Published online Dec 21, 2017. doi: 10.3748/wjg.v23.i47.8345
Peer-review started: November 4, 2017
First decision: November 14, 2017
Revised: November 17, 2017
Accepted: November 27, 2017
Article in press: November 27, 2017
Published online: December 21, 2017
Processing time: 46 Days and 9.9 Hours
To analyze the diagnostic value of a circular RNA (circRNA), circ-LDLRAD3, in pancreatic cancer.
Expression levels of circ-LDLRAD3 were tested in both cells and clinical samples; the latter included 30 paired pancreatic cancer tissues and adjacent non-tumorous tissues, 31 plasma samples from patients with pancreatic cancer, and 31 plasma samples from healthy volunteers. Real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed to measure expression levels of circ-LDLRAD3 in cells and clinical samples; then, the relationship between clinicopathological factors of patient samples and expression of circ-LDLRAD3 in pancreatic cancer was analyzed. The diagnostic value of circ-LDLRAD3 was verified by receiver operating characteristic (ROC) curve analysis.
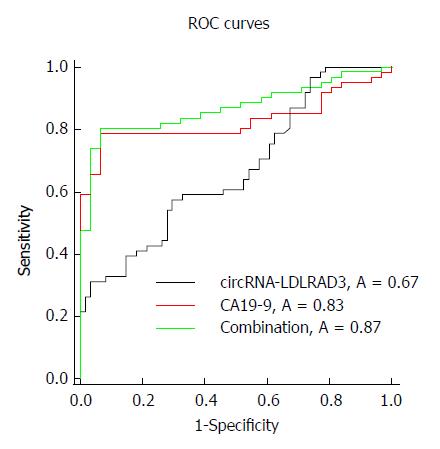
Circ-LDLRAD3 was up-regulated in pancreatic cancer cell lines (P < 0.01), pancreatic cancer tissues (P < 0.01), and plasma samples from patients with pancreatic cancer (P < 0.01). High expression of circ-LDLRAD3 was significantly associated with venous invasion, lymphatic invasion, and metastasis. The area under the ROC curve of circ-LDLRAD3 alone or combination with CA19-9 was 0.67 and 0.87, respectively, with a sensitivity and specificity of 0.5738 (alone) and 0.7049 (alone), and 0.8033 (combination) and 0.9355 (combination), respectively.
These data suggest that circ-LDLRAD3 may be a biomarker in the diagnosis of pancreatic cancer.
Core tip: Circular RNAs (circRNAs), a novel class of stable endogenous RNAs, play important roles in the occurrence and progression of cancer; however, little is known about their diagnostic value in pancreatic cancer. Our study focused on a novel circRNA, circ-LDLRAD3. Expression levels of circ-LDLRAD3 were tested in both cells and clinical samples, including tissue samples and plasma samples. Then, the relationship between clinicopathological factors of patient samples and expression of circ-LDLRAD3 in pancreatic cancer was analyzed. The diagnostic value of circ-LDLRAD3 was verified by ROC curve analysis. Our study suggests that circ-LDLRAD3 may be a new biomarker in the diagnosis of pancreatic cancer.
- Citation: Yang F, Liu DY, Guo JT, Ge N, Zhu P, Liu X, Wang S, Wang GX, Sun SY. Circular RNA circ-LDLRAD3 as a biomarker in diagnosis of pancreatic cancer. World J Gastroenterol 2017; 23(47): 8345-8354
- URL: https://www.wjgnet.com/1007-9327/full/v23/i47/8345.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i47.8345
Pancreatic cancer is a malignancy of the digestive system with insidious onset and rapid development, resulting in delayed and difficult early diagnoses and poor prognosis[1,2]. The incidence and mortality of pancreatic cancer are rising every year worldwide, and it is the 7th and 4th leading cause of mortality from all malignant tumors in China[3] and the United States[4], respectively. Surgical resection remains the major means of treatment for pancreatic cancer; however, the 5-year survival rate for patients undergoing a complete resection remains as low as 6%[5]. The key to improving the prognosis of pancreatic cancer mostly lies in early diagnosis and early treatment, which can be achieved by detection of relevant molecular markers among patients with high risk, followed by early and timely interventions[6-8].
Circular RNAs (circRNAs) are a class of noncoding RNAs with continuous, covalently closed circular structures, which have been further found to exhibit species conservation and tissue specificity[9]. With the emergence of next-generation sequencing, especially RNA sequencing technology, circRNAs have been found to be extensively expressed in the cytoplasm. In addition, they have been garnering attention because of their specificity of expression, complexity of regulation, and important role in pathogenesis of many diseases, especially cancer[10]. Unlike their linear counterparts, circRNAs are characterized by stable ring structure formed by a covalently closed continuous loop. Without free 3’ and 5’ ends, these molecules are not easily degraded by nucleases, which makes them ideal biomarkers for detection of disease[11]. Investigators have identified disease-specific patterns of circRNA expression, which can serve as biomarkers for diseases[12], especially cancer[10,13]. However, there has been little investigation into the association of circRNAs with pancreatic cancer.
In this study, we focused our investigation on circRNA-hsa_circ_0006988, whose gene is located at chr11:36248634-36248980. Its gene symbol is LDLRAD3 (low density lipoprotein receptor class A domain containing 3), therefore we will refer to circRNA-hsa_circ_0006988 as circ-LDLRAD3 instead of its original name in circBase[14] (http://www.circbase.org). We chose circ-LDLRAD3 as a target for further study because we previously identified that it may be up-regulated in a previous microarray screening (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE69362)[15] and associated with pancreatic cancer in circBase[14] and circ2Traits[16]. By expanding the sample size, we found that the expression levels of circ-LDLRAD3 were higher in both pancreatic cancer tissues and plasma from patients with pancreatic cancer as compared to control samples. Moreover, up-regulated expression of circ-LDLRAD3 was significantly related to major clinicopathological factors of patients with pancreatic cancer. Our results make clear that circ-LDLRAD3 may serve as a biomarker in the diagnosis of pancreatic cancer.
Thirty samples of pancreatic cancer and their paired adjacent pancreatic tissues were obtained from patients with pancreatic cancer treated at Shengjing Hospital of China Medical University (Shenyang, China) from September 2016 to June 2017. Paired normal tissue samples were obtained 5 cm from the pancreatic cancer tissue and were confirmed to contain no tumor cells after evaluation by two experienced pathologists. All specimens were immediately stored in liquid nitrogen until use.
Peripheral blood samples (4 mL) were collected from another 31 patients with pancreatic cancer and 31 healthy volunteers prior to any medical interventions at Shengjing Hospital of China Medical University (Shenyang, China) from October 2016 to July 2017. Plasma samples were isolated as previously described. The anti-coagulant for peripheral blood samples was ethylenediaminetetraacetic acid (EDTA). Clinical information was collected for all patients and healthy volunteers.
Tumors were staged according to the 8th tumor-node-metastasis (TNM) staging system drafted by the International Union Against Cancer. No patients received radiotherapy, chemotherapy, or targeted therapy before surgery. All patients and healthy volunteers provided written informed consent before the procedure. The Institutional Review Board of China Medical University approved this study based on the Helsinki Declaration.
The normal pancreatic cell lines, HPC-Y5 and HPDE6-C7, were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The pancreatic cancer cell lines, Capan-2, Panc-1, SW1990, and AsPC-1, were obtained from ATCC (Manassas, United States). HPC-Y5, HPDE6-C7, and Panc-1 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Gaithersburg, MD, United States); Capan-2 and AsPC-1 cells were cultured in RPMI-1640 medium (Gibco, Gaithersburg, MD, United States); and SW1990 cells were cultured in Leibovitz's L-15 medium (Gibco, Gaithersburg, MD, United States). All media contained 10% fetal bovine serum (FBS) (Gibco, Gaithersburg, MD, United States) and all cells were cultured in a humidified atmosphere consisting of 5% CO2 and 95% air at 37 °C.
Total RNA from all cell lines, pancreatic cancer tissues, and paired adjacent tissues was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, United States). Total RNA in plasma samples was extracted using a mirVana PARIS Kit (Ambion, Carlsbad, CA, United States) following the manufacturer’s instructions. Quantity and quality of RNA were determined spectrophotometrically at 260 nm and 280 nm. The integrity and contamination were confirmed using denaturing agarose gel electrophoresis.
Total RNA was reverse transcribed using a PrimeScript reagent kit with gRNA Eraser (Random primers) (TaKaRa, Dalian, China) according to the manufacturer’s instructions.
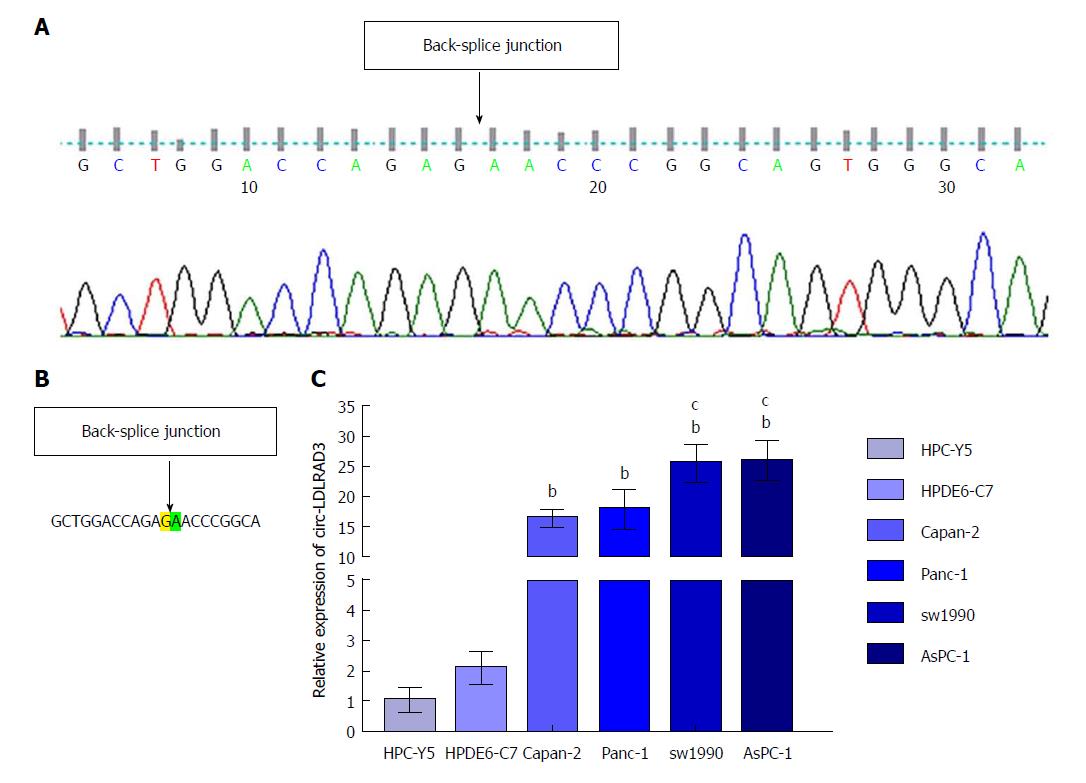
To precisely examine the primer sequences of circ-LDLRAD3, Sanger sequencing was utilized. In brief, a T vector carrying the target fragment was utilized for Sanger sequencing in order to determine the back-spliced junction of circ-LDLRAD3. The following divergent primers were synthesized by Geneseed Biotech (Guangzhou, China): 5’-CTTGCTGGACCAGAGAAC-3’ (forward) and 5’-CATGAGGTTGTTCCGCTTC-3’ (reverse). Sanger sequencing was performed by the same company.
Real-time quantitative reverse transcription polymerase chain reactions (qRT-PCR) was performed using a Roche 480II system (Roche, Basel, Switzerland) utilizing SYBR Premix Ex Taq II (Tli RNaseH Plus) (Takara, Dalian, China), following the manufacturer-provided instructions. Primers for GAPDH were synthesized by Sangon Biotech (Shanghai, China) as follows: 5’-GCACCGTCAAGGCTGAGAAC-3’ (forward) and 5’-TGGTGAAGACGCCAGTGGA-3’ (reverse). The data were analyzed using the comparative cycle threshold (ΔCT) method after three independent experiments. All results are expressed as the mean ± SD.
Serum carbohydrate antigen 19-9 (CA19-9) and carcinoembryonic antigen (CEA) were measured using a Roche E601 machine (Roche, Basel, Switzerland) with a cutoff value of 40 U/mL and 5 ng/mL, respectively.
All statistical data were analyzed using SPSS 23.0 (SPSS, Chicago, IL, United States), GraphPad 7.0 (GraphPad Software, La Jolla, CA, United States), and SigmaPlot 12.5 (SigmaPlot Software, La Jolla, CA, United States). Differences in expression levels of circ-LDLRAD3 between pancreatic cancer tissues and paired adjacent non-tumorous tissues were compared by using paired t-tests, and differences in expression levels of circ-LDLRAD3 between plasma samples from patients with pancreatic cancer and those from healthy volunteers were compared by Student’s t-tests. A Fisher’s exact test was used to analyze the association between circ-LDLRAD3 expression and patients’ clinicopathological factors. A Spearman’s rank correlation coefficient was introduced to further calculate bivariate correlations. The receiver operating characteristics (ROC) curve was established to evaluate the diagnostic value of circ-LDLRAD3; the cutoff value of circ-LDLRAD3 was calculated using Youden index (specificity + sensitivity-1). The comparison of the area under the ROC curve (AUC) was analyzed by Z-test. P values < 0.05 were considered statistically significant.
Sanger sequencing of circLDLRAD3 qRT-PCR product was first conducted to determine the back-junction of circ-LDLRAD3. The results of the back-splice junction of circ-LDLRAD3 indicated there was no difference between our product and that found in CircBase (Figure 1A and B). Next, expression levels of circ-LDLRAD3 were tested in normal pancreatic cell lines (HPC-Y5 and HPDE6-C7) and pancreatic cancer cell lines (Capan-2, Panc-1, SW1990, and AsPC-1). These results indicate that the relative expression levels of circ-LDLRAD3 were higher in pancreatic cancer cell lines than in normal pancreatic cell lines (P < 0.01). In addition, the relative expression levels of circ-LDLRAD3 in metastatic pancreatic cancer cell lines (SW1990 and AsPC-1) were higher than those in primary pancreatic cancer cell lines (Capan-2 and Panc-1) (P < 0.05) (Figure 1C).
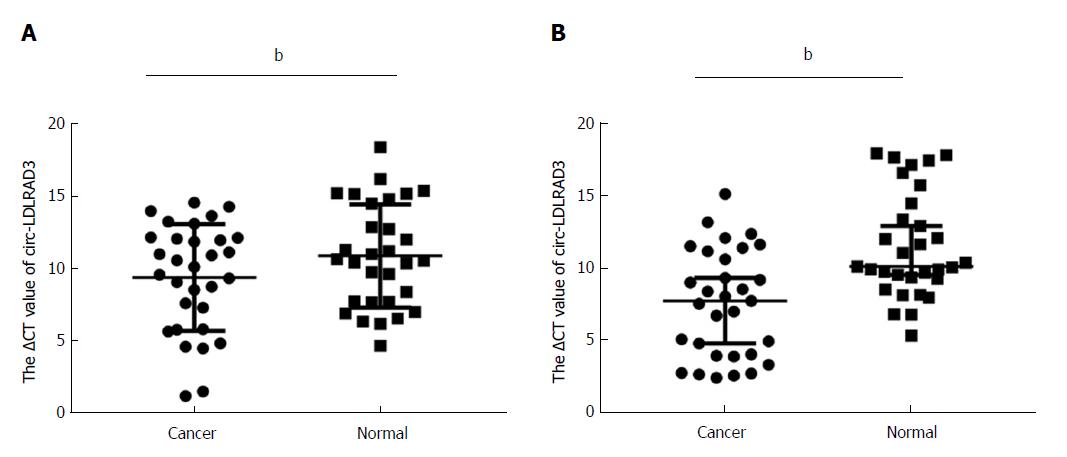
Expression of circ-LDLRAD3 was measured via qRT-PCR in 30 pancreatic cancer tissues compared with paired adjacent non-tumorous tissues and in plasma samples of patients with pancreatic cancer compared with healthy volunteers. Lower ΔCT values indicate higher expression of circ-LDLRAD3. As shown in Figure 2A, expression of circ-LDLRAD3 was up-regulated in pancreatic cancer tissues (P < 0.01), while expression of circ-LDLRAD3 in plasma samples with pancreatic cancer were higher than those in healthy volunteers (P < 0.01, Figure 2B).
The above data demonstrated that circ-LDLRAD3 expression was significantly up-regulated in pancreatic cancer tissues and plasma samples of patients with pancreatic cancer; hence, we analyzed the association between circ-LDLRAD3 and clinicopathological factors of patients with pancreatic cancer.
As shown in Tables 1-3, in pancreatic cancer tissues, a strong association was observed between circ-LDLRAD3 expression and venous invasion (P = 0.025) and lymphatic invasion (P = 0.014). However, no association was found between circ-LDLRAD3 expression and other clinicopathological factors including age (P = 0.279), gender (P = 0.255), tumor diameter (P = 0.279), CA19-9 (P = 0.643), CEA (P = 0.88), clinical stage (P = 0.256), T classification (P = 0.274), N classification (P = 0.429), and metastasis (none). A Spearman analysis of correlation between circ-LDLRAD3 and various clinicopathological factors indicated that expression of circ-LDLRAD3 was correlated with clinical stage (P = 0.022), T classification (P = 0.003), venous invasion (P = 0.025), and lymphatic invasion (P = 0.008).
| Characteristic | n (%) |
| Age (yr) | |
| ≥ 60 | 19 (63.3) |
| < 60 | 11 (36.7) |
| Gender | |
| Male | 9 (30) |
| Female | 21 (70) |
| Tumor diameter (cm) | |
| ≤ 4 | 19 (63.3) |
| > 4 | 11 (36.7) |
| CA19-9 | |
| Positive | 19 (63.3) |
| Negative | 11 (36.7) |
| CEA | |
| Positive | 16 (56.7) |
| Negative | 13 (43.3) |
| Clinical stage | |
| IA | 3 (10) |
| IB | 10 (33.3) |
| IIA | 7 (23.3) |
| IIB | 9 (30) |
| III | 1 (3.3) |
| IV | 0 (0) |
| T classification | |
| T1 | 3 (10) |
| T2 | 15 (50) |
| T3 | 11 (36.7) |
| T4 | 1 (3.3) |
| N classification | |
| N0 | 20 (66.7) |
| N1 | 10 (33.3) |
| N2 | 0 (0) |
| Metastasis | |
| M0 | 30 (100) |
| M1 | 0 (0) |
| Venous invasion | |
| No | 24 (80) |
| Yes | 6 (20) |
| Lymphatic invasion | |
| No | 23 (76.7) |
| Yes | 7 (23.3) |
| Expression of circ-LDLRAD3 | |
| Low expression | 12 (40) |
| High expression | 18 (60) |
| Characteristic | Circ-LDLRAD3 | P value | |
| Low or none, n | High, n | ||
| Age (yr) | |||
| ≥ 60 | 9 | 10 | 0.279 |
| < 60 | 3 | 8 | |
| Gender | |||
| Male | 7 | 14 | 0.255 |
| Female | 5 | 4 | |
| Tumor diameter (cm) | |||
| ≤ 4 | 9 | 10 | 0.279 |
| > 4 | 3 | 8 | |
| CA19-9 | |||
| Positive | 5 | 6 | 0.643 |
| Negative | 7 | 12 | |
| CEA | |||
| Positive | 5 | 8 | 0.88 |
| Negative | 7 | 10 | |
| Clinical stage | |||
| IA | 2 | 1 | 0.256 |
| IB | 6 | 4 | |
| IIA | 1 | 6 | |
| IIB | 3 | 6 | |
| III | 0 | 1 | |
| IV | 0 | 0 | |
| T classification | |||
| T1 | 2 | 1 | 0.274 |
| T2 | 8 | 8 | |
| T3 | 2 | 8 | |
| T4 | 0 | 1 | |
| N classification | |||
| N0 | 9 | 11 | 0.429 |
| N1 | 3 | 7 | |
| N2 | 0 | 0 | |
| Metastasis | |||
| M0 | 12 | 18 | None |
| M1 | 0 | 0 | |
| Venous invasion | |||
| No | 12 | 12 | 0.025 |
| Yes | 0 | 6 | |
| Lymphatic invasion | |||
| No | 12 | 11 | 0.014 |
| Yes | 0 | 7 | |
| Variable | Circ-LDLRAD3 expression level | |
| Spearman correlation | P value | |
| Age (yr) | -0.22 | 0.243 |
| Gender | -0.122 | 0.521 |
| Tumor diameter (cm) | -0.303 | 0.104 |
| CA19-9 | 0.028 | 0.883 |
| CEA | 0.019 | 0.919 |
| Clinical stage | -0.415 | 0.022 |
| T classification | -0.519 | 0.003 |
| N classification | -0.196 | 0.299 |
| Metastasis | None | None |
| Venous invasion | -0.607 | < 0.001 |
| Lymphatic invasion | -0.478 | 0.008 |
In the plasma of patients with pancreatic cancer (Tables 4-6), circ-LDLRAD3 levels were significantly associated with CA19-9 (P = 0.03), N classification (P = 0.049), venous invasion (P = 0.005), and lymphatic invasion (0.014). No association was found between circ-LDLRAD3 and age, gender, tumor diameter, CEA, clinical stage, T classification, or metastasis. In Spearman analysis, circ-LDLRAD3 expression was correlated with clinical stage (P < 0.001), metastasis (P = 0.004), venous invasion (P = 0.029), and lymphatic invasion (P < 0.001).
| Characteristic | n (%) |
| Age (yr) | |
| ≥ 60 | 15 (48.4) |
| < 60 | 16 (51.6) |
| Gender | |
| Male | 19 (61.3) |
| Female | 12 (38.7) |
| Tumor diameter (cm) | |
| ≤ 4 | 21 (67.7) |
| > 4 | 10 (32.3) |
| CA19-9 | |
| Positive | 25 (87.1) |
| Negative | 4 (12.9) |
| CEA | |
| Positive | 10 (32.3) |
| Negative | 21 (67.7) |
| Clinical stage | |
| IA | 5 (16.1) |
| IB | 6 (19.4) |
| IIA | 6 (19.4) |
| IIB | 5 (16.1) |
| III | 6 (19.4) |
| IV | 3 (9.7) |
| T classification | |
| T1 | 5 (16.1) |
| T2 | 15 (48.4) |
| T3 | 9 (29.0) |
| T4 | 2 (6.5) |
| N classification | |
| N0 | 21 (67.7) |
| N1 | 6 (19.4) |
| N2 | 4 (12.9) |
| Metastasis | |
| M0 | 28 (90.3) |
| M1 | 3 (9.7) |
| Venous invasion | |
| No | 19 (61.3) |
| Yes | 12 (38.7) |
| Lymphatic invasion | |
| No | 21 (67.7) |
| Yes | 10 (32.3) |
| Expression of circ-LDLRAD3 | |
| Low expression | 9 (29) |
| High expression | 22 (71) |
| Characteristic | Circ-LDLRAD3 | P value | |
| Low or one, n | High, n | ||
| Age (yr) | |||
| ≥ 60 | 5 | 10 | 0.609 |
| < 60 | 4 | 12 | |
| Gender | |||
| Male | 5 | 14 | 0.675 |
| Female | 4 | 8 | |
| Tumor diameter (cm) | |||
| ≤ 4 | 6 | 15 | 0.935 |
| > 4 | 3 | 7 | |
| CA19-9 | |||
| Positive | 6 | 21 | 0.030 |
| Negative | 3 | 1 | |
| CEA | |||
| Positive | 5 | 5 | 0.076 |
| Negative | 4 | 17 | |
| Clinical stage | |||
| IA | 3 | 2 | 0.060 |
| IB | 3 | 3 | |
| IIA | 3 | 3 | |
| IIB | 0 | 5 | |
| III | 0 | 6 | |
| IV | 0 | 3 | |
| T classification | |||
| T1 | 3 | 2 | 0.282 |
| T2 | 3 | 12 | |
| T3 | 3 | 6 | |
| T4 | 0 | 2 | |
| N classification | |||
| N0 | 9 | 12 | 0.049 |
| N1 | 0 | 6 | |
| N2 | 0 | 4 | |
| Metastasis | |||
| M0 | 9 | 19 | 0.244 |
| M1 | 0 | 3 | |
| Venous invasion | |||
| No | 9 | 10 | 0.005 |
| Yes | 0 | 12 | |
| Lymphatic invasion | |||
| No | 9 | 12 | 0.014 |
| Yes | 0 | 10 | |
| Variable | Circ-LDLRAD3 expression level | |
| Spearman correlation | P value | |
| Age (yr) | -0.108 | 0.562 |
| Gender | 0.059 | 0.752 |
| Tumor diameter (cm) | -0.102 | 0.584 |
| CA19-9 | -0.398 | 0.027 |
| CEA | -0.085 | 0.650 |
| Clinical stage | -0.603 | < 0.001 |
| T classification | -0.129 | 0.491 |
| N classification | -0.271 | 0.140 |
| Metastasis | -0.5 | 0.004 |
| Venous invasion | -0.392 | 0.029 |
| Lymphatic invasion | -0.611 | < 0.001 |
To identify whether circ-LDLRAD3 can serve as a biomarker in pancreatic cancer, ΔCT values were further evaluated. The area under the ROC curve (AUC) was 0.67; the cutoff value, sensitivity, and specificity were 9.315, 0.5738, and 0.7049, respectively. When combined with CA19-9, the AUC was increased to 0.87 and the sensitivity and specificity were 0.8033 and 0.9355, respectively (Figure 3).
There have been few recent therapeutic advances in the treatment of pancreatic cancer. For more than 10 years, surgery and chemotherapy with gemcitabine have been the standard treatment methods[17-19]; yet, only 13%-15% of patients with pancreatic cancer are likely to undergo pancreaticoduodenectomy[20]. Furthermore, patients with pancreatic cancer are prone to experience multidrug chemotherapy resistance[21]. There are several challenges in the diagnosis and treatment of pancreatic cancer. First, there is difficulty in making an early diagnosis. The pathological and biological characteristics of pancreatic cancer result in early symptoms which lack specificity[22]. Distant metastases have already occurred in roughly 50% of patients with pancreatic cancer at the time of treatment while the resection rate was only 15%[23]. Second, the heterogeneity of pancreatic cancer makes it difficult to treat. Whole genome analysis of pancreatic cancer shows that 12 core signaling pathways have genetic changes. Alterations in multiple genes and multiple pathways increase the difficulty of achieving effective treatment, resulting in poor prognoses[20,24]. Therefore, the key to the diagnosis and treatment of pancreatic cancer lies in early detection and diagnosis. Risk assessment of pancreatic cancer-relevant molecular markers in patients and early and timely intervention to prevent deterioration will have a positive effect on the diagnosis and treatment of pancreatic cancer[25,26].
CircRNAs are a novel class of RNAs with O-shaped closed structure that exist in the living cells. Unlike traditional linear RNA molecules, circRNAs are resistant to degradation by exonuclease and RNases because there are no 5’-end, 3’-end, or even poly(A) tail[27]. Hence, circRNA can stably exist in cells for a long period of time. Furthermore, circRNA molecules in human cells are ten-fold more numerous that the number of homogenetic linear isomer RNA molecules[28]. CircRNA molecules have highly conserved sequences, a stable existence, and tissue-specific expression; circRNAs have been demonstrated to regulate gene expression in post-transcriptional ways[29]. For example, circRNAs can act as microRNA (miRNA) sponges. Li et al[30] reported that circ-ITCH competitively sponged miRNA-7, miRNA-17, and miRNA-214, leading to higher expression of the ITCH gene. The ITCH gene product has been shown to inhibit Dvl2 phosphorylation and, furthermore, to inhibit the Wnt signaling pathway to prevent tumorigenesis in the esophagus[30]. In addition, many differentially expressed circRNAs have been investigated in tissue, blood[31], saliva[32], and other bodily fluid[33] samples, suggesting that circRNA molecules can serve as biomarkers in many diseases including diabetes mellitus[34], coronary artery disease[35], and cancer[10]. CircRNAs, together with other known biomarkers, may be able to improve the accuracy of specificity of diagnosis in certain diseases. However, little work has been published thus far regarding the role of circRNAs in pancreatic cancer.
This is the first study to report the expression pattern of circ-LDLRAD3 and its diagnostic value in pancreatic cancer. The expression of circ-LDLRAD3 was higher in pancreatic cancer cell lines, pancreatic cancer tissues, and plasma samples of patients with pancreatic cancer when compared to matched control samples. Moreover, the expression of circ-LDLRAD3 in metastatic pancreatic cell lines was higher than that in primary cell lines and there was a strong correlation between circ-LDLRAD3 expression and venous and lymphatic invasion in both tissues and plasma samples. Interestingly, in plasma samples, circ-LDLRAD3 was found to be associated with metastasis. Considering that there were no pancreatic cancer tissue samples with metastasis in the 30 patients tested, we strongly believe that the expression of circ-LDLRAD3 correlates with venous invasion, lymphatic invasion, and metastasis. These data indicate that circ-LDLRAD3 has potential to be a novel biomarker of metastatic pancreatic cancer with invasion potential.
This study provides a new avenue for the early diagnosis of pancreatic cancer, which has traditionally been clinically difficult[32]. The sensitivity and specificity of tumor marker CA19-9 in the diagnosis of pancreatic cancer are 79%-81% and 82%-90%, respectively. However, about 3%-7% of pancreatic cancer patients are Lewis antigen negative and also do not express CA19-9; abnormal CA19-9 levels are not detected in this type of patients[20,36,37]. In this study, serum levels of circ-LDLRAD3 were found to be closely related to blood CA19-9 levels. Compared with the diagnostic value of circ-LDLRAD3 alone in pancreatic cancer, whose AUC, sensitivity, and specificity were 0.67, 0.5738, and 0.7049, respectively, the combination of circ-LDLRAD3 and CA19-9 increased the diagnostic value, with corresponding values for AUC, sensitivity, and specificity were 0.87, 0.8033, and 0.9355, respectively. These results suggest that circ-LDLRAD3 has potential as a novel biomarker in the diagnosis of pancreatic cancer.
However, due to the limited number of available tissue and plasma samples from patients with pancreatic cancer, only 30 paired pancreatic cancer tissues and 31 matched plasma samples were analyzed. Studies utilizing a large number of samples in multiple centers should be implemented in future. The study of circ-LDLRAD3 function in pancreatic cancer is also likely to improve the understanding of the occurrence and progression mechanisms of pancreatic cancer.
In conclusion, our data indicate that circ-LDLRAD3 expression was significantly up-regulated in pancreatic cancer cell lines, pancreatic cancer tissues, and pancreatic cancer plasma samples. Furthermore, circ-LDLRAD3 expression was correlated with lymphatic invasion, venous invasion, and metastasis. Therefore, circ-LDLRAD3 has potential as a novel biomarker indicative of tumor invasion capacity in the diagnosis of pancreatic cancer.
Pancreatic cancer is a malignancy with a very poor prognosis. There have been few recent therapeutic advances in the treatment of pancreatic cancer for more than 10 years. The key to improving the prognosis of pancreatic cancer mostly lies in early diagnosis and early treatment. Circular RNAs (circRNAs) are a class of noncoding RNAs characterized by stable ring structure formed by a covalently closed continuous loop, which makes them stable in cells, tissues, and body fluid. Therefore, they can serve as ideal biomarkers for detection of diseases, especially cancer. This study indicates that circ-LDLRAD3 has potential as a novel biomarker indicative of tumor invasion capacity in the diagnosis of pancreatic cancer.
This study aimed to analyze and evaluate the diagnostic value of a new circular RNA, circ-LDLRAD3, in pancreatic cancer. And research data suggest that circ-LDLRAD3 may be used as a biomarker in pancreatic cancer diagnosis.
The main objectives in this study were pancreatic cancer and a new circular RNA, circ-LDLRAD3. The results showed that the expression level of circ-LDLRAD3 was up-regulated in pancreatic cancer and it can serve as a biomarker in pancreatic cancer.
The expression levels of circ-LDLRAD3 were detected using real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) in pancreatic cancer cell lines, normal pancreatic cell lines, paired pancreatic cancer tissues and adjacent non-tumorous tissues, and plasma samples from patients with pancreatic cancer and healthy volunteers. The relationship between circ-LDLRAD3 expression and patients’ clinicopathological factors was analyzed, the diagnostic value of circ-LDLRAD3 was further calculated alone and combined with CA19-9.
Our study found that expression levels of circ-LDLRAD3 were up-regulated in pancreatic cell lines, pancreatic cancer tissues, and plasma samples from pancreatic cancer patients. It may serve as a new biomarker in the diagnosis of pancreatic cancer. Studies utilizing a large number of samples in multiple centers should be implemented in future. The study of circ-LDLRAD3 function in pancreatic cancer is also likely to improve the understanding of the occurrence and progression mechanisms of pancreatic cancer.
This study indicated that the expression of a new circular RNA, circ-LDLRAD3, was significantly up-regulated in pancreatic cancer cell lines, pancreatic cancer tissues, and pancreatic cancer plasma samples. Furthermore, circ-LDLRAD3 expression was correlated with lymphatic invasion, venous invasion, and metastasis. Therefore, circ-LDLRAD3 has potential as a novel biomarker indicative of tumor invasion capacity in the diagnosis of pancreatic cancer. It is highly believed that the key to improving the prognosis of pancreatic cancer mostly lies in early diagnosis and early treatment. Therefore, searching for ideal biomarkers is essential. Circular RNAs are a class of non-coding RNAs which are stable because of their unique circular structure. Previous studies have confirmed that some circRNAs can serve as biomarkers in certain diseases. In this study, we focused a new circular RNA, circ-LDLRAD3, and hypothesized that expression levels of circ-LDLRAD3 were up-regulated in pancreatic cancer. Moreover, this study verified the hypothesis and found that expression levels of circ-LDLRAD3 were significantly up-regulated in pancreatic cancer cell lines, pancreatic cancer tissues, and pancreatic cancer plasma samples, whose expression levels were correlated with lymphatic invasion, venous invasion, and metastasis. Therefore, circ-LDLRAD3 may be a new biomarker in the diagnosis of pancreatic cancer.
This is the first study to report the expression pattern of circ-LDLRAD3 and its diagnostic value in pancreatic cancer and provides a new avenue for the early diagnosis of pancreatic cancer. However, due to the limited number of available tissue and plasma samples from patients with pancreatic cancer, studies utilizing a large number of samples in multiple centers should be implemented in future. The study of circ-LDLRAD3 function in pancreatic cancer is also likely to improve the understanding of the occurrence and progression mechanisms of pancreatic cancer. And more types of circular RNAs and their relationship with pancreatic cancer should be verified in the future research.
| 1. | Dimastromatteo J, Houghton JL, Lewis JS, Kelly KA. Challenges of Pancreatic Cancer. Cancer J. 2015;21:188-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 38] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 2. | Yamane S, Katada C, Tanabe S, Azuma M, Ishido K, Yano T, Wada T, Watanabe A, Kawanishi N, Furue Y. Clinical Outcomes in Patients with Cancer of Unknown Primary Site Treated By Gastrointestinal Oncologists. J Transl Int Med. 2017;5:58-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 3. | Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11444] [Cited by in RCA: 13321] [Article Influence: 1332.1] [Reference Citation Analysis (4)] |
| 4. | Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11065] [Cited by in RCA: 12228] [Article Influence: 1358.7] [Reference Citation Analysis (3)] |
| 5. | Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. 2016;388:73-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1397] [Cited by in RCA: 1786] [Article Influence: 178.6] [Reference Citation Analysis (1)] |
| 6. | Korenblit J, Tholey DM, Tolin J, Loren D, Kowalski T, Adler DG, Davolos J, Siddiqui AA. Effect of the time of day and queue position in the endoscopic schedule on the performance characteristics of endoscopic ultrasound-guided fine-needle aspiration for diagnosing pancreatic malignancies. Endosc Ultrasound. 2016;5:78-84. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 7. | Ozkan M, Cakiroglu M, Kocaman O, Kurt M, Yilmaz B, Can G, Korkmaz U, Dandil E, Eksi Z. Age-based computer-aided diagnosis approach for pancreatic cancer on endoscopic ultrasound images. Endosc Ultrasound. 2016;5:101-107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 70] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 8. | Urayama S. Pancreatic cancer early detection: expanding higher-risk group with clinical and metabolomics parameters. World J Gastroenterol. 2015;21:1707-1717. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 25] [Cited by in RCA: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 9. | Huang S, Yang B, Chen BJ, Bliim N, Ueberham U, Arendt T, Janitz M. The emerging role of circular RNAs in transcriptome regulation. Genomics. 2017;109:401-407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 161] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 10. | Wang Y, Mo Y, Gong Z, Yang X, Yang M, Zhang S, Xiong F, Xiang B, Zhou M, Liao Q. Circular RNAs in human cancer. Mol Cancer. 2017;16:25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 222] [Cited by in RCA: 314] [Article Influence: 34.9] [Reference Citation Analysis (0)] |
| 11. | Cortés-López M, Miura P. Emerging Functions of Circular RNAs. Yale J Biol Med. 2016;89:527-537. [PubMed] |
| 12. | Peng ZY. The biomarkers for acute kidney injury: A clear road ahead? J Transl Int Med. 2016;4:95-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 13. | Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol Rev. 2016;96:1297-1325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1378] [Cited by in RCA: 1364] [Article Influence: 136.4] [Reference Citation Analysis (2)] |
| 14. | Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666-1670. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1131] [Cited by in RCA: 1411] [Article Influence: 117.6] [Reference Citation Analysis (0)] |
| 15. | Qu S, Song W, Yang X, Wang J, Zhang R, Zhang Z, Zhang H, Li H. Microarray expression profile of circular RNAs in human pancreatic ductal adenocarcinoma. Genom Data. 2015;5:385-387. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 78] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 16. | Ghosal S, Das S, Sen R, Basak P, Chakrabarti J. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet. 2013;4:283. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 306] [Cited by in RCA: 379] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 17. | Strobel O, Büchler MW. Pancreatic cancer: Clinical practice guidelines - what is the evidence? Nat Rev Clin Oncol. 2016;13:593-594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 18. | Makohon-Moore A, Iacobuzio-Donahue CA. Pancreatic cancer biology and genetics from an evolutionary perspective. Nat Rev Cancer. 2016;16:553-565. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 281] [Cited by in RCA: 319] [Article Influence: 31.9] [Reference Citation Analysis (0)] |
| 19. | Van Ryckeghem F. Corticosteroids, the oldest agent in the prevention of chemotherapy-induced nausea and vomiting: What about the guidelines? J Transl Int Med. 2016;4:46-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 20. | Notta F, Chan-Seng-Yue M, Lemire M, Li Y, Wilson GW, Connor AA, Denroche RE, Liang SB, Brown AM, Kim JC. Erratum: A renewed model of pancreatic cancer evolution based on genomic rearrangement patterns. Nature. 2017;542:124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 21. | Roe JS, Hwang CI, Somerville TDD, Milazzo JP, Lee EJ, Da Silva B, Maiorino L, Tiriac H, Young CM, Miyabayashi K. Enhancer Reprogramming Promotes Pancreatic Cancer Metastasis. Cell. 2017;170:875-888.e20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 364] [Article Influence: 40.4] [Reference Citation Analysis (2)] |
| 22. | Michalski CW, Hackert T, Büchler MW. Targeting metabolism in pancreatic cancer. Lancet Oncol. 2017;18:699-700. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 6] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 23. | Tsutsumi H, Hara K, Mizuno N, Hijioka S, Imaoka H, Tajika M, Tanaka T, Ishihara M, Yoshimura K, Shimizu Y. Clinical impact of preoperative endoscopic ultrasound-guided fine-needle aspiration for pancreatic ductal adenocarcinoma. Endosc Ultrasound. 2016;5:94-100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 30] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 24. | Yamabe A, Irisawa A, Bhutani MS, Shibukawa G, Fujisawa M, Sato A, Yoshida Y, Arakawa N, Ikeda T, Igarashi R. Efforts to improve the diagnostic accuracy of endoscopic ultrasound-guided fine-needle aspiration for pancreatic tumors. Endosc Ultrasound. 2016;5:225-232. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 25. | Zhu P, Sun S. Endoscopic ultrasound pin-points the precision medicine for pancreatic cancer. Endosc Ultrasound. 2016;5:1-3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 26. | Kleeff J, Michl P. Targeted therapy of pancreatic cancer: biomarkers are needed. Lancet Oncol. 2017;18:421-422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 27. | Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1024] [Cited by in RCA: 1384] [Article Influence: 125.8] [Reference Citation Analysis (6)] |
| 28. | Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1007] [Cited by in RCA: 1336] [Article Influence: 133.6] [Reference Citation Analysis (0)] |
| 29. | Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4631] [Cited by in RCA: 6216] [Article Influence: 478.2] [Reference Citation Analysis (0)] |
| 30. | Li F, Zhang L, Li W, Deng J, Zheng J, An M, Lu J, Zhou Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget. 2015;6:6001-6013. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 449] [Cited by in RCA: 570] [Article Influence: 57.0] [Reference Citation Analysis (0)] |
| 31. | Zhang YG, Yang HL, Long Y, Li WL. Circular RNA in blood corpuscles combined with plasma protein factor for early prediction of pre-eclampsia. BJOG. 2016;123:2113-2118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 93] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 32. | Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y, Wong DT, Xiao X. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem. 2015;61:221-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 447] [Cited by in RCA: 534] [Article Influence: 44.5] [Reference Citation Analysis (1)] |
| 33. | Qu S, Zhong Y, Shang R, Zhang X, Song W, Kjems J, Li H. The emerging landscape of circular RNA in life processes. RNA Biol. 2017;14:992-999. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 233] [Cited by in RCA: 330] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 34. | Zhao Z, Li X, Jian D, Hao P, Rao L, Li M. Hsa_circ_0054633 in peripheral blood can be used as a diagnostic biomarker of pre-diabetes and type 2 diabetes mellitus. Acta Diabetol. 2017;54:237-245. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 161] [Cited by in RCA: 194] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 35. | Zhao Z, Li X, Gao C, Jian D, Hao P, Rao L, Li M. Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci Rep. 2017;7:39918. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 141] [Cited by in RCA: 191] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 36. | Bhutani MS, Koduru P, Joshi V, Saxena P, Suzuki R, Irisawa A, Yamao K. The role of endoscopic ultrasound in pancreatic cancer screening. Endosc Ultrasound. 2016;5:8-16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 34] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 37. | Mohammad Alizadeh AH, Shahrokh S, Hadizadeh M, Padashi M, Zali MR. Diagnostic potency of EUS-guided FNA for the evaluation of pancreatic mass lesions. Endosc Ultrasound. 2016;5:30-34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
P- Reviewer: Chamberlain MC, Higuchi K, Jones G S- Editor: Gong ZM L- Editor: Wang TQ E- Editor: Huang Y