Published online Dec 7, 2017. doi: 10.3748/wjg.v23.i45.7965
Peer-review started: August 10, 2017
First decision: September 10, 2017
Revised: September 26, 2017
Accepted: October 26, 2017
Article in press: October 26, 2017
Published online: December 7, 2017
Processing time: 116 Days and 18.7 Hours
To investigate the potential role of microRNA-30a (miR-30a) in esophageal squamous cell carcinoma (ESCC).
Expression of miR-30a-3p/5p was analyzed using microarray data and fresh ESCC tissue samples. Both in vitro and in vivo assays were used to investigate the effects of miR-30a-3p/5p on ESCC cell proliferation. Furthermore, Kyoto Encyclopedia of Genes and Genomes analysis was performed to explore underlying mechanisms involved in ESCC, and then, assays were carried out to verify the potential molecular mechanism of miR-30a in ESCC.
Low expression of miR-30a-3p/5p was closely associated with advanced ESCC progression and poor prognosis of patients with ESCC. Knock-down of miR-30a-3p/5p promoted ESCC cell proliferation. Increased miR-30a-3p/5p expression inhibited the Wnt signaling pathway by targeting Wnt2 and Fzd2.
Down-regulation of miR-30a-3p/5p promotes ESCC cell proliferation by activating the Wnt signaling pathway through inhibition of Wnt2 and Fzd2.
Core tip: In this work, we found that low expression of miR-30a-3p/5p was closely associated with advanced esophageal squamous cell carcinoma (ESCC) progression and poor prognosis of patients with ESCC. Down-regulation of miR-30a-3p/5p suppressed ESCC cell proliferation both in vitro and in vivo. Furthermore, miR-30a-3p and miR-30a-5p could inhibit the activity of the Wnt signaling pathway by targeting the 3’ untranslated regions of Wnt2 and Fzd2, respectively. This study provided further evidence suggesting that miR-30a-3p/5p are diagnostic and prognostic biomarkers for ESCC, as miR-30a-3p/5p participate in the activation of the Wnt signaling pathway and subsequently, the regulation of ESCC cell proliferation.
- Citation: Qi B, Wang Y, Chen ZJ, Li XN, Qi Y, Yang Y, Cui GH, Guo HZ, Li WH, Zhao S. Down-regulation of miR-30a-3p/5p promotes esophageal squamous cell carcinoma cell proliferation by activating the Wnt signaling pathway. World J Gastroenterol 2017; 23(45): 7965-7977
- URL: https://www.wjgnet.com/1007-9327/full/v23/i45/7965.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i45.7965
Esophageal cancer is one of the most common human malignancies, ranking sixth among cancer-related deaths worldwide[1]. Esophageal squamous cell carcinoma (ESCC) is the major histological type and the leading cause of death from all esophageal cancer types in Asian countries, especially in China[2,3]. Because of the lack of early detection, the majority of patients with ESCC are diagnosed at advanced stages with high risk of metastasis and recurrence[4]. The 5-year overall survival rate of ESCC is less than 20%[1]; therefore, further investigation of the molecular mechanisms involved in ESCC is urgent and essential for developing early diagnostic and therapeutic strategies.
The development and progression of ESCC involve synergic effects of various pathogenic factors, including particular dietary factors (chemical and physical), human papillomavirus infection, and genetic susceptibility[5]. To further investigate genetic susceptibility and develop personalized targeted therapy for ESCC, high-throughput techniques have been used. Genetic landscapes of ESCC obtained by whole genome and exome sequencing have illustrated that genomic alterations in ESCC include single nucleotide variants, copy number alterations, and alterations in multiple signaling pathways, such as cell cycle regulation, DNA damage control, RTK-Ras-MAPK-PI3K-Akt, Notch, and Wnt[6,7]. Many researchers have recently reported the significance of both canonical and non-canonical Wnt signaling pathways in ESCC, thereby indicating the potential of the Wnt signaling pathway markers as prognostic and therapeutic targets[8-12]. However, the regulation of the Wnt signaling pathway in ESCC remains largely unknown.
MicroRNAs are a class of small (21-23 nt), single-stranded non-coding RNAs that regulate gene expression post-transcriptionally by binding to the 3’-untranslated region (UTR) of target mRNAs. This typically causes mRNA degradation or translation repression[13]. Highly conserved across species, microRNAs not only participate in biological processes[14], but also in the pathogenesis of human cancers[15].
MicroRNA-30 (miR-30) family is evolutionarily conserved and consist of five members, microRNA-30a (miR-30a) through miR-30e. miR-30 family members play different roles, as oncogenes or tumor suppressor genes, in different kinds of cancer. For instance, miR-30 family members inhibit non-small-cell lung cancer[16,17], breast cancer[18,19], and colorectal cancer[20,21], but promote glioma[22], gastric cancer[23], and pancreatic cancer[24]. The miR-30 family is involved in the regulation of cancer cell apoptosis, proliferation, invasiveness, and metastasis, as well as in epithelial-mesenchymal transition. In particular, miR-30 targets oncogenes and tumor suppressor genes under different circumstances, the detailed/complete mechanism of which remains to be explored.
Emerging evidence has indicated that the two strands of miR-30a (miR-30a-3p and miR-30a-5p) are involved in various kinds of cancer. Recent studies have revealed that miR-30a-3p/5p are closely associated with the Wnt signaling pathway in breast cancer, multiple myeloma, and glioma[19,25,26]; however, little has been reported about the expression and roles of miR-30a-3p/5p in ESCC progression. Analysis of public microarray data along with our previous experiment results has shown that miR-30a-3p/5p are down-regulated in ESCC in comparison with matched adjacent normal tissues. Additionally, bioinformatics analyses have indicated that the target genes of miR-30a-3p/5p were significantly enriched in the Wnt signaling pathway. Based on these findings, we sought to investigate the relationship between miR-30a-3p/5p expression and ESCC prognosis and the underlying mechanisms of the Wnt signaling pathway in ESCC.
This study was conducted on 99 pairs of fresh ESCC tissue biopsies and matched adjacent normal tissues from the operating room of our hospital. Medical records of corresponding patients provided information on gender, age, differentiation, TNM stage, survival time. The fresh biopsies were stored in liquid nitrogen before usage. Prior patient consent and approval from the Institutional Research Ethics Committee were obtained.
The human ESCC cell lines KYSE30 and KYSE150 and normal esophageal epithelial cell line Het-1A were cultured in DMEM medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, United States), supplemented with 10% fetal bovine serum (FBS) (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, United States) at 37 °C with 5% CO2.
Total RNA from cultured cells and ESCC tissues was isolated using the mirVana miRNA Isolation Kit (Ambion, Austin, TX, United States) according to the manufacturer’s instruction. Taqman miRNA reverse transcription kit (Applied Biosystems, Foster City, CA, United States) was then used to synthesize cDNA from total RNA. Using the iQ™ SYBR Green Supermix (BioRad Laboratories, Hercules, CA, United States) and the Applied Biosystems 7500 Sequence Detection System, quantitative polymerase chain reaction (qPCR) was performed. The positive control (genomic DNA) and negative controls (PBS and samples processed without the RT step) were included. Data were normalized to the geometric mean of the housekeeping gene GAPDH or U6 values (internal control of small nuclear RNA expression) and analyzed using the 2-ΔΔCT method. Sequences of the primers for qPCR are summarized in Table S1.
Proteins were isolated, subjected to SDS-PAGE, transferred onto polyvinylidene fluoride (PVDF) membranes, and incubated with anti-Cyclin D1 (Abcam, Cambridge, MA, United States), anti-p27 (Abcam, Cambridge, MA, United States), anti-p21 (Abcam, Cambridge, MA, United States), anti-WNT2 (Bioworld Technology Inc. St. Louis Park, MN, United States), and anti-FZD2 (Bioworld Technology Inc. St. Louis Park, MN, United States) antibodies. α-tubulin (Sigma-Aldrich; Merck Millipore) was used as a loading control. Immunoreactive proteins were then detected by chemiluminescence. All the above operations were performed according to standard methods[27].
To construct the plasmids expressing miR-30a-3p or miR-30-5p, the fragments of pri-miR-30a were amplified using PCR, and then, respectively, cloned into the lentiviral vector pLVTHM (Addgene Inc., Cambridge, MA, United States). The mimics, negative controls, and inhibitors of miR-30a-3p or miR-30a-5p were purchased from Genecopoeia (Guangzhou, Guangdong, China), and transfected into cells with Lipofectamine 2000 reagent (Invitrogene) according to the manufacturer’s instructions. To perform luciferase assay, small regions containing the target sequences of miR-30a-3p or miR-30a-5p in 3’-UTR were generated by PCR amplification, and cloned into psi-CHECK luciferase reporter plasmid (Promega). Two concentrations of miR-30a-3p or miR-30a-5p-mimics (10 nmol/L and 20 nmol/L) plus wild-type or mutant 3’-UTR of the target genes were applied. The primers used are listed in Table S2.
Cells (1 × 103) were seeded on 96-well plates and cultured for 24 h. Twenty microliters of 5 g/L of 3-(4,5-dimethylthiazol-z-yl)-2,5-diphenyltetrazolium bromide (MTT, Sigma, St Louis, MO, United States) was added to each well and incubated for 4 h. After MTT was removed, 150 μL of dimethyl sulphoxide DMSO (sigma, St, Louis, MO, United States) was added to the wells. Absorbance was measured at 490 nm with a Microplate Autoreader (Bio-Rad, Hercules, CA, United States). The experiment was repeated three times. Data are presented as the mean ± SD.
Cells were trypsinized, plated on 6-well plates (200 cells/well), and cultured for 2 wk. The colonies were stained with 1% crystal violet for 30 s after fixation with 4% paraformaldehyde for 5 min. The number of colonies, defined as > 50 cells/colony, were counted. Data are presented as the mean ± SD for three dependent experiments.
Five hundred cells were suspended in 2 mL of complete medium containing 0.3% agar (Sigma, St Louis, MO). Then, the agar-cell mixture was plated on top of a bottom layer with 1% complete medium-agar mixture. Ten days later, the colonies were measured with an ocular micrometer. Colonies larger than 0.1 mm in diameter were counted. The experiment was repeated independently three times for each cell line. Data are presented as the mean ± SD.
Cells (4 × 104) were seeded in 24-well plates and settled for 24 h. Then, 1.5 μg of the TOPFlash (b-catenin/TCF reporter) and its mutant control FOPFlash luciferase reporter plasmid, plus 0.15 μg of pRL-TK Renilla plasmid (Promega), were, respectively, transfected into cells using the Lipofectamine 2000 reagent according to the manufacturer’s recommendation. Luciferase and Renilla signal was measured 24 h after transfection using the Dual Luciferase Reporter Assay Kit (Promega corp., Madison, WI, United States) according to the protocol provided by the manufacturer. All the experiments were performed in triplicate.
Xenograft tumours were generated by subcutaneous injection of ESCC cells (2 × 106) into the hindlimbs of 4-6 week-old Balb/C athymic nude mice (nu/nu; Animal Center of XinXiang Medical University, HeNan China. n = 6 for each group). All mice were housed and maintained under specific pathogen-free conditions, and all experiments were approved by the Animal Care and Use Committee and performed in accordance with institutional guidelines. Tumour size was measured using a slide calliper and tumour volume was determined by the formula 0.44 × A × B2, where A represents the diameter of the base of the tumour and B represents the corresponding perpendicular value. After euthanasia, the tumours were excised, fixed in 10% neutral buffered formalin, embedded in paraffin, cut into 4 μm sections, and stained with haematoxylin.
The public microarray data for analysis of miR-30a-3p/5p expression in this study was downloaded from the GEO database (GSE43732).
All statistical analyses were performed using SPSS version 20.0 for Windows (IBM, Armonk, NY, United States). Statistical tests included log-rank test, χ2 test, and the Student’s t-test. The two-tailed Student’s t-test was used to compare intergroup differences. Survival data were analyzed by the Kaplan-Meier method and were compared using the log-rank test. P < 0.05 was considered statistically significant.
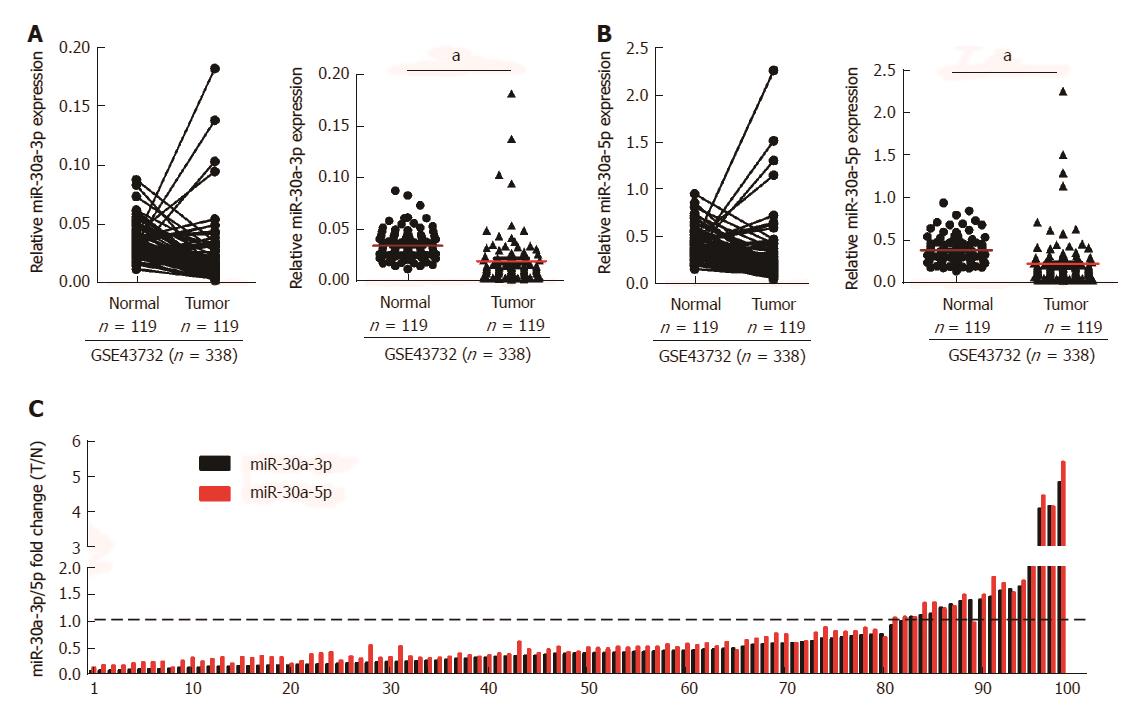
To investigate the role of miR-30a-3p/5p in ESCC, we initially analyzed the expression of miR-30a-3p/5p in ESCC tissues and normal tissues, as well as using public microarray data (GSE43732, n = 338) from GEO database (https://http://www.ncbi.nlm.nih.gov/geo/). Results showed that both miR-30a-3p and miR-30a-5p were down-regulated in ESCC tissues when compared with adjacent normal tissues (Figures 1A and B). Additionally, we detected the expression of miR-30a-3p/5p in 99 cases of fresh ESCC biopsies and their paired adjacent normal tissues by qPCR. Consistent with the public microarray data, miR-30a-3p/5p were down-regulated in 81.82% (81/99) of ESCC tissues compared to their expression in the matched adjacent normal tissues (Figure 1C).
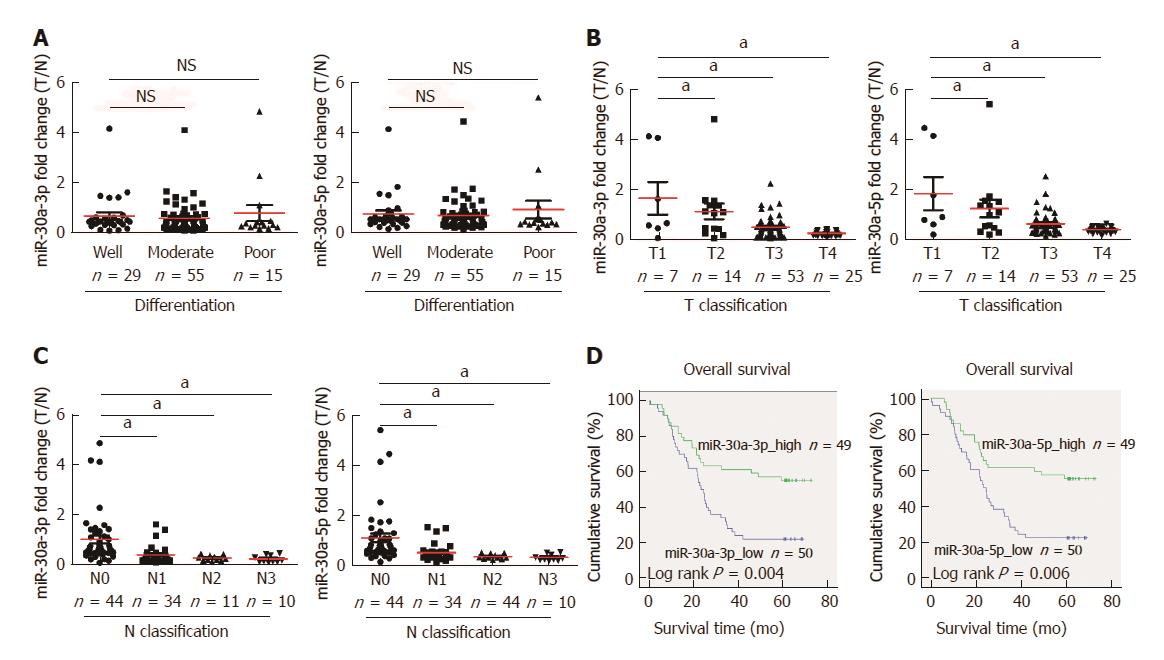
Statistical analyses further revealed no difference in the miR-30a-3p/5p expression between different differentiation statuses of ESCC (Figure 2A), but miR-30a-3p/5p expression was inversely correlated with classifications of primary tumor and lymphatic metastasis (Figures 2B and C). Most importantly, survival analysis indicated that the group of lower miR-30a-3p/5p expression had shorter 5-year overall survival, and was associated with poorer prognosis of patients with ESCC (Figure 2D, log-rank, P < 0.05).
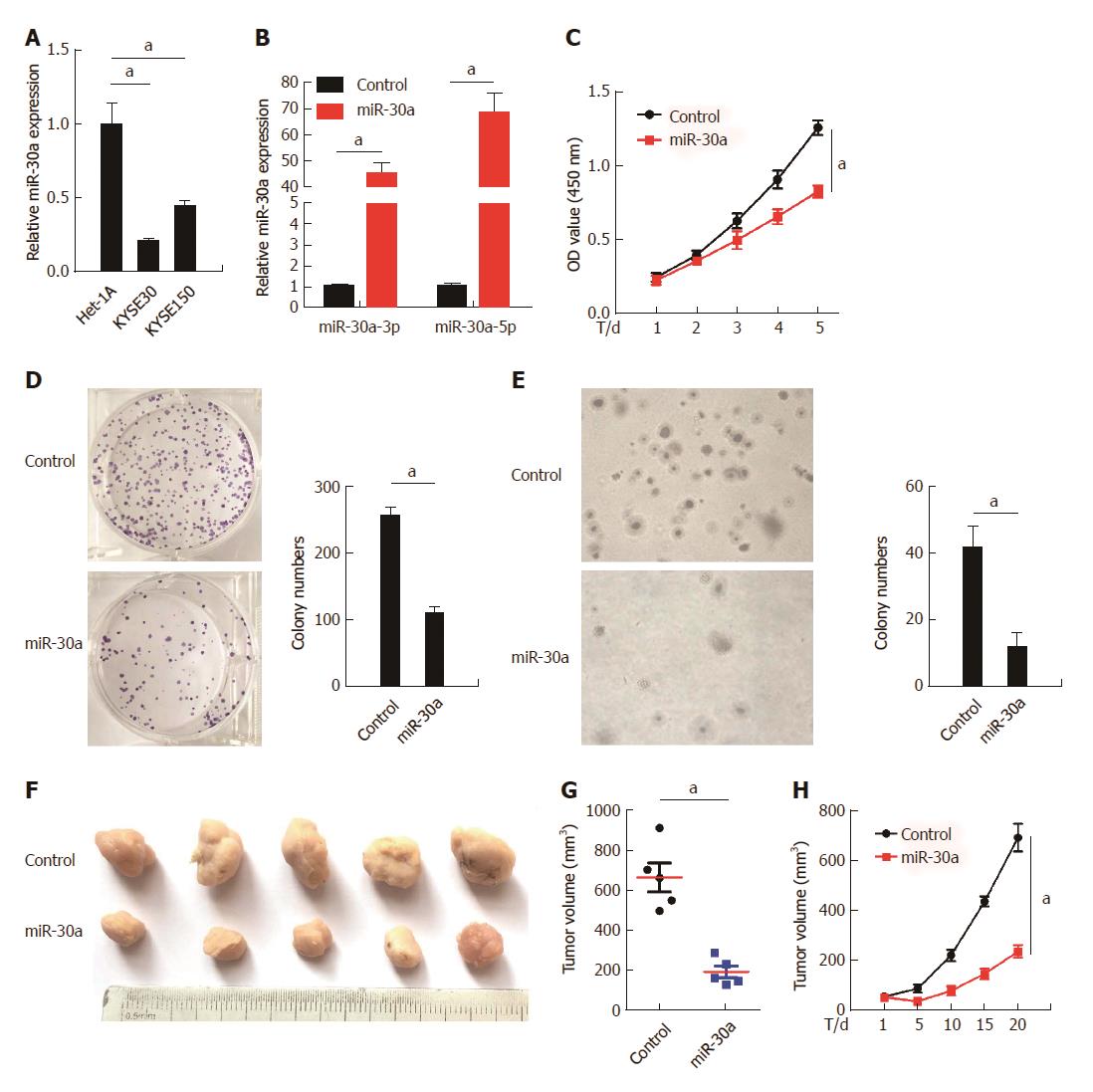
As shown in Figure 3A, expression of miR-30a-3p/5p was significantly lower in ESCC cell lines KYSE30 and KYSE150 than in normal esophageal epithelial cell line Het-1A. To evaluate the potential roles of miR-30a-3p/5p in ESCC pathogenesis, we over-expressed miR-30a-3p/5p in KYSE30 cells by transfecting miR-30a-3p/5p-mimic oligonucleotides (Figure 3B). MTT and colony formation assays indicated that over-expression of miR-30a-3p/5p significantly repressed the proliferation of HYSE30 cells in comparison with the control group. It should be noted that the anchorage-independent growth ability of KYSE30 cells was also attenuated by over-expression of miR-30a-3p/5p in the soft-agar assays. To confirm this effect in vivo, KYSE30 cells were engineered to stably over-express miR-30a-3p/5p, and then, subcutaneously injected into the nude mice to perform tumorigenesis assays. Results showed that the miR-30a-3p/5p over-expression group exhibited remarkably smaller tumor volume and slower tumor growth rate in comparison with the control group (Figures 3F, G, and H).
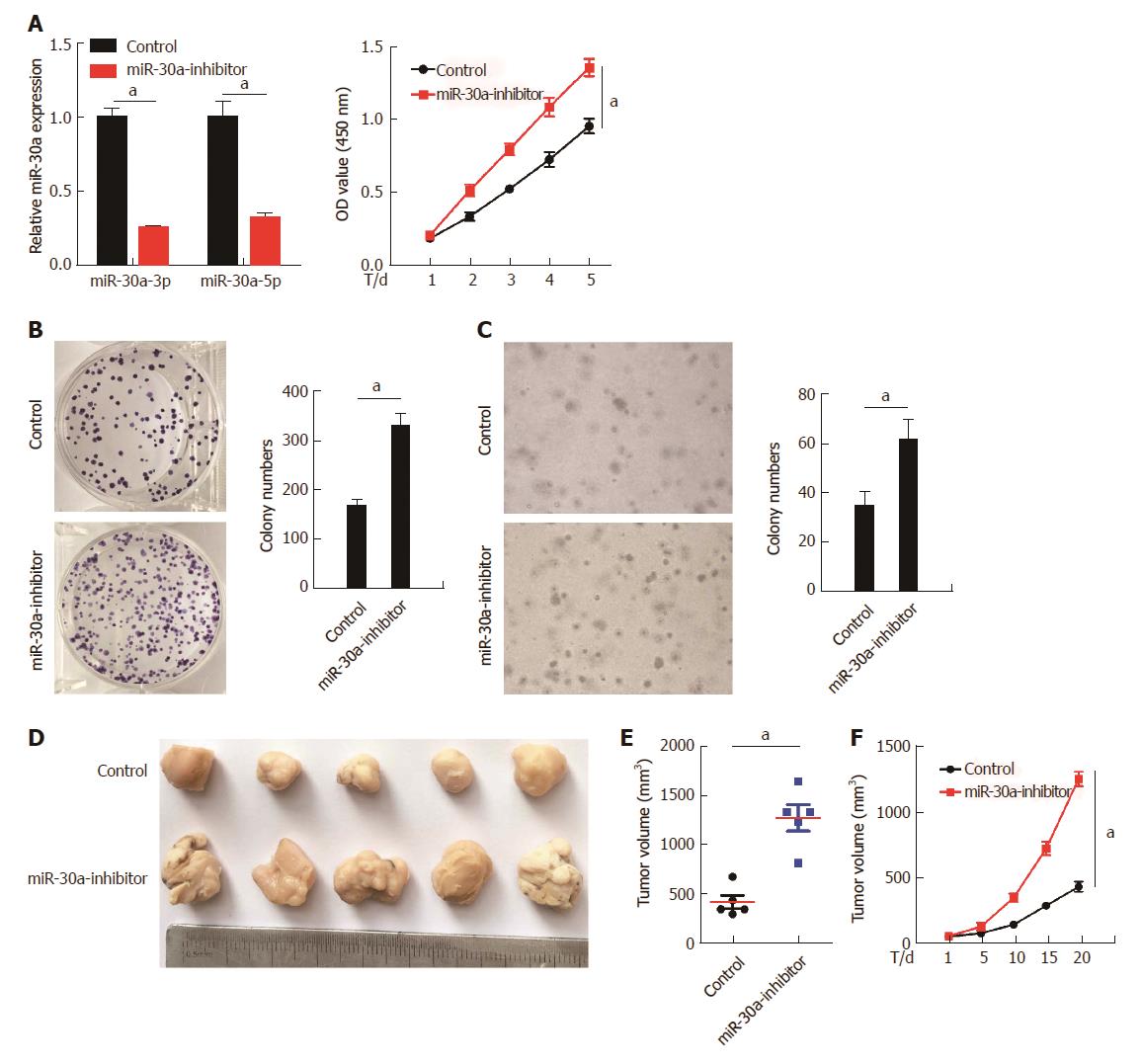
To further confirm the role of miR-30a-3p/5p in ESCC cell proliferation, we suppressed the expression of miR-30a-3p/5p in KYSE150 cells by expressing miR-30a inhibitor (Figure 4A, top). As indicated by the MTT and colony formation assays (Figure 4A bottom and B), inhibition of miR-30a-3p/5p significantly increased the proliferation of KYSE150 cells in comparison with the control groups. In addition, the anchorage-independent growth ability of KYSE150 cells was enhanced by inhibition of miR-30a-3p/5p in the soft-agar assays (Figure 4C). Tumorigenesis assays performed in the nude mice showed that inhibition of miR-30a-3p/5p increased the tumor volume and tumor growth rate in comparison with the control group (Figure 4D, E, and F).
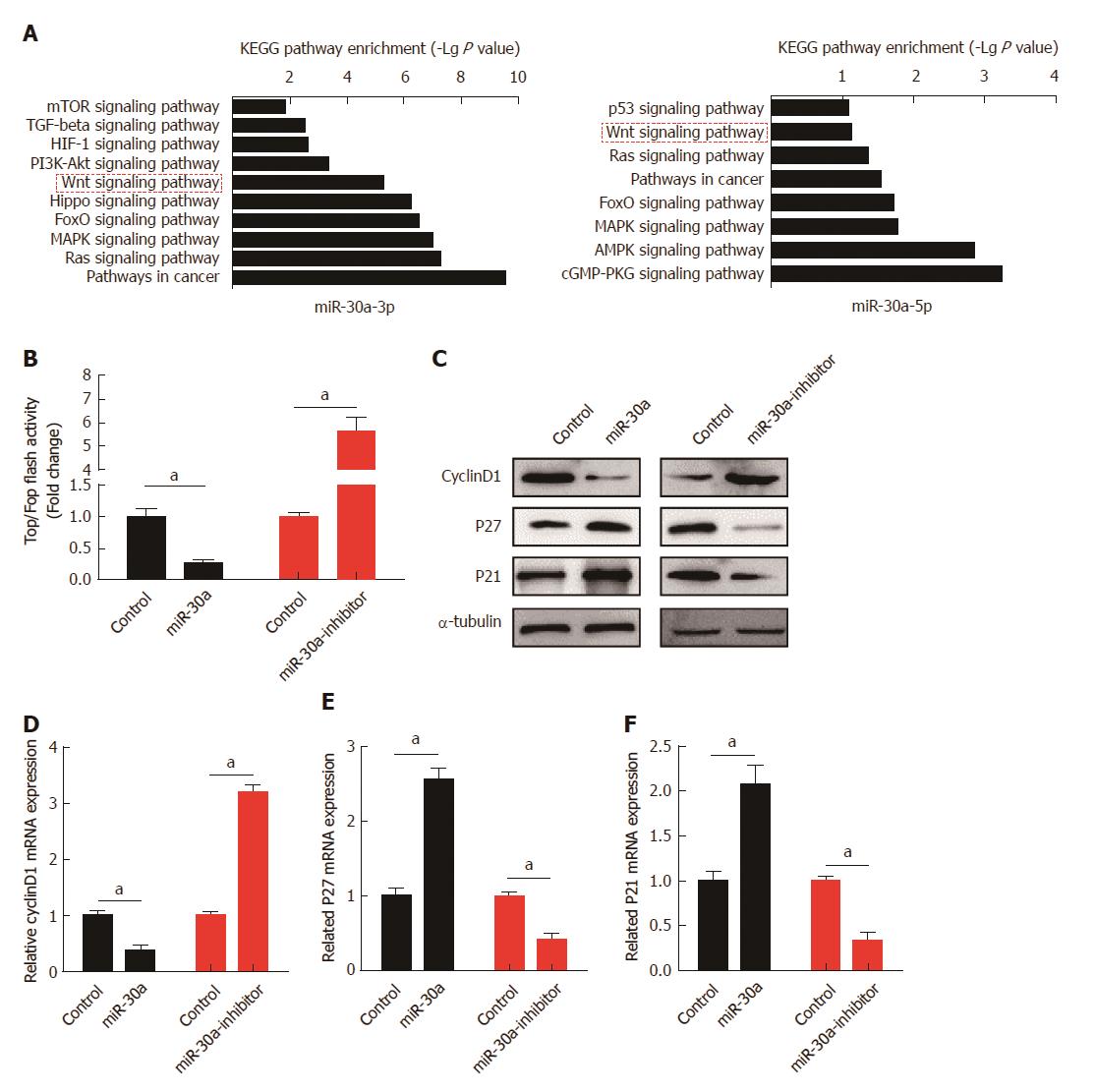
Both in vitro and in vivo experiments indicated that miR-30a-3p/5p served as tumor suppressors in ESCC. To figure out the underlying mechanisms, target genes of miR-30a-3p and miR-30a-5p were predicted using public algorithm miRWalk2.0 (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/index.html), and KEGG pathway enrichment analyses were performed. Results suggested that target genes of miR-30a-3p or miR-30a-5p were involved in the Wnt signaling pathway (Figure 5A). The luciferase reporter assay showed that over-expression of miR-30a significantly attenuated the activity of the Wnt signaling pathway, while inhibition of miR-30a remarkably enhanced it (Figure 5B). Analyses of downstream genes of the Wnt signaling pathway further indicated that over-expression of miR-30a decreased the expression of Cyclin D1 and increased the expression of p27 and p21. In contrast, inhibition of miR-30a increased the expression of Cyclin D1 and decreased the expression of p27 and p21, both at protein and mRNA levels, as indicated by the results of Western blot and qPCR analyses, respectively (Figures 5C, D, E, and F).
We next performed qPCR analyses to screen the potential target genes of miR-30a-3p/5p that are related to Wnt signaling. Results showed that mRNA expression of Wnt2 was repressed by transfecting miR-30a-3p-mimics, and Fzd2 was inhibited by miR-30a-5p-mimics (Figure 6A). Analyses using public algorithms miRWalk2.0 indicated that Wnt2 and Fzd2 might be the respective targets of miR-30a-3p and miR-30a-5p (Figure 6B). Then, we respectively validated the effects of miR-30a-3p and miR-3a-5p on Wnt2 and Fzd2 expression. As shown in Figure 6C, over-expression of miR-30a-3p decreased the expression of Wnt2, while inhibition of miR-30a-3p increased it, at both protein and mRNA levels. The miR-30a-5p showed the same effects on Fzd2 expression. Luciferase reporter assays also demonstrated that over-expression of miR-30a-3p significantly reduced luciferase activity of the wild-type Wnt2-3’-UTR in a dose-dependent pattern, while it had no effects on the mutant type. Likewise, miR-30a-5p had the same effects on the wild-type FZD2-3’-UTR and the mutant one (Figure 6D).
MicroRNAs regulate protein translation at the post-transcriptional level by binding to mRNAs of target genes. Emerging evidence has shown that the dysregulation of microRNAs plays an important role in multiple tumor-related processes, such as cell differentiation, proliferation, apoptosis, autophagy, angiogenesis, invasion, and metastasis. In this study, we found that, according to analyses of public microarray data and validation in ESCC biopsies, miR-30a-3p/5p were down-regulated in ESCC tissues in comparison with matched adjacent normal tissues. It was interesting that aberrant expression of miR-30a-3p/5p was found in many kinds of cancer and showed opposite tendencies. miR-30a-3p/5p were down-regulated in bladder cancer[28], breast cancer[29,30], lung cancer[31,32], hepatocellular carcinoma[33], cutaneous squamous cell carcinoma[34], and ESCC[35], but were up-regulated in glioma[36], nasopharyngeal carcinoma[37], papillary thyroid carcinoma[38], ovarian serous adenocarcinoma[39], and head and neck squamous cell carcinoma[40]. These opposite tendencies of miR-30a-3p/5p in different cancers may lie in the different cell types and different genetic background.
It has been found that because of the particular expression patterns of microRNAs in cancers, some signatures consisting of microRNAs are linked with cancer progression and prognosis. For instance, the 5-microRNA classifier, including miR-210, miR-182, miR-486-5p, miR-30a, and miR-140-3p, could distinguish lung squamous cell carcinoma from normal lung tissues[41]. Another miRNA signature, including miR-451, miR-221, miR-30a, miR-10b, and miR-29a, has been identified to distinguish between metastatic and non-metastatic clear cell renal cell carcinoma (ccRCC)[42]. In our study, down-regulation of miR-30a-3p/5p expression was found in ESCC tissues, and it was significantly correlated with advanced status of primary tumor and lymph node metastasis, as well as poor prognosis of patients with ESCC. The abnormal expression pattern of miR-30a-3p/5p in ESCC might also serve as potential diagnostic and prognostic markers in ESCC.
Apart from the expression pattern, it has been observed that miR-30a-3p/5p exhibited multiple roles in the regulation of tumor progression. miR-30a could suppress breast cancer cell migration and invasion[19]. In non-small cell lung cancer (NSCLC), miR-30a has been found to be inversely associated with invasive ability, and it suppressed epithelial to mesenchymal transition (EMT) of NSCLC cells[16]. In a similar fashion, down-regulation of miR-30a expression in hepatocellular carcinoma accelerated tumor cell migration, invasion, and EMT[43]. In contrast, miR-30a-5p has been identified as an oncogenic factor in glioma, and knock-down of miR-30a-5p inhibited glioma cell growth and cell invasion, while over-expression of miR-30a-5p had had opposite effects[44]. In this study, we revealed that over-expression of miR-30a-3p/5p suppressed the proliferation of ESCC cells, while down-regulation of miR-30a-3p/5p expression had the opposite effect both in in vitro and in vivo assays.
Our functional approach showed that miR-30a-3p/5p function as tumor suppressors in ESCC. The published microarray analysis, displaying that the down-regulation of miR-30a-3p/5p expression was correlated with the activation of Wnt signaling in ESCC, supplemented our findings. There are still gaps in the literature, as the molecular mechanisms by which miR-30a-3p/5p regulate the activation of Wnt signaling are unclear. Wnt signaling plays an essential role in various diseases and is considered a hallmark of colorectal tumorigenesis. Colorectal tumorigenesis is initiated by mutations in the Wnt signaling pathway (e.g., APC or beta-catenin) that constitutively activate the pathway, and also by binding a Wnt-protein ligand to a Frizzled family receptor, passing the biological signal to the dishevelled protein inside the cell, and leading to the regulation of gene transcription[45]. Accumulating literature has recently indicated that miRNAs regulate components of Wnt signaling in various cancers[46]. For example, miR-200a directly targets beta-catenin (beta-carotene) to inhibit cell proliferation in meningiomas[47], while miR-34 mediates suppression of Axin-2 to increase nuclear GSK3-beta and decrease Snai1 in colorectal cancer progression[48]. In addition, miR-30a-5p has been found to directly suppress PRDM1, resulting in activation of Wnt/beta-catenin (carotene) signaling in glioma[49].
We demonstrated that miR-30a-3p and miR-30a-5p directly target the 3’-UTRs of Wnt2 and Fzd2 and inhibit their expression, respectively. This leads to the inhibition of the Wnt signaling pathway, which might be the dominant component in miR-30a-3p/5p in regulating ESCC cell proliferation.
In conclusion, low expression of miR-30a-3p/5p was closely associated with advanced ESCC progression and poor prognosis of patients with ESCC. Down-regulation of miR-30a-3p/5p suppressed ESCC cell proliferation both in vitro and in vivo. Furthermore, miR-30a-3p and miR-30a-5p could inhibit the activity of the Wnt signaling pathway by targeting the 3’UTRs of WNT2 and FZD2, respectively. This study provided further evidence suggesting that miR-30a-3p/5p are diagnostic and prognostic biomarkers for ESCC, as miR-30a-3p/5p participate in the activation of the Wnt signaling pathway and subsequently, the regulation of ESCC cell proliferation.
MicroRNA-30a (miR-30a) serves as a post-transcriptional regulator by directly targeting mRNAs in many biological processes, and it shows multiple roles in different kinds of cancer. Wnt signaling pathway is well known in the development and progression of various cancers. MiR-30a was recently found to be closely associated with Wnt signaling pathway in cancers; however, the potential role and underlying mechanism of miR-30a in esophageal squamous cell carcinoma (ESCC) have not been illustrated.
To investigate the potential role of microRNA-30a (miR-30a) in ESCC.
The study investigated the potential role of microRNA-30a (miR-30a) in ESCC, which is urgent and essential for developing early diagnostic and therapeutic strategies.
Expression of miR-30a-3p/5p was analyzed using microarray data and fresh ESCC tissue samples. Both in vitro and in vivo assays were used to investigate the effects of miR-30a-3p/5p on ESCC cell proliferation. Furthermore, Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was performed to explore underlying mechanisms involved in ESCC, and then, assays were carried out to verify the potential molecular mechanism of microRNA-30a (miR-30a) in ESCC.
Low expression of miR-30a-3p/5p was closely associated with advanced ESCC progression and poor prognosis of patients with ESCC. Knock-down of miR-30a-3p/5p promoted ESCC cell proliferation. We further demonstrated that increased miR-30a-3p/5p expression inhibited the Wnt signaling pathway by targeting Wnt2 and Fzd2.
Down-regulation of miR-30a-3p/5p promotes ESCC cell proliferation by activating the Wnt signaling pathway through inhibition of Wnt2 and Fzd2.
This study will provide an example for investigating the relationship between the expression of microRNAs and ESCC prognosis and the underlying mechanisms of the Wnt signaling pathway in ESCC. The direction of the future research is to provide more evidence for developing novel strategies by targeting microRNA-30a in ESCC. In our future research, the long-acting microRNA-30a will be used to treat the ESCC cells or animal models, and to observe the inhibitory effect of microRNA-30a on ESCC cells.
| 1. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12135] [Cited by in RCA: 13046] [Article Influence: 1304.6] [Reference Citation Analysis (3)] |
| 2. | Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11444] [Cited by in RCA: 13323] [Article Influence: 1332.3] [Reference Citation Analysis (4)] |
| 3. | Rustgi AK, El-Serag HB. Esophageal carcinoma. N Engl J Med. 2014;371:2499-2509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 817] [Cited by in RCA: 1035] [Article Influence: 86.3] [Reference Citation Analysis (0)] |
| 4. | Headrick JR, Nichols FC 3rd, Miller DL, Allen MS, Trastek VF, Deschamps C, Schleck CD, Thompson AM, Pairolero PC. High-grade esophageal dysplasia: long-term survival and quality of life after esophagectomy. Ann Thorac Surg. 2002;73:1697-1702; discussion 1702-3. [PubMed] |
| 5. | Yang CS, Chen X, Tu S. Etiology and Prevention of Esophageal Cancer. Gastrointest Tumors. 2016;3:3-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 69] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 6. | Gao YB, Chen ZL, Li JG, Hu XD, Shi XJ, Sun ZM, Zhang F, Zhao ZR, Li ZT, Liu ZY. Genetic landscape of esophageal squamous cell carcinoma. Nat Genet. 2014;46:1097-1102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 557] [Cited by in RCA: 569] [Article Influence: 47.4] [Reference Citation Analysis (1)] |
| 7. | Zeng H, Zheng R, Guo Y, Zhang S, Zou X, Wang N, Zhang L, Tang J, Chen J, Wei K. Cancer survival in China, 2003-2005: a population-based study. Int J Cancer. 2015;136:1921-1930. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 415] [Cited by in RCA: 508] [Article Influence: 46.2] [Reference Citation Analysis (0)] |
| 8. | Ai R, Sun Y, Guo Z, Wei W, Zhou L, Liu F, Hendricks DT, Xu Y, Zhao X. NDRG1 overexpression promotes the progression of esophageal squamous cell carcinoma through modulating Wnt signaling pathway. Cancer Biol Ther. 2016;17:943-954. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 39] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 9. | Deng F, Zhou K, Cui W, Liu D, Ma Y. Clinicopathological significance of wnt/β-catenin signaling pathway in esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2015;8:3045-3053. [PubMed] |
| 10. | Wang P, Li L, Li T. Positive correlation of cysteine-rich 61 and target genes of Wnt/β-catenin pathway in esophageal squamous cell carcinoma. J Cancer Res Ther. 2016;12:19-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 11. | Wang X, Liu D, Zhou K, Wang B, Liu Q, Deng F, Li Q, Ma Y. Expression of Wnt11 and Rock2 in esophageal squamous cell carcinoma by activation of the WNT/PCP pathway and its clinical significance. Pathol Res Pract. 2016;212:880-885. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 12. | He J, Zhou M, Chen X, Yue D, Yang L, Qin G, Zhang Z, Gao Q, Wang D, Zhang C. Inhibition of SALL4 reduces tumorigenicity involving epithelial-mesenchymal transition via Wnt/β-catenin pathway in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2016;35:98. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 73] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 13. | Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14460] [Cited by in RCA: 16303] [Article Influence: 959.0] [Reference Citation Analysis (2)] |
| 14. | He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522-531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4964] [Cited by in RCA: 5377] [Article Influence: 244.4] [Reference Citation Analysis (0)] |
| 15. | Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta. 2016;1859:169-176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 329] [Cited by in RCA: 429] [Article Influence: 39.0] [Reference Citation Analysis (0)] |
| 16. | Kumarswamy R, Mudduluru G, Ceppi P, Muppala S, Kozlowski M, Niklinski J, Papotti M, Allgayer H. MicroRNA-30a inhibits epithelial-to-mesenchymal transition by targeting Snai1 and is downregulated in non-small cell lung cancer. Int J Cancer. 2012;130:2044-2053. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 235] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 17. | Zhong K, Chen K, Han L, Li B. MicroRNA-30b/c inhibits non-small cell lung cancer cell proliferation by targeting Rab18. BMC Cancer. 2014;14:703. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 96] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 18. | Bockhorn J, Yee K, Chang YF, Prat A, Huo D, Nwachukwu C, Dalton R, Huang S, Swanson KE, Perou CM. MicroRNA-30c targets cytoskeleton genes involved in breast cancer cell invasion. Breast Cancer Res Treat. 2013;137:373-382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 87] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 19. | Cheng CW, Wang HW, Chang CW, Chu HW, Chen CY, Yu JC, Chao JI, Liu HF, Ding SL, Shen CY. MicroRNA-30a inhibits cell migration and invasion by downregulating vimentin expression and is a potential prognostic marker in breast cancer. Breast Cancer Res Treat. 2012;134:1081-1093. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 175] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 20. | Zhang Q, Yu L, Qin D, Huang R, Jiang X, Zou C, Tang Q, Chen Y, Wang G, Wang X. Role of microRNA-30c targeting ADAM19 in colorectal cancer. PLoS One. 2015;10:e0120698. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 46] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 21. | Zhong M, Bian Z, Wu Z. miR-30a suppresses cell migration and invasion through downregulation of PIK3CD in colorectal carcinoma. Cell Physiol Biochem. 2013;31:209-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 83] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 22. | Quintavalle C, Donnarumma E, Iaboni M, Roscigno G, Garofalo M, Romano G, Fiore D, De Marinis P, Croce CM, Condorelli G. Effect of miR-21 and miR-30b/c on TRAIL-induced apoptosis in glioma cells. Oncogene. 2013;32:4001-4008. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 92] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 23. | Wang J, Jiao Y, Cui L, Jiang L. miR-30 functions as an oncomiR in gastric cancer cells through regulation of P53-mediated mitochondrial apoptotic pathway. Biosci Biotechnol Biochem. 2017;81:119-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 30] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 24. | Tsukasa K, Ding Q, Miyazaki Y, Matsubara S, Natsugoe S, Takao S. miR-30 family promotes migratory and invasive abilities in CD133(+) pancreatic cancer stem-like cells. Hum Cell. 2016;29:130-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 25. | Wang Z, Dai X, Chen Y, Sun C, Zhu Q, Zhao H, Liu G, Huang Q, Lan Q. MiR-30a-5p is induced by Wnt/β-catenin pathway and promotes glioma cell invasion by repressing NCAM. Biochem Biophys Res Commun. 2015;465:374-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 36] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 26. | Zhao JJ, Lin J, Zhu D, Wang X, Brooks D, Chen M, Chu ZB, Takada K, Ciccarelli B, Admin S. miR-30-5p functions as a tumor suppressor and novel therapeutic tool by targeting the oncogenic Wnt/β-catenin/BCL9 pathway. Cancer Res. 2014;74:1801-1813. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 158] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 27. | Liao WT, Ye YP, Zhang NJ, Li TT, Wang SY, Cui YM, Qi L, Wu P, Jiao HL, Xie YJ. MicroRNA-30b functions as a tumour suppressor in human colorectal cancer by targeting KRAS, PIK3CD and BCL2. J Pathol. 2014;232:415-427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 122] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 28. | Ichimi T, Enokida H, Okuno Y, Kunimoto R, Chiyomaru T, Kawamoto K, Kawahara K, Toki K, Kawakami K, Nishiyama K. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer. 2009;125:345-352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 300] [Cited by in RCA: 318] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 29. | Baffa R, Fassan M, Volinia S, O’Hara B, Liu CG, Palazzo JP, Gardiman M, Rugge M, Gomella LG, Croce CM. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J Pathol. 2009;219:214-221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 394] [Cited by in RCA: 389] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 30. | Tang J, Ahmad A, Sarkar FH. The role of microRNAs in breast cancer migration, invasion and metastasis. Int J Mol Sci. 2012;13:13414-13437. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 145] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 31. | Xie K, Wang C, Qin N, Yang J, Zhu M, Dai J, Jin G, Shen H, Ma H, Hu Z. Genetic variants in regulatory regions of microRNAs are associated with lung cancer risk. Oncotarget. 2016;7:47966-47974. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 32. | Yang Y, Li X, Yang Q, Wang X, Zhou Y, Jiang T, Ma Q, Wang YJ. The role of microRNA in human lung squamous cell carcinoma. Cancer Genet Cytogenet. 2010;200:127-133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 71] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 33. | Wang W, Lin H, Zhou L, Zhu Q, Gao S, Xie H, Liu Z, Xu Z, Wei J, Huang X. MicroRNA-30a-3p inhibits tumor proliferation, invasiveness and metastasis and is downregulated in hepatocellular carcinoma. Eur J Surg Oncol. 2014;40:1586-1594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 63] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 34. | Sand M, Skrygan M, Georgas D, Sand D, Hahn SA, Gambichler T, Altmeyer P, Bechara FG. Microarray analysis of microRNA expression in cutaneous squamous cell carcinoma. J Dermatol Sci. 2012;68:119-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 84] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 35. | Kano M, Seki N, Kikkawa N, Fujimura L, Hoshino I, Akutsu Y, Chiyomaru T, Enokida H, Nakagawa M, Matsubara H. miR-145, miR-133a and miR-133b: Tumor-suppressive miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int J Cancer. 2010;127:2804-2814. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 351] [Cited by in RCA: 393] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 36. | Wang K, Jia Z, Zou J, Zhang A, Wang G, Hao J, Wang Y, Yang S, Pu P. Analysis of hsa-miR-30a-5p expression in human gliomas. Pathol Oncol Res. 2013;19:405-411. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 36] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 37. | Wang HY, Li YY, Fu S, Wang XP, Huang MY, Zhang X, Shao Q, Deng L, Zeng MS, Zeng YX. MicroRNA-30a promotes invasiveness and metastasis in vitro and in vivo through epithelial-mesenchymal transition and results in poor survival of nasopharyngeal carcinoma patients. Exp Biol Med (Maywood). 2014;239:891-898. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 38. | Igci YZ, Ozkaya M, Korkmaz H, Bozgeyik E, Bayraktar R, Ulasli M, Erkilic S, Eraydin A, Oztuzcu S. Expression Levels of miR-30a-5p in Papillary Thyroid Carcinoma: A Comparison Between Serum and Fine Needle Aspiration Biopsy Samples. Genet Test Mol Biomarkers. 2015;19:418-423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 39. | Zhou J, Gong G, Tan H, Dai F, Zhu X, Chen Y, Wang J, Liu Y, Chen P, Wu X. Urinary microRNA-30a-5p is a potential biomarker for ovarian serous adenocarcinoma. Oncol Rep. 2015;33:2915-2923. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 145] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 40. | Saad MA, Kuo SZ, Rahimy E, Zou AE, Korrapati A, Rahimy M, Kim E, Zheng H, Yu MA, Wang-Rodriguez J. Alcohol-dysregulated miR-30a and miR-934 in head and neck squamous cell carcinoma. Mol Cancer. 2015;14:181. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 51] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 41. | Tan X, Qin W, Zhang L, Hang J, Li B, Zhang C, Wan J, Zhou F, Shao K, Sun Y. A 5-microRNA signature for lung squamous cell carcinoma diagnosis and hsa-miR-31 for prognosis. Clin Cancer Res. 2011;17:6802-6811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 149] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 42. | Heinzelmann J, Henning B, Sanjmyatav J, Posorski N, Steiner T, Wunderlich H, Gajda MR, Junker K. Specific miRNA signatures are associated with metastasis and poor prognosis in clear cell renal cell carcinoma. World J Urol. 2011;29:367-373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 159] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 43. | Liu Z, Tu K, Liu Q. Effects of microRNA-30a on migration, invasion and prognosis of hepatocellular carcinoma. FEBS Lett. 2014;588:3089-3097. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 60] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 44. | Jia Z, Wang K, Wang G, Zhang A, Pu P. MiR-30a-5p antisense oligonucleotide suppresses glioma cell growth by targeting SEPT7. PLoS One. 2013;8:e55008. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 51] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 45. | Teo JL, Kahn M. The Wnt signaling pathway in cellular proliferation and differentiation: A tale of two coactivators. Adv Drug Deliv Rev. 2010;62:1149-1155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 199] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 46. | Schepeler T. Emerging roles of microRNAs in the Wnt signaling network. Crit Rev Oncog. 2013;18:357-371. [PubMed] |
| 47. | Saydam O, Shen Y, Würdinger T, Senol O, Boke E, James MF, Tannous BA, Stemmer-Rachamimov AO, Yi M, Stephens RM. Downregulated microRNA-200a in meningiomas promotes tumor growth by reducing E-cadherin and activating the Wnt/beta-catenin signaling pathway. Mol Cell Biol. 2009;29:5923-5940. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 216] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 48. | Kim NH, Cha YH, Kang SE, Lee Y, Lee I, Cha SY, Ryu JK, Na JM, Park C, Yoon HG. p53 regulates nuclear GSK-3 levels through miR-34-mediated Axin2 suppression in colorectal cancer cells. Cell Cycle. 2013;12:1578-1587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 92] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 49. | Wang X, Wang K, Han L, Zhang A, Shi Z, Zhang K, Zhang H, Yang S, Pu P, Shen C. PRDM1 is directly targeted by miR-30a-5p and modulates the Wnt/β-catenin pathway in a Dkk1-dependent manner during glioma growth. Cancer Lett. 2013;331:211-219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 57] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
P- Reviewer: Bordonaro M, Pandi NS S- Editor: Chen K L- Editor: Wang TQ E- Editor: Huang Y