactobacillus acidophilus and Enterococcus faecalis significantly reduced Fusobacterium levels by nearly 5-fold in CRC surgery patients when compared with placebo probiotics (10.08% vs 1.91%, respectively; P = 0.03)[113]. Limitations of that study include the variable length of probiotic treatment and the presurgery use of antibiotics and bowel preparation, which can alter the microbiome[113]. Berberine (BBR) is an isoquinoline alkaloid and a component of the Chinese herb Coptis chinensis. BBR was shown to prevent insulin resistance and obesity in mice fed a high-fat diet, in association with an impact on the intestinal microbiome[114]. Administration of BBR to APCMin/+ and DMH mice inoculated with F. nucleatum led to reduced tumorigenesis and Fusobacterium-induced activation of the JAK/STAT and MAPK/ERK pathways[55]. Both probiotics and herbals may provide tactics for modulating F. nucleatum, but the implications are still under investigation.
Fusobacteria are significantly more abundant in colorectal tissues and stools of patients with CRC than in healthy controls. The histopathology of these findings is ambiguous, but the few available data suggest that Fusobacteria have been observed within the colonic biofilms, the colonic mucus layer, colonic crypts, and inside the colonic epithelium. F. nucleatum has been associated with proximal CRCs and CRCs with MSI-high features, a finding warranting additional investigation. Findings also suggest temporality and a biological gradient with presence of fusobacteria in CRC precursors. Further, researchers have observed increased detection and quantity of F. nucleatum rDNA in the visually normal mucosa of colorectal neoplasia patients when compared with healthy controls. The pathophysiology and significance of this finding is unclear, as is its relation to cancer progression. Fusobacteria are usually indigenous to healthy mouth microbiota, highly adherent to teeth and oropharyngeal epithelium in the presence of a low viscous saliva environment, and unspecialized for viscous environment. Therefore, they are normally only transient in the colon, which is protected by a mucus layer. Disruption of the colonic mucus layer or coinfection with other invasive bacteria may facilitate the presence of Fusobacterial species in CRC tissue. Furthermore, some Fusobacterial strains, specifically F. nucleatum, are considered active invaders, giving them the potential to disrupt an intact colonic mucosal barrier and potentiate colorectal disease. The presence of a host lectin (Gal-GalNAc) in the colon may also mediate F. nucleatum blood-borne transmission and attachment to CRC and precursors through interaction with an F. nucleatum protein, FAP2. F. nucleatum was demonstrated to have cancer-promoting properties in several rodent models, supporting its role in the human colon cancer cascade. This is thought to be due to its activation of inflammatory and oncogenic pathways associated with colon carcinogenesis, as well as its modulation of the immune microenvironment in a manner that favors cancer progression. The lack of prospective human studies is a large limitation of current literature regarding the temporality of Fusobacterium and cancer; most human studies to date were cross-sectional case-control studies. Thus, more evidence is needed to confirm causality and inform future detection and therapeutic efforts targeting F. nucleatum and other microbiota involved in CRC.
The presence of Fusobacterium, specifically Fusobacterium nucleatum (F. nucleatum), in the colon is increasingly linked to colorectal cancer (CRC). However, significant heterogeneity in study methods and findings poses challenges to interpretation. An evaluation of this rapidly expanding literature will help direct future studies to answer unresolved questions and to avoid previous design pitfalls in order to further our knowledge in this exciting field.
A critical evaluation of the scientific literature regarding the link between Fusobacterium/F. nucleatum and CRC may contribute to the development of more comprehensive and novel studies to better define this relationship and its potential applications in CRC treatment and prevention.
This systematic review evaluated the clinical and experimental evidence linking Fusobacterium and CRC. The authors reviewed studies investigating the relationship between Fusobacterium and the following variables: CRC, CRC patients’ characteristics and dietary patterns, CRC anatomic location, CRC molecular features, and CRC stage and prognosis. The authors also reviewed studies looking at presence of Fusobacterium in pre-neoplastic lesions, as well as experimental evidence testing the procarcinogenic potential of Fusobacterium. Finally, the authors looked at the implications of Fusobacterium for CRC detection and treatment. Elucidating these heterogeneous studies may impact our understanding of the relationship between Fusobacterium and CRC, as well as improve detection and chemoprevention tactics for CRC.
This is, to our knowledge, the first systematic review of the scientific evidence surrounding the link between Fusobacterium and CRC. Using PubMed, Embase, and Medline, the authors systematically reviewed all original studies investigating Fusobacterium/F. nucleatum and CRC published between January 1st, 2000, and July 1st, 2017. All abstracts were screened to identify original human, animal, and in vitro research. Out of the 355 articles that were screened, 90 articles were included in this review. Articles were excluded if diseases other than CRC were included and if they were written in languages other than English. All review articles and citations including only an abstract were excluded from analysis.
An accumulating body of evidence supports the hypothesis that Fusobacterium, especially F. nucleatum is more frequently detected in colorectal neoplasia, especially the microsatellite instability neoplastic pathway and proximal CRC. Studies investigating F. nucleatum in colorectal precancerous tissue suggest temporality and a biological gradient; however, ambiguity still exists on whether this increased detection of Fusobacterium is a cause or consequence of colorectal neoplasia. Diet may have a differential impact on colonic F. nucleatum enrichment, high fiber diet potentially reducing the risk of a subset of CRCs that are F. nucleatum-positive. Evidence also suggests a shorter CRC disease-specific survival in the presence of F. nucleatum, albeit with no relations between F. nucleatum and CRC staging. The homing of Fusobacteria and F. nucleatum to the colonic epithelium maybe partly due to increased Gal-GalNAc expression on colonic cells, virulence factors of F. nucleatum and other Fusobacteria, and changes to the local colonic environment with disruption of the protective mucus layer. Experimental evidence suggests that Fusobacterium nucleatum has a procarcinogenic potential that is likely mediated by activation of oncogenic and inflammatory pathways, as well as modulation of the tumor immune environment. The lack of prospective human studies is a large limitation of current literature. Furthermore, it will be essential to further delineate mechanisms and timing of Fusobacterium homing to the colonic mucosa, as well as its relation to cancer progression. This review may be used to develop hypotheses for novel strategies targeting colorectal cancer detection and prevention. Future robust analysis would also benefit from adjusting for confounders, such as Fusobacterial strain virulence factors, colonic preparation, antibiotic use, and diet.
Accumulating evidence supports the hypothesis that F. nucleatum may enhance colorectal carcinogenesis, especially the neoplastic pathway involving defects in microsatellite instability. Virulence factors of F. nucleatum may contribute to its procarcinogenic effect. The lack of prospective human studies is a large limitation of current literature regarding the link between Fusobacterium and CRC. This review may be used to guide novel strategies targeting colorectal cancer detection and prevention.
This review gathers an ample evidence implicating Fusobacterium in CRC etiology and highlights the promising global efforts aimed at testing the role of Fusobacterium in CRC detection, chemoprevention and outcomes. There are multiple gaps in knowledge and the current evidence lacks prospective human studies. To advance our knowledge, future prospective studies need to clarify the timing and mechanisms of Fusobacterium transmission to the colon in relation to colorectal carcinogenesis and histopathology of these findings. In order to potentially design future CRC therapies, additional investigations are also warranted to delineate the relationships between virulent Fusobacterium strains, specifically F. nucleatum, and induction of inflammatory, pro carcinogenic and immune mechanisms involved in early colorectal carcinogenesis. Furthermore, future efforts need to prospectively test the impact of diet, probiotics and other chemopreventative agents on colonic Fusobacterium and whether modulation of colonic presence/concentrations of Fusobacterium will alter the risk or outcomes of CRC. Finally the role of Fusobacterium in CRC screening is intriguing and studies combing Fusobacterium testing with other CRC screening methods such as stool DNA testing or even colonoscopy may potentially improve CRC detection and preventative efforts. This study outlines the significant heterogeneity in the methods used and the need for more consistent design. In order to attain more robust results, the authors suggest future studies to: (1) use a blinded prospective randomized controlled design and/or large sample size when possible; (2) perform better sampling by collecting unprepared colonic tissue stored as fresh frozen samples; (3) have more detailed microbiome sequencing using whole-genome shotgun metagenomic sequencing, FISH and other methods in order to assess Fusobacterium sub-species concentrations, the presence of virulence factors and location of Fusobacterium in relation to the colonic epithelium; and (4) adjust for potential dietary, geographic and racial variables that may have an impact on Fusobacterium/F. nucleatum presence or concentration within specimens.
| 1. | DeSantis CE, Lin CC, Mariotto AB, Siegel RL, Stein KD, Kramer JL, Alteri R, Robbins AS, Jemal A. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin. 2014;64:252-271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1848] [Cited by in RCA: 2083] [Article Influence: 173.6] [Reference Citation Analysis (2)] |
| 2. | Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643-649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2881] [Cited by in RCA: 2831] [Article Influence: 101.1] [Reference Citation Analysis (0)] |
| 3. | Worthley DL, Leggett BA. Colorectal cancer: molecular features and clinical opportunities. Clin Biochem Rev. 2010;31:31-38. [PubMed] |
| 4. | Pino MS, Chung DC. The chromosomal instability pathway in colon cancer. Gastroenterology. 2010;138:2059-2072. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 576] [Cited by in RCA: 625] [Article Influence: 39.1] [Reference Citation Analysis (0)] |
| 5. | Hampel H, Frankel WL, Martin E, Arnold M, Khanduja K, Kuebler P, Nakagawa H, Sotamaa K, Prior TW, Westman J. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer). N Engl J Med. 2005;352:1851-1860. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1013] [Cited by in RCA: 1035] [Article Influence: 49.3] [Reference Citation Analysis (0)] |
| 6. | Aaltonen LA, Salovaara R, Kristo P, Canzian F, Hemminki A, Peltomäki P, Chadwick RB, Kääriäinen H, Eskelinen M, Järvinen H. Incidence of hereditary nonpolyposis colorectal cancer and the feasibility of molecular screening for the disease. N Engl J Med. 1998;338:1481-1487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 767] [Cited by in RCA: 767] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 7. | Devaud N, Gallinger S. Chemotherapy of MMR-deficient colorectal cancer. Fam Cancer. 2013;12:301-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 8. | Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA. 1999;96:8681-8686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1941] [Cited by in RCA: 1889] [Article Influence: 70.0] [Reference Citation Analysis (1)] |
| 9. | Nakatsu G, Li X, Zhou H, Sheng J, Wong SH, Wu WK, Ng SC, Tsoi H, Dong Y, Zhang N. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun. 2015;6:8727. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 558] [Cited by in RCA: 538] [Article Influence: 48.9] [Reference Citation Analysis (0)] |
| 10. | Flemer B, Lynch DB, Brown JM, Jeffery IB, Ryan FJ, Claesson MJ, O’Riordain M, Shanahan F, O’Toole PW. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut. 2017;66:633-643. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 427] [Cited by in RCA: 591] [Article Influence: 65.7] [Reference Citation Analysis (0)] |
| 11. | Borges-Canha M, Portela-Cidade JP, Dinis-Ribeiro M, Leite-Moreira AF, Pimentel-Nunes P. Role of colonic microbiota in colorectal carcinogenesis: a systematic review. Rev Esp Enferm Dig. 2015;107:659-671. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 124] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 12. | Weiss EI, Shaniztki B, Dotan M, Ganeshkumar N, Kolenbrander PE, Metzger Z. Attachment of Fusobacterium nucleatum PK1594 to mammalian cells and its coaggregation with periodontopathogenic bacteria are mediated by the same galactose-binding adhesin. Oral Microbiol Immunol. 2000;15:371-377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 60] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 13. | Han YW, Shi W, Huang GT, Kinder Haake S, Park NH, Kuramitsu H, Genco RJ. Interactions between periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infect Immun. 2000;68:3140-3146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 354] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 14. | Krisanaprakornkit S, Kimball JR, Weinberg A, Darveau RP, Bainbridge BW, Dale BA. Inducible expression of human beta-defensin 2 by Fusobacterium nucleatum in oral epithelial cells: multiple signaling pathways and role of commensal bacteria in innate immunity and the epithelial barrier. Infect Immun. 2000;68:2907-2915. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 319] [Cited by in RCA: 328] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 15. | Peyret-Lacombe A, Brunel G, Watts M, Charveron M, Duplan H. TLR2 sensing of F. nucleatum and S. sanguinis distinctly triggered gingival innate response. Cytokine. 2009;46:201-210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 43] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 16. | Signat B, Roques C, Poulet P, Duffaut D. Fusobacterium nucleatum in periodontal health and disease. Curr Issues Mol Biol. 2011;13:25-36. [PubMed] |
| 17. | Könönen E, Kanervo A, Takala A, Asikainen S, Jousimies-Somer H. Establishment of oral anaerobes during the first year of life. J Dent Res. 1999;78:1634-1639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 88] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 18. | Moore WE, Moore LV. The bacteria of periodontal diseases. Periodontol 2000. 1994;5:66-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 740] [Cited by in RCA: 728] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 19. | Gharbia SE, Shah HN, Lawson PA, Haapasalo M. Distribution and frequency of Fusobacterium nucleatum subspecies in the human oral cavity. Oral Microbiol Immunol. 1990;5:324-327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 55] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 20. | Kim HS, Lee DS, Chang YH, Kim MJ, Koh S, Kim J, Seong JH, Song SK, Shin HS, Son JB. Application of rpoB and zinc protease gene for use in molecular discrimination of Fusobacterium nucleatum subspecies. J Clin Microbiol. 2010;48:545-553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 52] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 21. | Ohkusa T, Yoshida T, Sato N, Watanabe S, Tajiri H, Okayasu I. Commensal bacteria can enter colonic epithelial cells and induce proinflammatory cytokine secretion: a possible pathogenic mechanism of ulcerative colitis. J Med Microbiol. 2009;58:535-545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 128] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 22. | Ohkusa T, Okayasu I, Ogihara T, Morita K, Ogawa M, Sato N. Induction of experimental ulcerative colitis by Fusobacterium varium isolated from colonic mucosa of patients with ulcerative colitis. Gut. 2003;52:79-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 218] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 23. | Lourenço TG, Heller D, Silva-Boghossian CM, Cotton SL, Paster BJ, Colombo AP. Microbial signature profiles of periodontally healthy and diseased patients. J Clin Periodontol. 2014;41:1027-1036. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 137] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 24. | HILL AB. The Environment And Disease: Association Or Causation? Proc R Soc Med. 1965;58:295-300. [PubMed] |
| 25. | Ahn J, Sinha R, Pei Z, Dominianni C, Wu J, Shi J, Goedert JJ, Hayes RB, Yang L. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst. 2013;105:1907-1911. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 566] [Cited by in RCA: 734] [Article Influence: 56.5] [Reference Citation Analysis (1)] |
| 26. | Vogtmann E, Hua X, Zeller G, Sunagawa S, Voigt AY, Hercog R, Goedert JJ, Shi J, Bork P, Sinha R. Colorectal Cancer and the Human Gut Microbiome: Reproducibility with Whole-Genome Shotgun Sequencing. PLoS One. 2016;11:e0155362. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 202] [Cited by in RCA: 236] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 27. | Gao Z, Guo B, Gao R, Zhu Q, Qin H. Microbiota disbiosis is associated with colorectal cancer. Front Microbiol. 2015;6:20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 242] [Cited by in RCA: 400] [Article Influence: 36.4] [Reference Citation Analysis (1)] |
| 28. | Park CH, Han DS, Oh YH, Lee AR, Lee YR, Eun CS. Role of Fusobacteria in the serrated pathway of colorectal carcinogenesis. Sci Rep. 2016;6:25271. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 56] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 29. | Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, Lan Z, Zhang D, Xia H, Xu X, Jie Z. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun. 2015;6:6528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 688] [Cited by in RCA: 1036] [Article Influence: 94.2] [Reference Citation Analysis (0)] |
| 30. | Burns MB, Lynch J, Starr TK, Knights D, Blekhman R. Virulence genes are a signature of the microbiome in the colorectal tumor microenvironment. Genome Med. 2015;7:55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 190] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 31. | Viljoen KS, Dakshinamurthy A, Goldberg P, Blackburn JM. Quantitative profiling of colorectal cancer-associated bacteria reveals associations between fusobacterium spp., enterotoxigenic Bacteroides fragilis (ETBF) and clinicopathological features of colorectal cancer. PLoS One. 2015;10:e0119462. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 201] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 32. | Zhou Y, He H, Xu H, Li Y, Li Z, Du Y, He J, Zhou Y, Wang H, Nie Y. Association of oncogenic bacteria with colorectal cancer in South China. Oncotarget. 2016;7:80794-80802. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 65] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 33. | Dejea CM, Wick EC, Hechenbleikner EM, White JR, Mark Welch JL, Rossetti BJ, Peterson SN, Snesrud EC, Borisy GG, Lazarev M. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc Natl Acad Sci USA. 2014;111:18321-18326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 418] [Cited by in RCA: 549] [Article Influence: 45.8] [Reference Citation Analysis (0)] |
| 34. | Marchesi JR, Dutilh BE, Hall N, Peters WH, Roelofs R, Boleij A, Tjalsma H. Towards the human colorectal cancer microbiome. PLoS One. 2011;6:e20447. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 442] [Cited by in RCA: 434] [Article Influence: 28.9] [Reference Citation Analysis (1)] |
| 35. | Thomas AM, Jesus EC, Lopes A, Aguiar S Jr, Begnami MD, Rocha RM, Carpinetti PA, Camargo AA, Hoffmann C, Freitas HC, Silva IT, Nunes DN, Setubal JC, Dias-Neto E. Tissue-Associated Bacterial Alterations in Rectal Carcinoma Patients Revealed by 16S rRNA Community Profiling. Front Cell Infect Microbiol. 2016;6:179. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 114] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 36. | Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, Jia W, Cai S, Zhao L. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012;6:320-329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 738] [Cited by in RCA: 972] [Article Influence: 64.8] [Reference Citation Analysis (0)] |
| 37. | Gao R, Kong C, Huang L, Li H, Qu X, Liu Z, Lan P, Wang J, Qin H. Mucosa-associated microbiota signature in colorectal cancer. Eur J Clin Microbiol Infect Dis. 2017;36:2073-2083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 94] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 38. | Allali I, Delgado S, Marron PI, Astudillo A, Yeh JJ, Ghazal H, Amzazi S, Keku T, Azcarate-Peril MA. Gut microbiome compositional and functional differences between tumor and non-tumor adjacent tissues from cohorts from the US and Spain. Gut Microbes. 2015;6:161-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 101] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 39. | Zackular JP, Rogers MA, Ruffin MT 4th, Schloss PD. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev Res (Phila). 2014;7:1112-1121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 337] [Cited by in RCA: 407] [Article Influence: 33.9] [Reference Citation Analysis (1)] |
| 40. | Zeller G, Tap J, Voigt AY, Sunagawa S, Kultima JR, Costea PI, Amiot A, Böhm J, Brunetti F, Habermann N. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol Syst Biol. 2014;10:766. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 651] [Cited by in RCA: 926] [Article Influence: 77.2] [Reference Citation Analysis (8)] |
| 41. | Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299-306. [PubMed] |
| 42. | Chen Y, Peng Y, Yu J, Chen T, Wu Y, Shi L, Li Q, Wu J, Fu X. Invasive Fusobacterium nucleatum activates beta-catenin signaling in colorectal cancer via a TLR4/P-PAK1 cascade. Oncotarget. 2017;8:31802-31814. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 135] [Cited by in RCA: 140] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 43. | McCoy AN, Araújo-Pérez F, Azcárate-Peril A, Yeh JJ, Sandler RS, Keku TO. Fusobacterium is associated with colorectal adenomas. PLoS One. 2013;8:e53653. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 406] [Cited by in RCA: 410] [Article Influence: 31.5] [Reference Citation Analysis (0)] |
| 44. | Fukugaiti MH, Ignacio A, Fernandes MR, Ribeiro Júnior U, Nakano V, Avila-Campos MJ. High occurrence of Fusobacterium nucleatum and Clostridium difficile in the intestinal microbiota of colorectal carcinoma patients. Braz J Microbiol. 2015;46:1135-1140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 104] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 45. | Warren RL, Freeman DJ, Pleasance S, Watson P, Moore RA, Cochrane K, Allen-Vercoe E, Holt RA. Co-occurrence of anaerobic bacteria in colorectal carcinomas. Microbiome. 2013;1:16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 224] [Cited by in RCA: 259] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 46. | Ito M, Kanno S, Nosho K, Sukawa Y, Mitsuhashi K, Kurihara H, Igarashi H, Takahashi T, Tachibana M, Takahashi H. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int J Cancer. 2015;137:1258-1268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 230] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 47. | Nosho K, Sukawa Y, Adachi Y, Ito M, Mitsuhashi K, Kurihara H, Kanno S, Yamamoto I, Ishigami K, Igarashi H. Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer. World J Gastroenterol. 2016;22:557-566. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 292] [Cited by in RCA: 280] [Article Influence: 28.0] [Reference Citation Analysis (3)] |
| 48. | Mima K, Sukawa Y, Nishihara R, Qian ZR, Yamauchi M, Inamura K, Kim SA, Masuda A, Nowak JA, Nosho K. Fusobacterium nucleatum and T Cells in Colorectal Carcinoma. JAMA Oncol. 2015;1:653-661. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 346] [Cited by in RCA: 536] [Article Influence: 53.6] [Reference Citation Analysis (0)] |
| 49. | Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1199] [Cited by in RCA: 1552] [Article Influence: 103.5] [Reference Citation Analysis (0)] |
| 50. | Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, Liska V, Bruha J, Neary P, Dezeeuw N, Tommasino M. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis. 2014;33:1381-1390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 285] [Cited by in RCA: 402] [Article Influence: 33.5] [Reference Citation Analysis (1)] |
| 51. | Wu N, Yang X, Zhang R, Li J, Xiao X, Hu Y, Chen Y, Yang F, Lu N, Wang Z. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb Ecol. 2013;66:462-470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 349] [Article Influence: 26.8] [Reference Citation Analysis (1)] |
| 52. | Li YY, Ge QX, Cao J, Zhou YJ, Du YL, Shen B, Wan YJ, Nie YQ. Association of Fusobacterium nucleatum infection with colorectal cancer in Chinese patients. World J Gastroenterol. 2016;22:3227-3233. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 141] [Cited by in RCA: 150] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 53. | Mira-Pascual L, Cabrera-Rubio R, Ocon S, Costales P, Parra A, Suarez A, Moris F, Rodrigo L, Mira A, Collado MC. Microbial mucosal colonic shifts associated with the development of colorectal cancer reveal the presence of different bacterial and archaeal biomarkers. J Gastroenterol. 2015;50:167-179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 199] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 54. | Amitay EL, Werner S, Vital M, Pieper DH, Höfler D, Gierse IJ, Butt J, Balavarca Y, Cuk K, Brenner H. Fusobacterium and colorectal cancer: causal factor or passenger? Results from a large colorectal cancer screening study. Carcinogenesis. 2017;38:781-788. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 98] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 55. | Yu YN, Yu TC, Zhao HJ, Sun TT, Chen HM, Chen HY, An HF, Weng YR, Yu J, Li M. Berberine may rescue Fusobacterium nucleatum-induced colorectal tumorigenesis by modulating the tumor microenvironment. Oncotarget. 2015;6:32013-32026. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 121] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 56. | Ye X, Wang R, Bhattacharya R, Boulbes DR, Fan F, Xia L, Adoni H, Ajami NJ, Wong MC, Smith DP. Fusobacterium Nucleatum Subspecies Animalis Influences Proinflammatory Cytokine Expression and Monocyte Activation in Human Colorectal Tumors. Cancer Prev Res (Phila). 2017;10:398-409. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 120] [Article Influence: 13.3] [Reference Citation Analysis (1)] |
| 57. | Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One. 2012;7:e39743. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 788] [Cited by in RCA: 751] [Article Influence: 53.6] [Reference Citation Analysis (1)] |
| 58. | Kasai C, Sugimoto K, Moritani I, Tanaka J, Oya Y, Inoue H, Tameda M, Shiraki K, Ito M, Takei Y. Comparison of human gut microbiota in control subjects and patients with colorectal carcinoma in adenoma: Terminal restriction fragment length polymorphism and next-generation sequencing analyses. Oncol Rep. 2016;35:325-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 96] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 59. | Manson McGuire A, Cochrane K, Griggs AD, Haas BJ, Abeel T, Zeng Q, Nice JB, MacDonald H, Birren BW, Berger BW. Evolution of invasion in a diverse set of Fusobacterium species. MBio. 2014;5:e01864. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 95] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 60. | Allen-Vercoe E, Strauss J, Chadee K. Fusobacterium nucleatum: an emerging gut pathogen? Gut Microbes. 2011;2:294-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 106] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 61. | Tahara T, Yamamoto E, Suzuki H, Maruyama R, Chung W, Garriga J, Jelinek J, Yamano HO, Sugai T, An B. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 2014;74:1311-1318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 293] [Cited by in RCA: 375] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 62. | Yazici C, Wolf PG, Kim H, Cross TL, Vermillion K, Carroll T, Augustus GJ, Mutlu E, Tussing-Humphreys L, Braunschweig C. Race-dependent association of sulfidogenic bacteria with colorectal cancer. Gut. 2017;66:1983-1994. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 164] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 63. | O’Keefe SJ, Li JV, Lahti L, Ou J, Carbonero F, Mohammed K, Posma JM, Kinross J, Wahl E, Ruder E. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun. 2015;6:6342. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 694] [Cited by in RCA: 716] [Article Influence: 65.1] [Reference Citation Analysis (0)] |
| 64. | Mehta RS, Nishihara R, Cao Y, Song M, Mima K, Qian ZR, Nowak JA, Kosumi K, Hamada T, Masugi Y. Association of Dietary Patterns With Risk of Colorectal Cancer Subtypes Classified by Fusobacterium nucleatum in Tumor Tissue. JAMA Oncol. 2017;3:921-927. [PubMed] |
| 65. | Lüder Ripoli F, Mohr A, Conradine Hammer S, Willenbrock S, Hewicker-Trautwein M, Hennecke S, Murua Escobar H, Nolte I. A Comparison of Fresh Frozen vs. Formalin-Fixed, Paraffin-Embedded Specimens of Canine Mammary Tumors via Branched-DNA Assay. Int J Mol Sci. 2016;17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 27] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 66. | Jalanka J, Salonen A, Salojärvi J, Ritari J, Immonen O, Marciani L, Gowland P, Hoad C, Garsed K, Lam C. Effects of bowel cleansing on the intestinal microbiota. Gut. 2015;64:1562-1568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 197] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 67. | Kato I, Vasquez A, Moyerbrailean G, Land S, Djuric Z, Sun J, Lin HS, Ram JL. Nutritional Correlates of Human Oral Microbiome. J Am Coll Nutr. 2017;36:88-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 80] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 68. | Kato I, Vasquez AA, Moyerbrailean G, Land S, Sun J, Lin HS, Ram JL. Oral microbiome and history of smoking and colorectal cancer. J Epidemiol Res. 2016;2:92-101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 65] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 69. | Park HE, Kim JH, Cho NY, Lee HS, Kang GH. Intratumoral Fusobacterium nucleatum abundance correlates with macrophage infiltration and CDKN2A methylation in microsatellite-unstable colorectal carcinoma. Virchows Arch. 2017; Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 84] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 70. | Wei Z, Cao S, Liu S, Yao Z, Sun T, Li Y, Li J, Zhang D, Zhou Y. Could gut microbiota serve as prognostic biomarker associated with colorectal cancer patients’ survival? A pilot study on relevant mechanism. Oncotarget. 2016;7:46158-46172. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 174] [Cited by in RCA: 167] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 71. | Yu J, Chen Y, Fu X, Zhou X, Peng Y, Shi L, Chen T, Wu Y. Invasive Fusobacterium nucleatum may play a role in the carcinogenesis of proximal colon cancer through the serrated neoplasia pathway. Int J Cancer. 2016;139:1318-1326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 130] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 72. | Mima K, Cao Y, Chan AT, Qian ZR, Nowak JA, Masugi Y, Shi Y, Song M, da Silva A, Gu M. Fusobacterium nucleatum in Colorectal Carcinoma Tissue According to Tumor Location. Clin Transl Gastroenterol. 2016;7:e200. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 168] [Cited by in RCA: 228] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 73. | Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, Yang J, Dou R, Masugi Y, Song M. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65:1973-1980. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 530] [Cited by in RCA: 819] [Article Influence: 81.9] [Reference Citation Analysis (0)] |
| 74. | Ogino S, Kawasaki T, Kirkner GJ, Loda M, Fuchs CS. CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: possible associations with male sex and KRAS mutations. J Mol Diagn. 2006;8:582-588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 253] [Article Influence: 13.3] [Reference Citation Analysis (7)] |
| 75. | Dalerba P, Kalisky T, Sahoo D, Rajendran PS, Rothenberg ME, Leyrat AA, Sim S, Okamoto J, Johnston DM, Qian D. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol. 2011;29:1120-1127. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 569] [Cited by in RCA: 551] [Article Influence: 36.7] [Reference Citation Analysis (0)] |
| 76. | Lennard KS, Goosen RW, Blackburn JM. Bacterially-Associated Transcriptional Remodelling in a Distinct Genomic Subtype of Colorectal Cancer Provides a Plausible Molecular Basis for Disease Development. PLoS One. 2016;11:e0166282. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 38] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 77. | Sadanandam A, Lyssiotis CA, Homicsko K, Collisson EA, Gibb WJ, Wullschleger S, Ostos LC, Lannon WA, Grotzinger C, Del Rio M. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19:619-625. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 683] [Cited by in RCA: 775] [Article Influence: 59.6] [Reference Citation Analysis (0)] |
| 78. | Yoon H, Kim N, Park JH, Kim YS, Lee J, Kim HW, Choi YJ, Shin CM, Park YS, Lee DH. Comparisons of Gut Microbiota Among Healthy Control, Patients With Conventional Adenoma, Sessile Serrated Adenoma, and Colorectal Cancer. J Cancer Prev. 2017;22:108-114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 43] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 79. | Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1659] [Cited by in RCA: 2043] [Article Influence: 157.2] [Reference Citation Analysis (0)] |
| 80. | Belshaw NJ, Pal N, Tapp HS, Dainty JR, Lewis MP, Williams MR, Lund EK, Johnson IT. Patterns of DNA methylation in individual colonic crypts reveal aging and cancer-related field defects in the morphologically normal mucosa. Carcinogenesis. 2010;31:1158-1163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 61] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 81. | De Paoli-Iseppi R, Johansson PA, Menzies AM, Dias KR, Pupo GM, Kakavand H, Wilmott JS, Mann GJ, Hayward NK, Dinger ME. Comparison of whole-exome sequencing of matched fresh and formalin fixed paraffin embedded melanoma tumours: implications for clinical decision making. Pathology. 2016;48:261-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 34] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 82. | Zhu Q, Jin Z, Wu W, Gao R, Guo B, Gao Z, Yang Y, Qin H. Analysis of the intestinal lumen microbiota in an animal model of colorectal cancer. PLoS One. 2014;9:e90849. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 141] [Cited by in RCA: 150] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 83. | Abed J, Emgård JE, Zamir G, Faroja M, Almogy G, Grenov A, Sol A, Naor R, Pikarsky E, Atlan KA. Fap2 Mediates Fusobacterium nucleatum Colorectal Adenocarcinoma Enrichment by Binding to Tumor-Expressed Gal-GalNAc. Cell Host Microbe. 2016;20:215-225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 326] [Cited by in RCA: 644] [Article Influence: 71.6] [Reference Citation Analysis (0)] |
| 84. | Abed J, Maalouf N, Parhi L, Chaushu S, Mandelboim O, Bachrach G. Tumor Targeting by Fusobacterium nucleatum: A Pilot Study and Future Perspectives. Front Cell Infect Microbiol. 2017;7:295. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 56] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 85. | Yang GY, Shamsuddin AM. Gal-GalNAc: a biomarker of colon carcinogenesis. Histol Histopathol. 1996;11:801-806. [PubMed] |
| 86. | Sharma R, Schumacher U. Carbohydrate expression in the intestinal mucosa. Berlin: Springer-verlag 2001; 123-456. |
| 87. | Parikh ND, Gibson J, Nagar A, Ahmed AA, Aslanian HR. Confocal laser endomicroscopy features of sessile serrated adenomas/polyps. United European Gastroenterol J. 2016;4:599-603. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 88. | Sinha R, Ahn J, Sampson JN, Shi J, Yu G, Xiong X, Hayes RB, Goedert JJ. Fecal Microbiota, Fecal Metabolome, and Colorectal Cancer Interrelations. PLoS One. 2016;11:e0152126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 173] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 89. | Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1511] [Cited by in RCA: 1812] [Article Influence: 139.4] [Reference Citation Analysis (1)] |
| 90. | Schetter AJ, Heegaard NH, Harris CC. Inflammation and cancer: interweaving microRNA, free radical, cytokine and p53 pathways. Carcinogenesis. 2010;31:37-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 436] [Cited by in RCA: 498] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 91. | Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, Gao R, Liu M, Yin M, Pan C. Fusobacterium nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor-κB, and Up-regulating Expression of MicroRNA-21. Gastroenterology. 2017;152:851-866.e24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 455] [Cited by in RCA: 786] [Article Influence: 87.3] [Reference Citation Analysis (1)] |
| 92. | Kumar A, Thotakura PL, Tiwary BK, Krishna R. Target identification in Fusobacterium nucleatum by subtractive genomics approach and enrichment analysis of host-pathogen protein-protein interactions. BMC Microbiol. 2016;16:84. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 44] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 93. | Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51728] [Cited by in RCA: 48675] [Article Influence: 3245.0] [Reference Citation Analysis (12)] |
| 94. | Tomkovich S, Yang Y, Winglee K, Gauthier J, Mühlbauer M, Sun X, Mohamadzadeh M, Liu X, Martin P, Wang GP. Locoregional Effects of Microbiota in a Preclinical Model of Colon Carcinogenesis. Cancer Res. 2017;77:2620-2632. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 185] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 95. | Tang B, Wang K, Jia YP, Zhu P, Fang Y, Zhang ZJ, Mao XH, Li Q, Zeng DZ. Fusobacterium nucleatum-Induced Impairment of Autophagic Flux Enhances the Expression of Proinflammatory Cytokines via ROS in Caco-2 Cells. PLoS One. 2016;11:e0165701. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 49] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 96. | Shenker BJ, Listgarten MA, Taichman NS. Suppression of human lymphocyte responses by oral spirochetes: a monocyte-dependent phenomenon. J Immun. 1984;132:2039-2045. |
| 97. | Jewett A, Hume WR, Le H, Huynh TN, Han YW, Cheng G, Shi W. Induction of apoptotic cell death in peripheral blood mononuclear and polymorphonuclear cells by an oral bacterium, Fusobacterium nucleatum. Infect Immun. 2000;68:1893-1898. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 78] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 98. | Kinder Haake S, Lindemann RA. Fusobacterium nucleatum T18 aggregates human mononuclear cells and inhibits their PHA-stimulated proliferation. J Periodontol. 1997;68:39-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 99. | Shenker BJ, Datar S. Fusobacterium nucleatum inhibits human T-cell activation by arresting cells in the mid-G1 phase of the cell cycle. Infect Immun. 1995;63:4830-4836. [PubMed] |
| 100. | Huynh T, Kapur RV, Kaplan CW, Cacalano N, Kinder Haake S, Shi W, Sieling P, Jewett A. The role of aggregation in Fusobacterium nucleatum- induced immune cell death. J Endod. 2011;37:1531-1535. [PubMed] |
| 101. | Kaplan CW, Ma X, Paranjpe A, Jewett A, Lux R, Kinder-Haake S, Shi W. Fusobacterium nucleatum outer membrane proteins Fap2 and RadD induce cell death in human lymphocytes. Infect Immun. 2010;78:4773-4778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 152] [Article Influence: 9.5] [Reference Citation Analysis (1)] |
| 102. | Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012;12:253-268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2369] [Cited by in RCA: 2948] [Article Influence: 210.6] [Reference Citation Analysis (0)] |
| 103. | Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, Enk J, Bar-On Y, Stanietsky-Kaynan N, Coppenhagen-Glazer S. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42:344-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1015] [Cited by in RCA: 1083] [Article Influence: 98.5] [Reference Citation Analysis (1)] |
| 104. | Saito T, Nishikawa H, Wada H, Nagano Y, Sugiyama D, Atarashi K, Maeda Y, Hamaguchi M, Ohkura N, Sato E. Two FOXP3(+)CD4(+) T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat Med. 2016;22:679-684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 471] [Cited by in RCA: 686] [Article Influence: 68.6] [Reference Citation Analysis (0)] |
| 105. | Thiele Orberg E, Fan H, Tam AJ, Dejea CM, Destefano Shields CE, Wu S, Chung L, Finard BB, Wu X, Fathi P. The myeloid immune signature of enterotoxigenic Bacteroides fragilis-induced murine colon tumorigenesis. Mucosal Immunol. 2017;10:421-433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 168] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 106. | Liang Q, Chiu J, Chen Y, Huang Y, Higashimori A, Fang J, Brim H, Ashktorab H, Ng SC, Ng SSM. Fecal Bacteria Act as Novel Biomarkers for Noninvasive Diagnosis of Colorectal Cancer. Clin Cancer Res. 2017;23:2061-2070. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 239] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 107. | Suehiro Y, Sakai K, Nishioka M, Hashimoto S, Takami T, Higaki S, Shindo Y, Hazama S, Oka M, Nagano H. Highly sensitive stool DNA testing of Fusobacterium nucleatum as a marker for detection of colorectal tumours in a Japanese population. Ann Clin Biochem. 2017;54:86-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 59] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 108. | Baxter NT, Ruffin MT 4th, Rogers MA, Schloss PD. Microbiota-based model improves the sensitivity of fecal immunochemical test for detecting colonic lesions. Genome Med. 2016;8:37. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 261] [Cited by in RCA: 237] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 109. | Wong SH, Kwong TNY, Chow TC, Luk AKC, Dai RZW, Nakatsu G, Lam TYT, Zhang L, Wu JCY, Chan FKL. Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut. 2017;66:1441-1448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 157] [Cited by in RCA: 242] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 110. | Huang S, Yang Z, Zou D, Dong D, Liu A, Liu W, Huang L. Rapid detection of nusG and fadA in Fusobacterium nucleatum by loop-mediated isothermal amplification. J Med Microbiol. 2016;65:760-769. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 111. | Yu J, Feng Q, Wong SH, Zhang D, Liang QY, Qin Y, Tang L, Zhao H, Stenvang J, Li Y. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. 2017;66:70-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 917] [Cited by in RCA: 838] [Article Influence: 93.1] [Reference Citation Analysis (0)] |
| 112. | Wang HF, Li LF, Guo SH, Zeng QY, Ning F, Liu WL, Zhang G. Evaluation of antibody level against Fusobacterium nucleatum in the serological diagnosis of colorectal cancer. Sci Rep. 2016;6:33440. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 84] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 113. | Gao Z, Guo B, Gao R, Zhu Q, Wu W, Qin H. Probiotics modify human intestinal mucosa-associated microbiota in patients with colorectal cancer. Mol Med Rep. 2015;12:6119-6127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 83] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 114. | Zhang X, Zhao Y, Zhang M, Pang X, Xu J, Kang C, Li M, Zhang C, Zhang Z, Zhang Y. Structural changes of gut microbiota during berberine-mediated prevention of obesity and insulin resistance in high-fat diet-fed rats. PLoS One. 2012;7:e42529. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 342] [Cited by in RCA: 410] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: United States
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
P- Reviewer: Lim SC, Swidsinski A S- Editor: Ma YJ L- Editor: A E- Editor: Lu YJ