Published online Nov 21, 2017. doi: 10.3748/wjg.v23.i43.7705
Peer-review started: August 15, 2017
First decision: August 30, 2017
Revised: September 27, 2017
Accepted: September 28, 2017
Article in press: September 28, 2017
Published online: November 21, 2017
Processing time: 97 Days and 18.5 Hours
To determine the role of G0/G1 switch gene 2 (G0S2) and its transcriptional regulation in palmitate-induced hepatic lipid accumulation.
HepG2 cells were treated with palmitate, or palmitate in combination with CCAAT/enhancer binding protein (C/EBP)β siRNA or G0S2 siRNA. The mRNA expression of C/EBPβ, peroxisome proliferator-activated receptor (PPAR)γ and PPARγ target genes (G0S2, GPR81, GPR109A and Adipoq) was examined by qPCR. The protein expression of C/EBPβ, PPARγ, and G0S2 was determined by Western blotting. Lipid accumulation was detected with Oil Red O staining and quantified by absorbance value of the extracted Oil Red O dye. Lipolysis was evaluated by measuring the amount of glycerol released into the medium.
Palmitate caused a dose-dependent increase in lipid accumulation and a dose-dependent decrease in lipolysis in HepG2 cells. In addition, palmitate increased the mRNA expression of C/EBPβ, PPARγ, and PPARγ target genes (G0S2, GPR81, GPR109A, and Adipoq) and the protein expression of C/EBPβ, PPARγ, and G0S2 in a dose-dependent manner. Knockdown of C/EBPβ decreased palmitate-induced PPARγ and its target genes (G0S2, GPR81, GPR109A, and Adipoq) mRNA expression and palmitate-induced PPARγ and G0S2 protein expression in HepG2 cells. Knockdown of C/EBPβ also attenuated lipid accumulation and augmented lipolysis in palmitate-treated HepG2 cells. G0S2 knockdown attenuated lipid accumulation and augmented lipolysis, while G0S2 knockdown had no effects on the mRNA expression of C/EBPβ, PPARγ, and PPARγ target genes (GPR81, GPR109A and Adipoq) in palmitate-treated HepG2 cells.
Palmitate can induce lipid accumulation in HepG2 cells by activating C/EBPβ-mediated G0S2 expression.
Core tip: Obesity-associated nonalcoholic fatty liver disease is characterized by excessive deposition of fat in hepatocytes. The saturated free fatty acid palmitate, the concentration of which is often elevated in obesity, is a major contributor to an increase in intrahepatic triglyceride. G0/G1 switch gene 2 (G0S2) is a critical regulator of hepatic lipid accumulation. However, the role of G0S2 and its transcriptional regulation in palmitate-induced hepatic lipid accumulation is not clear. We found that palmitate can induce lipid accumulation in HepG2 cells by activating C/EBPβ-mediated G0S2 expression.
- Citation: Zhao NQ, Li XY, Wang L, Feng ZL, Li XF, Wen YF, Han JX. Palmitate induces fat accumulation by activating C/EBPβ-mediated G0S2 expression in HepG2 cells. World J Gastroenterol 2017; 23(43): 7705-7715
- URL: https://www.wjgnet.com/1007-9327/full/v23/i43/7705.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i43.7705
Nonalcoholic fatty liver disease (NAFLD) is characterized by excessive deposition of fat in hepatocytes in the absence of excessive alcohol intake. It is one of the most common emerging liver diseases throughout the world, coinciding with the global obesity epidemic[1]. Elevated plasma free fatty acid (FFA) levels are a common feature of obesity[2] and play an etiological role in the pathogenesis of NAFLD[3]. In particular, the saturated fatty acid palmitate, which makes up 30%-40% of high plasma FFA concentration[4], is a major contributor to an increase in intrahepatic triglyceride[5]. However, the molecular mechanism by which palmitate contributes to the accumulation of excess triglyceride in hepatocytes is not entirely clear.
Several studies of NAFLD have demonstrated that a decreased rate of triglyceride mobilization promotes triglyceride accumulation in the liver[6,7]. The rate-limiting step of intracellular triacylglycerol mobilization is cleavage of the first ester bond in triglycerides, which is catalyzed by adipose triglyceride lipase (ATGL)[8]. In adipocytes, the protein product of G0/G1 switch gene 2 (G0S2) is a dominant inhibitor of ATGL[9]. It binds directly to ATGL and attenuates ATGL-mediated lipolysis via inhibiting the triglyceride hydrolase activity of ATGL[9-11]. G0S2 is also abundantly expressed in the liver, suggesting that the regulatory function of G0S2 is not limited to adipose tissue[9]. Notably, G0S2 overexpression in the liver increases the accumulation of triglycerides and promotes fatty liver formation[12,13]. Conversely, loss of G0S2 in the liver results in a marked decrease in hepatic triacylglycerol levels and protects against high-fat-diet-induced liver steatosis[13]. These findings implicate an important role for G0S2 as a regulator of triglyceride content in the liver and as a contributor to obesity-associated liver steatosis.
G0S2 expression is regulated by a complex transcriptional mechanism that involves proliferator-activated receptor (PPAR)γ. Transactivation, gel shift and chromatin immunoprecipitation assays have identified G0S2 as a direct target gene of PPARγ[14]. The transcription factor CCAAT/enhancer binding protein (C/EBP)β is involved in adipogenesis and is crucial for inducing initial expression of PPARγ during adipogenesis[15,16]. Importantly, C/EBPβ overexpression increases PPARγ mRNA level and triglyceride content in the liver, whereas C/EBPβ RNA interference attenuates palmitate-induced PPARγ expression and triglyceride accumulation in hepatocytes[5].
Based on these observations, we propose the following hypothesis: palmitate stimulates C/EBPβ and its downstream target PPARγ and consequent G0S2 expression, and then G0S2 contributes to palmitate-induced fat accumulation in the liver. In this study, using human HepG2 hepatoma cells, a cellular model of hepatic steatosis[17], we examined lipolysis in hepatocytes, hepatocellular triglyceride accumulation, and the expression of C/EBPβ, PPARγ and PPARγ-regulated genes (G0S2, GPR81, GPR109A and Adipoq) in response to palmitate treatment. In addition, via siRNA-mediated gene knockdown experiments, we investigated the relationship between expression of the aforementioned proteins and hepatocyte lipolysis and lipid accumulation.
HepG2 cells (China Center for Type Culture Collection, Wuhan, China) were cultured in Dulbecco’s modified Eagle’s medium (Invitrogen, Carlsbad, CA, United States) containing 10% fetal bovine serum (Invitrogen), 100 U/mL penicillin, 100 μg/mL streptomycin, and 1% L-glutamine. Cells were grown at 37 °C in an atmosphere of 5% CO2/95% air in a cell culture flask. The effect of palmitate was examined by addition of this agent to the cells plated in six-well plates at 2 × 105 cells per well.
Palmitate (Sigma, St. Louis, MO, United States) stock solution was prepared by coupling palmitate to bovine serum albumin (BSA; Sigma) as previously described[18]. Palmitate was fully dissolved in pure ethanol for a concentration of 195 mmol/L, ensuring that the final concentration of ethanol in the palmitate stock solution did not exceed 1.5% by volume. This palmitate stock solution was then added to a prewarmed BSA solution (10% w/w, 37 °C) to achieve a final palmitate concentration of 3 mmol/L. The solution was dissolved by incubating at 37 °C in a water bath for a further 10 min. The final molar ratio of palmitate to BSA was 2:1. The control vehicle was prepared using a stock of 10% w/w BSA with an equivalent volume of ethanol added to match that contained in the final palmitate stock. The final concentration of ethanol was < 0.2% by volume in all experiments.
Total RNA was isolated from cultured HepG2 cells using TRIzol reagent (Invitrogen), and RNA quality was evaluated via electrophoresis. Reverse transcription (RT) was performed using Superscript II reverse transcriptase (Invitrogen). The RT conditions for each cDNA amplification were 42 °C for 15 min, 85 °C for 5 s, and the cDNAs amplified were stored at -20 °C. Gene expression analysis was performed by quantitative PCR (qPCR) on a StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA, United States) using SYBR Green as the detection dye. Primer sequences used for the detection of genes were designed as follows: C/EBPβ forward primer: 5′-CAAGCACAGCGACGAGTACAAGATCC-3′ and reverse primer: 5′- GCTTGAACAAGTTCCGCAGGGTG -3′; PPARγ forward primer: 5′-ACCACTCCCACTCC TTTG-3′ and reverse primer: 5′-GCAGGCTCCACTTT GATT-3′; G0S2 forward primer: 5′-CCTCTTCGGCG TGGTGCT-3′ and reverse prime: 5′-CTGCTGCTT GCCTTTCTCC-3′; GPR81 forward primer: 5′-CAGA CAGGCTCGGATGAAGAAG-3′ and reverse prime: 5′-TTGTAGAATTTGGGAAAGGAGGG-3′; GPR109A forward primer: 5′-TGGACCTGGCGTTC TTTA-3′ and reverse primer: 5′-GCTCGTGCTGCGGTTATT-3′; Adipoq forward primer: 5′-AGGAAAGGAGAACCTGGAGAAG-3′ and reverse prime: 5′-ATAGACTGTGATGTGGTAGGC AAA-3′; β-actin forward primer: 5′-TGGCACCCAGCA CAATGAA-3′ and reverse primer: 5′-CTAAGTCATAGT CCGCCTAGAA-3′. The expected size of the amplified products was 194 bp (C/EBPβ), 169 bp (PPARγ), 160 bp (G0S2), 240 bp (GPR81), 170 bp (GPR109A), 204 bp (Adipoq) and 186 bp (β-actin). β-Actin was used as a control housekeeping gene. Cycling conditions were 94 °C for 5 s and 60 °C for 30 s, followed by 45 cycles. The predicted size of the PCR products was confirmed by 2% agarose gel electrophoresis stained with ethidium bromide. Melting curve analysis was performed for each sample in direct connection to the PCR, to verify the specificity of the amplified PCR product. The results were stated as the fold difference in expression for each target gene compared to that of β-actin as an internal control in the same sample, using the 2-ΔΔCt method. All experiments were carried out in duplicate.
To measure the nuclear C/EBPβ protein level, nuclear protein extracts were isolated from HepG2 cells using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific, Waltham, MA, United States). Meanwhile, HepG2 cells were harvested and lysed with ice-cold RIPA lysis buffer containing protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany) and proteins were extracted from whole-cell lysates. The protein concentration was quantified using Bio-Rad DC Protein Assay Kit (Bio-Rad Laboratories, Hercules, CA, United States). After denaturation by boiling of protein, equal amounts of total protein (40 μg) were loaded and resolved on 10% SDS-PAGE for 2 h at room temperature. The proteins were subsequently transferred to polyvinylidene difluoride membranes (Atto Corporation, Tokyo, Japan). The membranes were blocked with 5% non-fat milk dissolved in Tris-buffered saline/Tween 20 buffer for 2 h and incubated with primary antibodies overnight, and then the secondary antibodies for 1 h. Primary antibodies used were C/EBPβ (1:200; Santa Cruz Biotechnology, Santa Cruz, CA, United States), PPARγ (1:1000; Cell Signaling, NEB, Vienna, Austria) and G0S2 (1:100; Sigma). The β-actin antibody (1:2000;) was used as a loading control. Secondary antibody was goat anti-rabbit IgG-horseradish peroxidase conjugate (1:2000; Bio-Rad Laboratories). The immunoreactive protein bands were visualized using an enhanced chemiluminescence detection system (Amersham Pharmacia Biotech, Piscataway, NJ, United States). The density of the band was quantified using ImageJ software (NIH, Bethesda, MD, United States), and the data were transformed and normalized relative to β-actin as the integral optical density ratio. All experiments were performed at least three times and representative data were shown.
RNA oligonucleotides directed against C/EBPβ (sense sequence: GCAATCGGTTTAAACATGGCT) and G0S2 (sense sequence: GCATCCACCAAAGGAGTTTGG) were purchased from GeneChem Co. Ltd. (Shanghai, China) to silence target proteins. A negative control siRNA was also purchased from GeneChem, which had no matches in the human genome. siRNA transfection to HepG2 cells was conducted using Lipofectamine 2000 (Invitrogen). HepG2 cells were seeded into six-well plates and allowed to culture overnight. The aforementioned siRNAs (2.5 μg) and 5 μL Lipofectamine 2000 Reagent were respectively diluted in 250 μL Opti-MEM Medium (Invitrogen) and incubated separately for 10 min at room temperature. After the 10-min incubation, equal volumes of diluted siRNA and Lipofectamine 2000 Reagent were mixed gently and incubated for 10 min at room temperature to form siRNA–lipid complexes. For transfection, the siRNA–lipid complexes were subsequently combined with HepG2 cells in six-well culture plates at 106 cells per well and incubated for 24 h. Knockdown efficiency of the siRNAs was determined by Western blotting. Transfected cells were treated with 200 μmol/L palmitate for 24 h before harvesting.
HepG2 cells were grown on six-well plates, washed three times with phosphate-buffered saline, and fixed with 10% formaldehyde for 30 min at room temperature. The fixed cells were washed with deionized distilled water, dipped in 60% isopropanol for 3 min, stained with 2 mg/mL of Oil Red O staining solution (Sigma) for 60 min, and washed with deionized distilled water three times to remove unbound dye. Cell nuclei were counterstained with hematoxylin for 3 min and washed with deionized distilled water. Images were obtained using an Axiovert 40 CFL microscope (Olympus, Tokyo, Japan). After microscopic examination, the Oil-Red-O-based amount of triglyceride was quantified in each well. After washing and drying completely, 200 μL isopropanol extraction solution was added to each staining well and the mixtures were incubated for 10 min, followed by gentle vibration to release Oil Red O for 10 min at room temperature. The extracted dye was removed by gentle pipetting, and its absorbance was measured at 500 nm by microplate reader (Versamax; Molecular Devices, Sunnyvale, CA, United States). All tests were performed in triplicate.
Lipolysis was evaluated by measuring the amount of glycerol released into the medium. Aliquots of culture medium were centrifuged to remove debris, and directly subjected to glycerol measurement. The amounts of glycerol released were quantified using a glycerol quantification kit (Biovision Inc., Milpitas, CA, United States). Released glycerol was determined using an autoanalyzer (Cobas-Mira; Roche Diagnostics, Basel, Switzerland) to detect the absorbance at 550 nm. All samples were measured in duplicates.
All experimental data were expressed as means ± SE. Statistical differences were evaluated by Student’s t test or one-way analysis of variance where appropriate using SPSS version 18.0 (SPSS, Chicago, IL, United States). Differences were considered as statistically significant when P values were < 0.05.
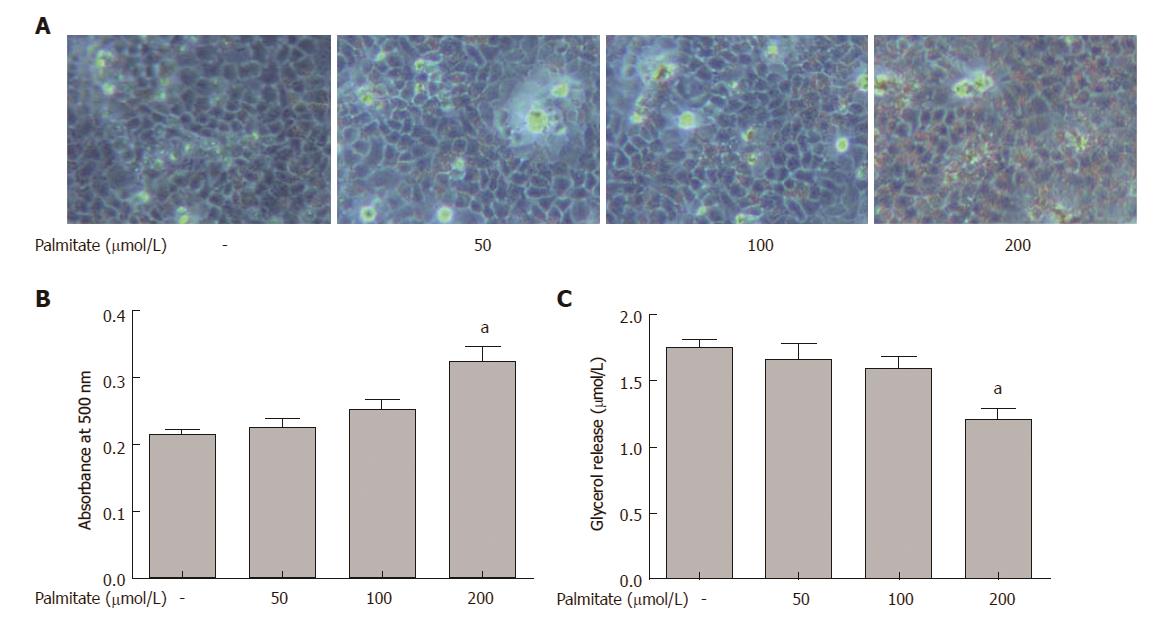
HepG2 cells were incubated with increasing amounts of palmitate for 24 h, and lipid accumulation was examined by Oil Red O staining. Palmitate caused a dose-dependent increase in lipid accumulation in HepG2 cells (Figure 1A and B). HepG2 cells with palmitate also caused a dose-dependent decrease in lipolysis, demonstrated by reduced glycerol release into the medium (Figure 1C). Palmitate at 200 μmol/L, which represents a high physiological level of circulating palmitate in obesity[2], caused a significant increase in lipid accumulation and a significant decrease in lipolysis. Therefore, this concentration of palmitate was used in all the following siRNA knockdown experiments.
Palmitate induced expression of C/EBPβ, PPARγ, and PPARγ target genes in HepG2 cells
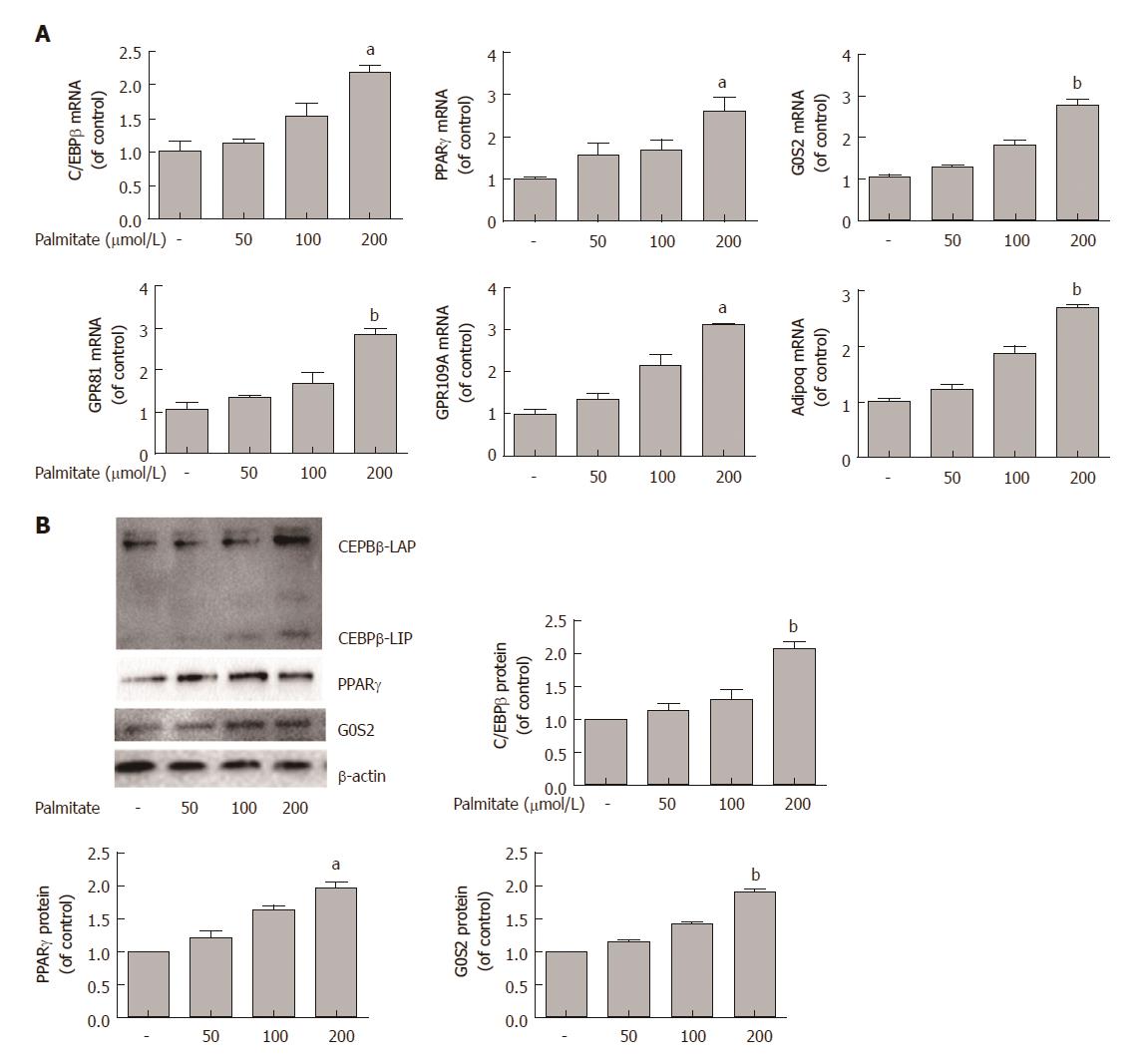
Lipid accumulation is controlled by a few key transcription factors, including C/EBPβ and PPARγ. We assessed the effects of palmitate on the expression of C/EBPβ, PPARγ, and several known PPARγ target genes in HepG2 cells. HepG2 cells were incubated with increasing amounts of palmitate for 24 h, and quantitative PCR analysis revealed that palmitate caused a dose-dependent increase in the mRNA expression of C/EBPβ, PPARγ, and PPARγ target genes (G0S2, GPR81, GPR109A, and Adipoq) (Figure 2A). Western blotting showed that incubation of HepG2 cells with palmitate also caused a dose-dependent increase in the protein expression of C/EBPβ, PPARγ, and G0S2 (Figure 2B).
C/EBPβ knockdown reduced palmitate-induced PPARγ and its target gene expression in HepG2 cells
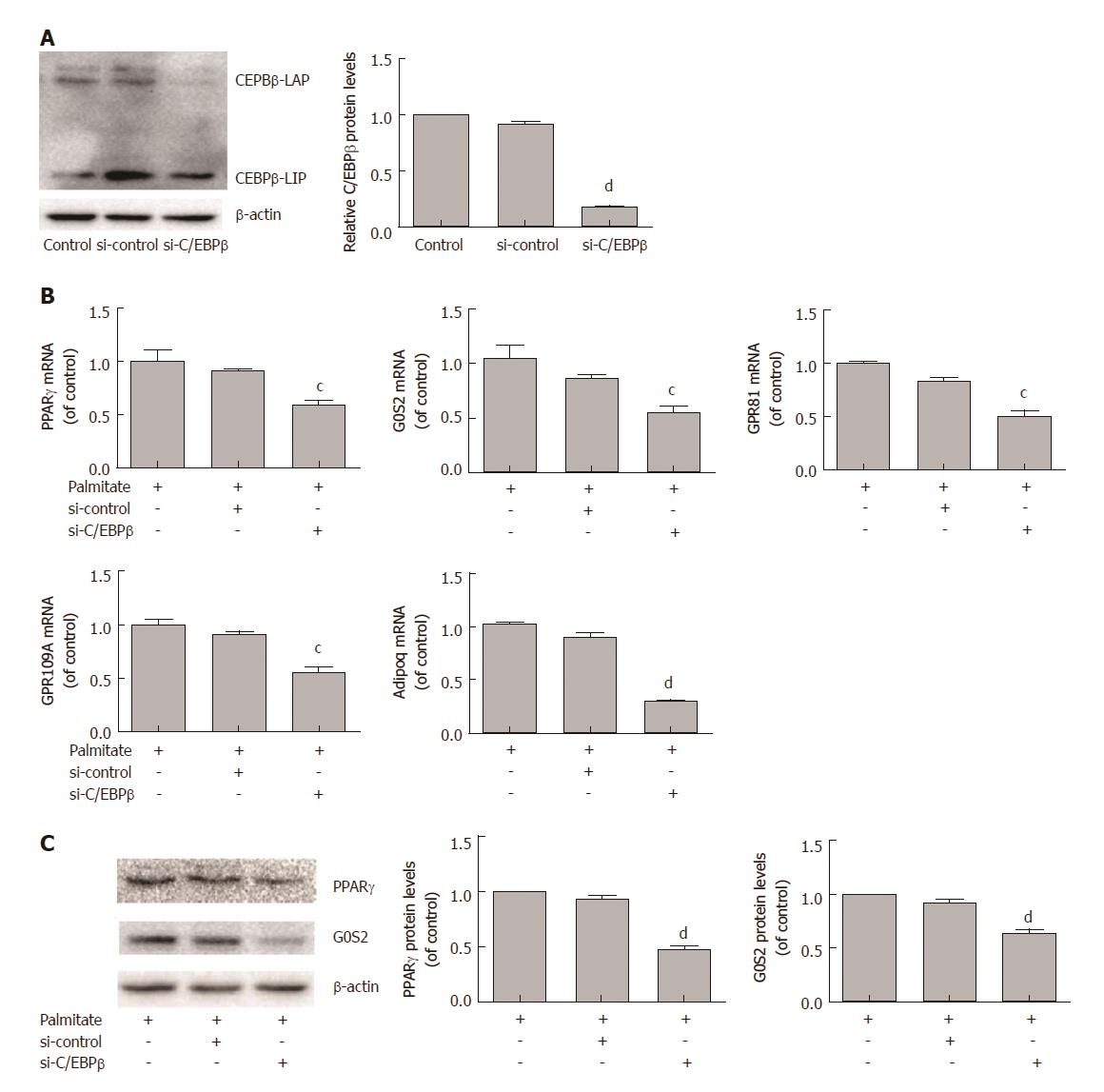
We next examined the role of C/EBPβ in palmitate-induced PPARγ and its target gene expression in HepG2 cells. HepG2 cells were transfected with C/EBPβ siRNA and treated with 200 μmol/L palmitate for 24 h. C/EBPβ siRNA efficiently decreased C/EBPβ protein expression (Figure 3A). qPCR analysis revealed that C/EBPβ knockdown significantly decreased palmitate-induced PPARγ and its target genes (G0S2, GPR81, GPR109A, and Adipoq) mRNA expression (Figure 3B). Western blotting showed that C/EBPβ knockdown significantly decreased palmitate-induced PPARγ and G0S2 protein expression (Figure 3C).
C/EBPβ knockdown attenuated lipid accumulation and augmented lipolysis in HepG2 cells treated with palmitate
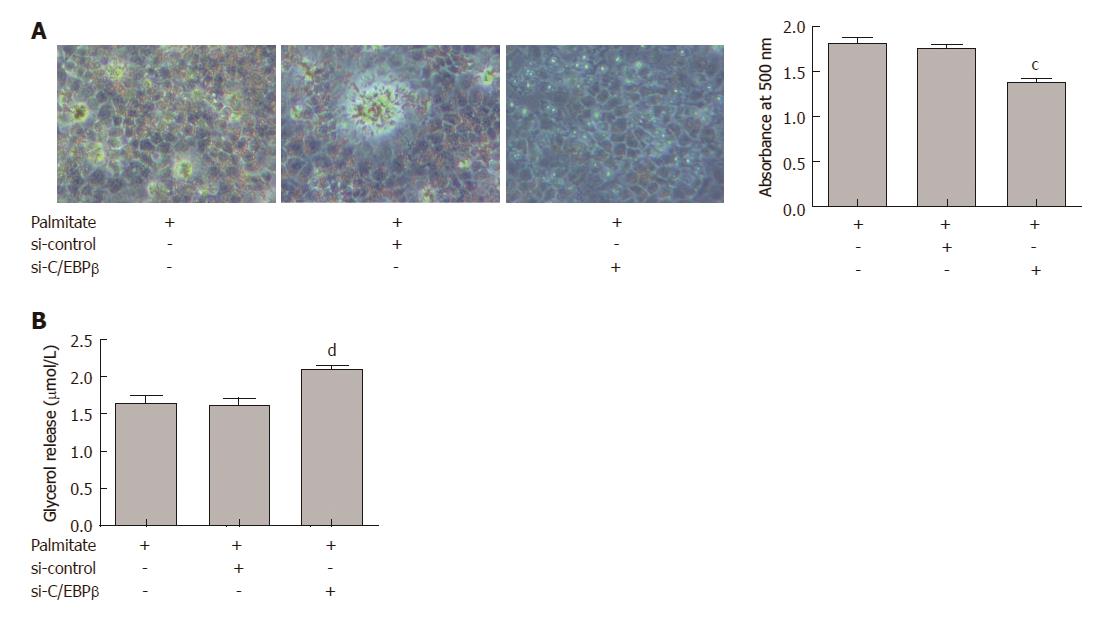
We investigated the effects of C/EBPβ on lipid accumulation and lipolysis in HepG2 cells treated with palmitate. HepG2 cells were transfected with C/EBPβ siRNA and treated with 200 μmol/L palmitate for 24 h. C/EBPβ knockdown significantly attenuated palmitate-induced lipid accumulation in HepG2 cells (Figure 4A). C/EBPβ knockdown significantly augmented lipolysis in HepG2 cells treated with palmitate (Figure 4B).
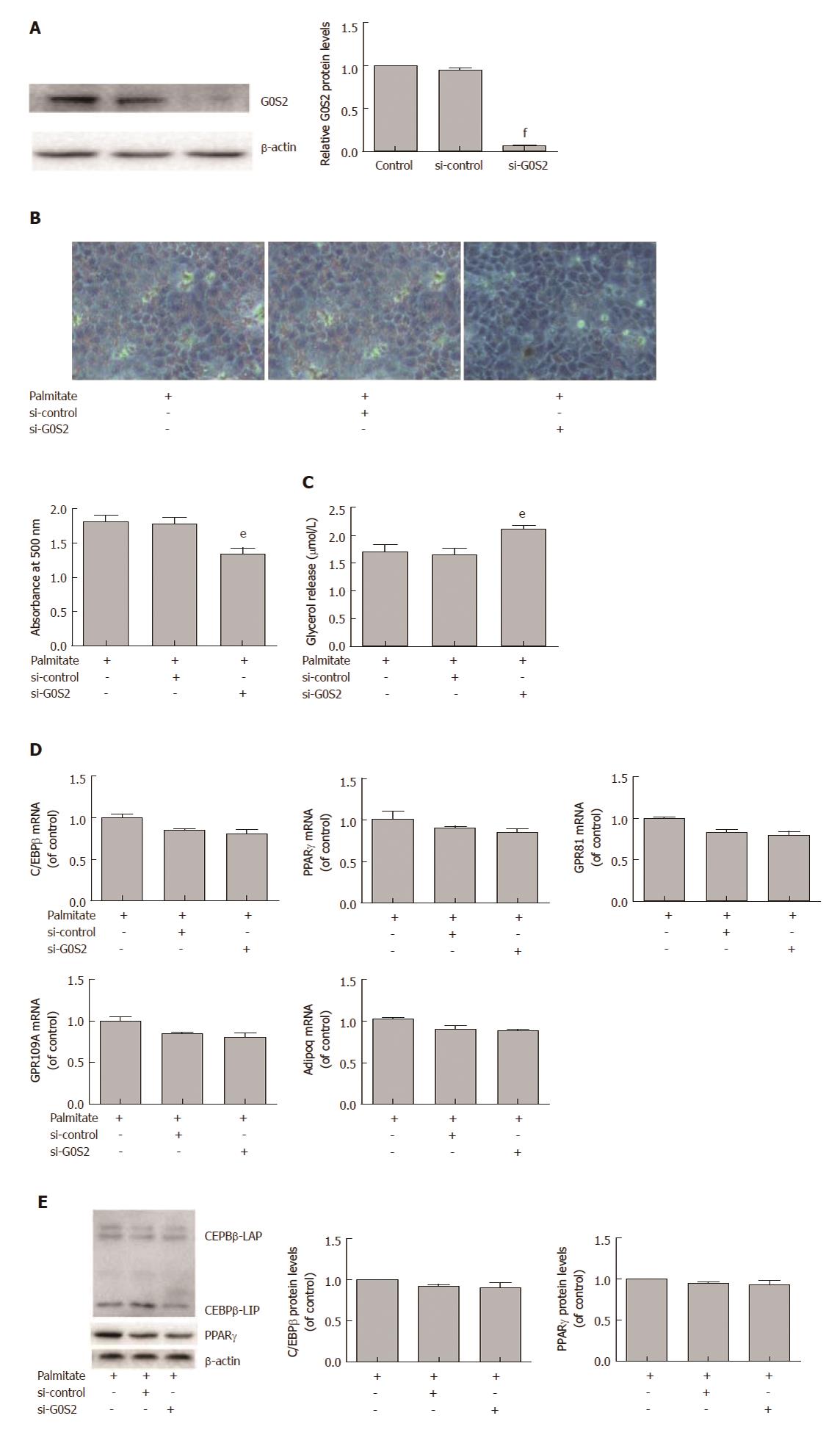
G0S2 plays an important role in regulating hepatic lipid accumulation and lipolysis. We explored the effects of G0S2 in lipid accumulation and lipolysis in HepG2 cells treated with palmitate. HepG2 cells were transfected with G0S2 siRNA and treated with 200 μmol/L palmitate for 24 h. G0S2 siRNA efficiently decreased G0S2 protein expression (Figure 5A). G0S2 knockdown significantly attenuated palmitate-induced lipid accumulation in HepG2 cells (Figure 5B). G0S2 knockdown significantly augmented lipolysis in HepG2 cells treated with palmitate (Figure 5C). However, G0S2 knockdown had no effects on palmitate-induced mRNA expression of C/EBPβ, PPARγ, and other PPARγ target genes (GPR81, GPR109A and Adipoq) (Figure 5D) and palmitate-induced protein expression of C/EBPβ and PPARγ in HepG2 cells (Figure 5E).
Obesity is associated with elevation of circulating FFA due to impaired lipid storage capacity in subcutaneous adipose tissue. The increased FFA supply that occurs as a result leads to lipid accumulation in the liver[19,20]. Previous studies showed that the saturated fatty acid palmitate induces lipid accumulation in HepG2 cells[21,22]. In the present study, we also demonstrated that palmitate induced lipid accumulation, and moreover, palmitate suppressed lipolysis in hepatocytes.
C/EBPβ is an important basic leucine zipper transcription factor whose mRNA can produce two C/EBPβ isoforms: liver-enriched activating protein (46 kDa) and liver-enriched inhibitory protein (21 kDa)[23]. C/EBPβ is involved in hepatic lipogenesis[5]. In hepatocytes, palmitate upregulates C/EBPβ expression, which in turn induces PPARγ expression and increases hepatic lipid accumulation[5]. Previous studies showed that C/EBPβ directly binds the PPARγ promoter prior to transcriptional activation during the early phase of adipogenesis[24,25]. PPARγ is a master adipogenic transcription factor, and it is necessary for adipogenesis[16]. G0S2, GPR81, GPR109A, and Adipoq are known PPARγ downstream target genes involved in lipolysis[26,27]. In the present study, we found that palmitate increased the mRNA expression of C/EBPβ, PPARγ and PPARγ target genes (G0S2, GPR81, GPR109A and Adipoq) and the protein expression of C/EBPβ, PPARγ and G0S2 in a dose-dependent manner in hepatocytes. We also found that knockdown of C/EBPβ significantly decreased PPARγ and its target genes (G0S2, GPR81, GPR109A and Adipoq) mRNA expression and PPARγ and G0S2 protein expression in palmitate-treated HepG2 cells. In addition, gene silencing of C/EBPβ attenuated lipid accumulation and augmented lipolysis in HepG2 cells treated with palmitate. These findings again demonstrate a critical role for C/EBPβ in palmitate-induced hepatic lipid accumulation.
In the above PPARγ target genes, G0S2 acts as an important regulator of triglyceride content in the liver[12,13]. Adipose-tissue-derived fatty acids upregulate fasting G0S2 expression in the liver to induce hepatic triglyceride accumulation[28]. G protein-coupled receptor (GPR)81 functions as a specific receptor for lactate and mediates insulin-induced antilipolytic effects in an autocrine and paracrine manner[27,29]. GPR109A is a receptor for the ketone body 3-hydroxy-butyric acid and functions as a metabolic sensor that regulates lipolytic activity during starvation to avoid excessive triglyceride degradation[30,31]. Its biological role is related to the ketone body 3-hydroxy-butyrate[31]. Adiponectin (Adipoq) is a adipokine that is downregulated in obesity[32]. In the liver, Adipoq can augment the oxidation of fatty acid to alleviate hepatic lipid accumulation[33]. Therefore, G0S2 may play a critical role in C/EBPβ-mediated hepatic lipid accumulation in palmitate-treated HepG2 cells. In this study, we found that knockdown of G0S2 significantly attenuated lipid accumulation and augmented lipolysis in HepG2 cells treated with palmitate. More importantly, inhibition of the G0S2 expression had no effects on mRNA expression of C/EBPβ, PPARγ, and PPARγ target genes (GPR81, GPR109A and Adipoq) and protein expression of C/EBPβ and PPARγ in palmitate-treated HepG2 cells. Together, these results indicate that palmitate induces lipid accumulation by activating C/EBPβ-mediated expression of G0S2.
In summary, we observed that palmitate can induce lipid accumulation in HepG2 cells by activating C/EBPβ-mediated G0S2 expression. The result provides novel evidence linking G0S2 expression to palmitate-induced hepatic lipogenesis. Considering that liver lipid accumulation is not only a hallmark of NAFLD, but also the first and critical step in the initiation and progression of NAFLD, interfering with G0S2 may represent an effective strategy for the treatment of obesity-related hepatic steatosis.
Obesity-associated nonalcoholic fatty liver disease (NAFLD) is characterized by excessive deposition of fat in hepatocytes. The saturated free fatty acid palmitate, the concentration of which is often elevated in obesity, is a major contributor to an increase in intrahepatic triglyceride. G0/G1 switch gene 2 (G0S2) plays an important role in regulating hepatic lipid metabolism. However, the role of G0S2 and its transcriptional regulation in palmitate-induced hepatic lipid accumulation has remained unclear.
This study was carried out to clarify the molecular mechanism connecting palmitate to obesity-associated NAFLD. As CCAAT/enhancer binding protein beta (C/EBPβ), proliferator-activated receptor gamma (PPARγ) and G0S2 all relate to obesity-associated NAFLD, we investigated their roles and interrelationships in palmitate-induced hepatic lipid accumulation. The results lead to important new insights into the molecular mechanism of NAFLD.
The goal of this study was to determine the role of G0S2 and its transcriptional regulation in palmitate-induced hepatic lipid accumulation. The results suggest a previously unknown link between C/EBPβ and G0S2 that contributes to hepatic steatosis.
In this study, we examined lipolysis, lipid accumulation, and the expression of C/EBPβ, PPARγ and PPARγ-regulated genes (G0S2, GPR81, GPR109A and Adipoq) in response to palmitate treatment in HepG2 cells. Specifically, we investigated the relationships between expression of the aforementioned proteins and hepatocyte lipolysis and lipid accumulation by using siRNA-mediated gene knockdown experiments.
Palmitate significantly facilitated lipid accumulation and suppressed lipolysis in HepG2 cells. Palmitate also significantly increased the expression of C/EBPβ, PPARγ, and PPARγ target genes (G0S2, GPR81, GPR109A and Adipoq). C/EBPβ knockdown significantly reduced palmitate-induced PPARγ and G0S2 expression. Moreover, C/EBPβ knockdown attenuated lipid accumulation and augmented lipolysis in palmitate-treated HepG2 cells. Importantly, G0S2 knockdown significantly attenuated lipid accumulation and augmented lipolysis in palmitate-treated HepG2 cells, while G0S2 knockdown had no efects on palmitate-induced expression of C/EBPβ, PPARγ, and PPARγ target genes (GPR81, GPR109A, and Adipoq).
Palmitate can induce lipid accumulation in HepG2 cells by activating C/EBPβ-mediated G0S2 expression. The result provides novel evidence linking G0S2 expression to palmitate-induced hepatic lipogenesis. Considering that liver lipid accumulation is not only a hallmark of NAFLD, but also the first and critical step in the initiation and progression of NAFLD, interfering with G0S2 may represent an effective strategy for the treatment of obesity-related hepatic steatosis.
| 1. | Fabbrini E, Sullivan S, Klein S. Obesity and nonalcoholic fatty liver disease: biochemical, metabolic, and clinical implications. Hepatology. 2010;51:679-689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1303] [Cited by in RCA: 1586] [Article Influence: 99.1] [Reference Citation Analysis (1)] |
| 2. | Mittendorfer B, Magkos F, Fabbrini E, Mohammed BS, Klein S. Relationship between body fat mass and free fatty acid kinetics in men and women. Obesity (Silver Spring). 2009;17:1872-1877. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 150] [Cited by in RCA: 145] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 3. | Song Z, Song M, Lee DY, Liu Y, Deaciuc IV, McClain CJ. Silymarin prevents palmitate-induced lipotoxicity in HepG2 cells: involvement of maintenance of Akt kinase activation. Basic Clin Pharmacol Toxicol. 2007;101:262-268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 21] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 4. | Cacicedo JM, Benjachareowong S, Chou E, Ruderman NB, Ido Y. Palmitate-induced apoptosis in cultured bovine retinal pericytes: roles of NAD(P)H oxidase, oxidant stress, and ceramide. Diabetes. 2005;54:1838-1845. [PubMed] |
| 5. | Schroeder-Gloeckler JM, Rahman SM, Janssen RC, Qiao L, Shao J, Roper M, Fischer SJ, Lowe E, Orlicky DJ, McManaman JL. CCAAT/enhancer-binding protein beta deletion reduces adiposity, hepatic steatosis, and diabetes in Lepr(db/db) mice. J Biol Chem. 2007;282:15717-15729. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 107] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 6. | Ong KT, Mashek MT, Bu SY, Greenberg AS, Mashek DG. Adipose triglyceride lipase is a major hepatic lipase that regulates triacylglycerol turnover and fatty acid signaling and partitioning. Hepatology. 2011;53:116-126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 299] [Cited by in RCA: 289] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 7. | Samuel VT, Choi CS, Phillips TG, Romanelli AJ, Geisler JG, Bhanot S, McKay R, Monia B, Shutter JR, Lindberg RA. Targeting foxo1 in mice using antisense oligonucleotide improves hepatic and peripheral insulin action. Diabetes. 2006;55:2042-2050. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 147] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 8. | Zechner R, Zimmermann R, Eichmann TO, Kohlwein SD, Haemmerle G, Lass A, Madeo F. FAT SIGNALS--lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 2012;15:279-291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 719] [Cited by in RCA: 902] [Article Influence: 64.4] [Reference Citation Analysis (0)] |
| 9. | Yang X, Lu X, Lombès M, Rha GB, Chi YI, Guerin TM, Smart EJ, Liu J. The G(0)/G(1) switch gene 2 regulates adipose lipolysis through association with adipose triglyceride lipase. Cell Metab. 2010;11:194-205. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 411] [Cited by in RCA: 398] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 10. | Cornaciu I, Boeszoermenyi A, Lindermuth H, Nagy HM, Cerk IK, Ebner C, Salzburger B, Gruber A, Schweiger M, Zechner R. The minimal domain of adipose triglyceride lipase (ATGL) ranges until leucine 254 and can be activated and inhibited by CGI-58 and G0S2, respectively. PLoS One. 2011;6:e26349. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 78] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 11. | Schweiger M, Paar M, Eder C, Brandis J, Moser E, Gorkiewicz G, Grond S, Radner FP, Cerk I, Cornaciu I. G0/G1 switch gene-2 regulates human adipocyte lipolysis by affecting activity and localization of adipose triglyceride lipase. J Lipid Res. 2012;53:2307-2317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 88] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 12. | Wang Y, Zhang Y, Qian H, Lu J, Zhang Z, Min X, Lang M, Yang H, Wang N, Zhang P. The g0/g1 switch gene 2 is an important regulator of hepatic triglyceride metabolism. PLoS One. 2013;8:e72315. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 42] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 13. | Zhang X, Xie X, Heckmann BL, Saarinen AM, Czyzyk TA, Liu J. Targeted disruption of G0/G1 switch gene 2 enhances adipose lipolysis, alters hepatic energy balance, and alleviates high-fat diet-induced liver steatosis. Diabetes. 2014;63:934-946. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 87] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 14. | Zandbergen F, Mandard S, Escher P, Tan NS, Patsouris D, Jatkoe T, Rojas-Caro S, Madore S, Wahli W, Tafuri S. The G0/G1 switch gene 2 is a novel PPAR target gene. Biochem J. 2005;392:313-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 187] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 15. | Feng S, Reuss L, Wang Y. Potential of Natural Products in the Inhibition of Adipogenesis through Regulation of PPARγ Expression and/or Its Transcriptional Activity. Molecules. 2016;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 86] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 16. | Xi Y, Shen W, Ma L, Zhao M, Zheng J, Bu S, Hino S, Nakao M. HMGA2 promotes adipogenesis by activating C/EBPβ-mediated expression of PPARγ. Biochem Biophys Res Commun. 2016;472:617-623. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 49] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 17. | Park JY, Kim Y, Im JA, Lee H. Oligonol suppresses lipid accumulation and improves insulin resistance in a palmitate-induced in HepG2 hepatocytes as a cellular steatosis model. BMC Complement Altern Med. 2015;15:185. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 44] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 18. | Egnatchik RA, Leamy AK, Noguchi Y, Shiota M, Young JD. Palmitate-induced activation of mitochondrial metabolism promotes oxidative stress and apoptosis in H4IIEC3 rat hepatocytes. Metabolism. 2014;63:283-295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 128] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 19. | Galgani JE, Moro C, Ravussin E. Metabolic flexibility and insulin resistance. Am J Physiol Endocrinol Metab. 2008;295:E1009-E1017. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 333] [Cited by in RCA: 382] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 20. | Lumeng CN, Saltiel AR. Inflammatory links between obesity and metabolic disease. J Clin Invest. 2011;121:2111-2117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1520] [Cited by in RCA: 1766] [Article Influence: 117.7] [Reference Citation Analysis (0)] |
| 21. | Yan C, Chen J, Chen N. Long noncoding RNA MALAT1 promotes hepatic steatosis and insulin resistance by increasing nuclear SREBP-1c protein stability. Sci Rep. 2016;6:22640. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 156] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 22. | Yahagi N, Shimano H, Hasty AH, Matsuzaka T, Ide T, Yoshikawa T, Amemiya-Kudo M, Tomita S, Okazaki H, Tamura Y. Absence of sterol regulatory element-binding protein-1 (SREBP-1) ameliorates fatty livers but not obesity or insulin resistance in Lep(ob)/Lep(ob) mice. J Biol Chem. 2002;277:19353-19357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 291] [Cited by in RCA: 296] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 23. | Descombes P, Schibler U. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell. 1991;67:569-579. [PubMed] |
| 24. | Zhang Q, Ramlee MK, Brunmeir R, Villanueva CJ, Halperin D, Xu F. Dynamic and distinct histone modifications modulate the expression of key adipogenesis regulatory genes. Cell Cycle. 2012;11:4310-4322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 64] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 25. | Zuo Y, Qiang L, Farmer SR. Activation of CCAAT/enhancer-binding protein (C/EBP) alpha expression by C/EBP beta during adipogenesis requires a peroxisome proliferator-activated receptor-gamma-associated repression of HDAC1 at the C/ebp alpha gene promoter. J Biol Chem. 2006;281:7960-7967. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 164] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 26. | Duszka K, Bogner-Strauss JG, Hackl H, Rieder D, Neuhold C, Prokesch A, Trajanoski Z, Krogsdam AM. Nr4a1 is required for fasting-induced down-regulation of Pparγ2 in white adipose tissue. Mol Endocrinol. 2013;27:135-149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 23] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 27. | Jeninga EH, Bugge A, Nielsen R, Kersten S, Hamers N, Dani C, Wabitsch M, Berger R, Stunnenberg HG, Mandrup S. Peroxisome proliferator-activated receptor gamma regulates expression of the anti-lipolytic G-protein-coupled receptor 81 (GPR81/Gpr81). J Biol Chem. 2009;284:26385-26393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 77] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 28. | Jaeger D, Schoiswohl G, Hofer P, Schreiber R, Schweiger M, Eichmann TO, Pollak NM, Poecher N, Grabner GF, Zierler KA. Fasting-induced G0/G1 switch gene 2 and FGF21 expression in the liver are under regulation of adipose tissue derived fatty acids. J Hepatol. 2015;63:437-445. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 47] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 29. | Ahmed K, Tunaru S, Tang C, Müller M, Gille A, Sassmann A, Hanson J, Offermanns S. An autocrine lactate loop mediates insulin-dependent inhibition of lipolysis through GPR81. Cell Metab. 2010;11:311-319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 355] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 30. | Davenport AP, Alexander SP, Sharman JL, Pawson AJ, Benson HE, Monaghan AE, Liew WC, Mpamhanga CP, Bonner TI, Neubig RR. International Union of Basic and Clinical Pharmacology. LXXXVIII. G protein-coupled receptor list: recommendations for new pairings with cognate ligands. Pharmacol Rev. 2013;65:967-986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 245] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 31. | Offermanns S, Colletti SL, Lovenberg TW, Semple G, Wise A, IJzerman AP. International Union of Basic and Clinical Pharmacology. LXXXII: Nomenclature and Classification of Hydroxy-carboxylic Acid Receptors (GPR81, GPR109A, and GPR109B). Pharmacol Rev. 2011;63:269-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 159] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 32. | Hu E, Liang P, Spiegelman BM. AdipoQ is a novel adipose-specific gene dysregulated in obesity. J Biol Chem. 1996;271:10697-10703. [PubMed] |
| 33. | Giby VG, Ajith TA. Role of adipokines and peroxisome proliferator-activated receptors in nonalcoholic fatty liver disease. World J Hepatol. 2014;6:570-579. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 86] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
Manuscript source: Unsolicited Manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
P- Reviewer: Kanda T, Tarantino G S- Editor: Wei LJ L- Editor: A E- Editor: Huang Y