Published online Sep 28, 2017. doi: 10.3748/wjg.v23.i36.6674
Peer-review started: February 28, 2017
First decision: April 28, 2017
Revised: May 17, 2017
Accepted: July 4, 2017
Article in press: July 4, 2017
Published online: September 28, 2017
Processing time: 209 Days and 13.2 Hours
To uncover novel genetic markers that could contribute to predicting hepatocellular carcinoma (HCC) susceptibility in Caucasians.
The present retrospective case-control study compared genotype frequencies between a cohort of HCC cases and two, independent, HCC-free, age/sex-matched control groups. The HCC cohort comprised 192 homogeneous patients that had undergone orthotopic liver transplantation. The first control group comprised 167 patients that were matched to the HCC cohort for the percentage of hepatitis B (HBV) and/or hepatitis C (HCV) infections. A second control group included 192 virus-free, healthy individuals that were used to evaluate the generalizability of the identified predictive markers. All cases and controls were Caucasian. The three study populations were characterized with a panel of 31 markers derived from 21 genes that encoded key proteins involved in hepatocarcinogenesis-related pathways. The study end-point was to assess the association between genetic variants and HCC onset.
Five genetic markers were identified as risk factors for HCC in high-risk patients infected with HBV/HCV. According to a dominant model, reduced HCC risk was associated with three polymorphisms: ERCC1 rs3212986 (OR = 0.46, 95%CI: 0.30-0.71, P = 0.0005), GST-P1 rs1138272 (OR = 0.41, 95%CI: 0.21-0.81, P = 0.0097), and CYP17A1 rs743572 (OR = 0.50, 95%CI: 0.31-0.79, P = 0.0032). Conversely, according to a recessive model, increased HCC risk was associated with two polymorphisms: XRCC3 rs1799794 (OR = 3.70, 95%CI: 1.02-13.39, P = 0.0461) and ABCB1 rs1128503 (OR = 2.06, 95%CI: 1.18-3.61, P = 0.0111). These associations remained significant in a subgroup analysis, where patients were stratified according to viral status (HBV- or HCV-positive serology). Two variants exhibited a serology-specific effect: ABCB1 rs1128503 (OR = 4.18, 95%CI: 1.55-11.29, P = 0.0048) showed an effect in the HBV-positive subgroup; and ERCC1 rs3212986 (OR = 0.33, 95%CI: 0.18-0.60, P = 0.0003) showed an effect in the HCV-positive subgroup. Among the five markers identified, ERCC1 rs3212986 (OR = 0.43, P < 0.0001) and CYP17A1 rs743572 (OR = 0.73, P = 0.0310) had a different distribution in patients with HCC compared to healthy individuals. With a recursive partitioning approach, we also demonstrated that significant gene-gene interactions between ERCC1 rs3212986, CYP17A1 rs743572, GST-P1 rs1138272, and the previously described UGT1A7*3 predictive marker, played a role in the complex trait of HCC susceptibility.
We identified five polymorphisms and interactions that contributed crucially to predicting HCC risk. These findings represented an important step towards improving HCC diagnosis and management.
Core tip: It is a great challenge to define new biomarkers of hepatocellular carcinoma (HCC) risk for HCC management. This work identified five genetic markers in key pathways linked to hepatocarcinogenesis (ERCC1 rs3212986, GSTP1 rs1138272, CYP17A1 rs743572, XRCC3 rs1799794, and ABCB1 rs112850), which could predict individual HCC susceptibility, particularly in a high-risk [hepatitis B viral/hepatitis C viral (HCV)-infected] Caucasian population. We also identified potential gene-gene interactions that should be included in the definition of HCC risk. These findings could contribute to improved HCC surveillance, early cancer diagnoses, and potential curative therapies. For patients with HCV, these markers could be considered selection criteria for the personalized use of recently developed, highly expensive anti-viral therapies.
- Citation: De Mattia E, Cecchin E, Polesel J, Bignucolo A, Roncato R, Lupo F, Crovatto M, Buonadonna A, Tiribelli C, Toffoli G. Genetic biomarkers for hepatocellular cancer risk in a caucasian population. World J Gastroenterol 2017; 23(36): 6674-6684
- URL: https://www.wjgnet.com/1007-9327/full/v23/i36/6674.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i36.6674
Hepatocellular carcinoma (HCC) is one of the most common malignancies worldwide, with a particularly high prevalence in some areas of Asia and Africa. HCC represents the second leading cause of cancer-related deaths[1]. Although liver cancer is less frequent in Western developed countries, recent data indicate that, due to dissemination of hepatitis B (HBV) and C (HCV) viral infections, the HCC incidence is also dramatically rising, in both the United States and Europe[1,2].
Unlike most malignancies, the major risk factors for HCC development are well-defined. These risk factors are chronic HCV and HBV infections, liver cirrhosis, heavy alcohol intake, tobacco smoking, exposure to environmental and dietary carcinogens (i.e., aflatoxin B1), genetic and metabolic liver disease (i.e., hereditary hemochromatosis, non-alcoholic steatohepatitis), and other conditions capable of inducing liver damage[2]. Moreover, the prevalence of HCC increases with age and male sex[1-3].
In addition to environmental factors, recent insights into the biology of HCC have demonstrated that accumulations of genetic and epigenetic abnormalities can also play an essential role in liver carcinogenesis. In particular, data derived from candidate-gene investigations and, more recently, genome-wide associations studies have highlighted the possible role that genetic variants might play as significant determinants of HCC susceptibility[4-6]. Additionally, host genetic factors were shown to affect the clinical course of HBV or HCV infections, because they influence the individual’s predisposition to disease progression and hepatocarcinogenesis[7,8]. However, the currently available studies are highly heterogeneous in study design, sample size, ethnicity, and clinical-demographic features of the investigated population; these differences make it difficult to draw solid conclusions[4,6]. Moreover, the majority of case-control investigations were performed with Asian populations; thus, replication studies with different ethnicities are warranted[4-8].
It is of great clinical interest to identify genetic traits that can modify liver carcinogenesis, because they can potentially be used to improve preventive, diagnostic, and therapeutic strategies in HCC management[3,9,10]. To date, the majority of patients with HCC are diagnosed at an advanced tumor stage, which precludes potentially curative therapies, including orthotopic liver transplantation (OLT), surgical resection, and local ablation[3,9,10]. The HCC surveillance programs that could lead to early cancer detection and effective treatment are currently based on highly heterogeneous scoring systems that require costly measures[3,9,10]. The identification of additional molecular biomarkers predictive of individual HCC susceptibility, particularly in a high-risk population (i.e., HBV/HCV-positive), could be important in improving current guidelines for genetics-based patient screening and preventive strategies. In some countries, including the United States and Europe, HCV-related HCC accounts for most liver cancer incidents[11]. Consequently, an increased understanding of the genetic contribution to the clinical course of viral infection could be essential in optimizing the use of newly developed, but highly expensive, direct acting antiviral (DAA) treatments[12,13].
We previously reported that the low-activity alleles, UGT1A1*28, UGT1A7*3, UGT1A9*22, and related haplotypes may play a protective role against HCC development in Caucasian populations. These alleles positively modulate the serum levels of beneficial molecules, like bilirubin[14]. The present study aimed to extend genetic analyses by identifying additional molecular markers that could be integrated into the HCC risk stratification algorithms used in liver cancer prevention strategies. To that end, we analyzed a set of potential candidate HCC-risk markers with low penetrance. These candidate markers were based on functionally relevant polymorphic variants in genes that encoded proteins involved in key pathways underlying carcinogenesis[6,15-17]. The pathways we investigated were: (1) membrane transporters that play an essential role in xenobiotic defense and prevent the entry of toxic environmental compounds that might damage liver tissue; (2) DNA synthesis and repair mechanisms that modulate the cell’s capacity to respond to DNA damage induced by toxic agents, and thus, these mechanisms affect the accumulation of mutations in DNA; (3) detoxification systems that facilitate the elimination of detrimental endo/exogenous compounds; and (4) systems that defend against oxidative stress and related cellular damage, which can play a crucial role in the development and progression of HCC. We designed a retrospective case-control study and enrolled a homogeneous group of patients with HCC that had received OLT and two age/sex-matched control groups (a group positive for HBV and/or HCV and a healthy population). We aimed to identify novel genetic markers that could contribute to the determination of individual HCC risk, particularly in Caucasian populations.
We studied 192 patients with histologically confirmed HCC that had undergone OLTs between May 1991 and March 2006 at the Liver Transplantation Center of the S. Giovanni Battista Hospital (Turin, Italy). All patients were infected with HBV and/or HCV and had a cirrhotic liver. The first control group included 167 subjects with HBV and/or HCV infections, but no evidence of cancer (including HCC). These subjects were followed up for hepatitis infections between November 2009 and September 2010 at the Department of Laboratory Medicine, S. Maria degli Angeli Hospital (Pordenone, Italy). This control group was matched to the HCC group to ensure equivalent percentages of patients with HBV and/or HCV and similar distributions of gender and age (in quinquennia), when possible.
The second control group included 192 healthy subjects that were matched to patients with HCC for gender and age (in quinquennia). For this control group, individuals under 70 years of age were randomly selected from a pool of blood donors that visited the Centro di Riferimento Oncologico National Cancer Institute in Aviano, Italy. This control group also included individuals aged ≥ 70 years that were cancer-free without HBV/HCV infections. These individuals had been approached during occasional blood testing at the S. Maria degli Angeli Hospital, and they consented to provide a 5-mL blood sample for research purposes.
All cases and controls were Italian Caucasian individuals, and they all provided written informed consent for genetic analysis and for inclusion in the study. The study protocol conformed to the ethics guidelines of the 1975 Declaration of Helsinki, and it was approved by the Institutional Review Board of each participating institution (Ethics Committee of Azienda Ospedaliera Universitaria, S. Giovanni Battista, Torino).
Genomic DNA was extracted from peripheral blood with the High Pure PCR Template Preparation Kit (Roche Diagnostics GmbH, Mannheim, Germany). The samples underwent standard proteinase K digestion and successive template purification. The genomic DNA solution was stored at 4 °C. Positive and negative control samples were included in each analysis.
We considered a set of 31 molecular markers that represented functionally relevant polymorphisms (PubMed-MEDLINE search) in genes that encoded key proteins involved in hepatocarcinogenesis-related pathways. In particular, the biological processes evaluated (and related genes) were: membrane transport (ABCB1, ABCC2), DNA synthesis and repair mechanisms (MTHFR, TYMS, ERCC1, ERCC2, XRCC1, XRCC3, APE1, hMLH1, hMSH2, hOOG1), detoxifying systems (CYP3A4, CYP3A5, CYP17A1, CYP2D6), and oxidative stress response (GST-M1, GST-T1, GST-P1; GSTA1, SOD2).
All the genes, variants, and assays applied are listed in Supplementary Table 1. For pyrosequencing, we used PSQ96MA (Qiagen, Hilden, Germany). PCR amplifications were performed in an Eppendorf Mastercycler gradient, with TaqGold DNA Polymerase (AB Applied Biosystems, Foster City, CA, United States). The details of the pyrosequencing assays, primer sequences, and PCR conditions are available upon request. Pre-designed TaqMan single-nucleotide polymorphism genotyping assays were conducted with the Applera TaqMan Universal Master mix on an ABI 7900HT Real-time PCR system (AB Applied Biosystems, Foster City, CA, United States), according to the manufacturer’s instructions for optimal allelic discrimination. All commercial TaqMan assays were purchased from the Applied Biosystems website (http://www.appliedbiosystems.com). Detailed protocols for genotyping methods, based on gel electrophoresis and enzymatic digestion, are available upon request.
| Characteristics | HCC group | HBV/HCV group | Healthy group |
| Sex | |||
| Men | 166 (86.5) | 141 (84.4) | 166 (86.5) |
| Women | 26 (13.5) | 26 (15.6) | 26 (13.5) |
| Age (yr) | |||
| < 55 | 85 (44.3) | 85 (50.9) | 96 (50.0) |
| 55-59 | 60 (31.3) | 19 (11.4) | 35 (18.2) |
| ≥ 60 | 47 (24.5) | 63 (37.7) | 61 (31.8) |
| Hepatitis infection | |||
| None | 0 (0.0) | 0 (0.0) | 192 (100.0) |
| HBV+ and HCV- | 74 (38.5) | 74 (44.3) | 0 (0.0) |
| HBV- and HCV+ | 109 (56.8) | 88 (52.7) | 0 (0.0) |
| HBV+ and HCV+ | 9 (4.7) | 5 (3.0) | 0 (0.0) |
| Years of orthotopic liver transplantation or resection | |||
| 1991-1995 | 27 (14.1) | ||
| 1996-2000 | 60 (31.3) | ||
| 2001-2006 | 102 (54.7) | ||
| Number of nodes | |||
| 1 | 110 (57.3) | ||
| ≥ 2 | 72 (37.5) | ||
| Unknown | 10 (5.2) | ||
| Maximal dimension of nodes (cm) | |||
| ≤ 2 | 79 (41.2) | ||
| 2.1 to ≤ 3 | 67 (34.9) | ||
| > 3 | 37 (19.3) | ||
| Unknown | 9 (4.7) | ||
In this study, we assessed associations between genetic markers and the individual risk of developing HCC. In healthy controls, deviation from Hardy-Weinberg equilibrium was tested for each polymorphism with the χ2 test, and no deviation was found (P < 0.05).
The odds ratio (OR) and 95%CI were estimated with unconditional logistic regression, which was controlled for matching variables. We also investigated three genetic models (dominant, recessive, and additive) for associations, and we reported the most statistically significant one, based on the Wald χ2-test. Statistical significance was set at P < 0.05 (two-sided). The analyses were performed with SAS 9.2, and a statistical review of the study was performed by a biomedical statistician.
Recursive partitioning was performed to elucidate high-order relationships among genetic factors, when stratifying patients between HCC cases and HBV/HCV group. The analysis was carried out with the five genetic variants that were identified in the present study as significant predictors of HCC risk and with the previously described UGT1A markers (i.e., UGT1A1*28, UGT1A9*22, UGT1A7*3)[14].
The demographic, clinical, and serological characteristics of the 192 patients with HCC and the two control groups are reported in Table 1. The predominant gender was male (86.5% of patients with HCC, 84.4% of patients in the HBV/HCV group, and 86.5% of healthy subjects). The median ages at the time of enrolment were 56, 54 and 55 years for the HCC, HBV/HCV, and healthy groups, respectively. The HCC cohort was clinicopathologically homogeneous; all patients fulfilled the surgical intervention criteria (i.e., T1/T2 primary tumor stage, evaluated with the TNM scale; well or moderately poorly differentiated grade at diagnosis; and no detectable vascular invasion).
The average genotype call rates were 0.98 (range: 0.92-1.00), 1.00 (range: 0.99-1.00), and 0.99 (range: 0.95-1.00) for the HCC, HBV/HCV, and healthy groups, respectively. All case and control samples were included in the study, because they all reached the fixed call rate threshold of 90%.
HCC vs HBV/HCV group: Considering that chronic HBV and/or HCV infections represent a major risk factor for HCC development, a case/control analysis was initially performed to compare the genotype frequency distribution between patients with HCC and individuals without HCC, but with matched HBV/HCV infections. The frequency distributions of variant genotypes were significantly different between cases and controls (Tables 2 and 3) for five polymorphisms. No Hardy-Weinberg disequilibrium among healthy controls was detected (P > 0.05), which suggested that it was a representative sampling of the investigated population. The observed genotype frequency distribution in healthy controls was consistent with data published in the literature for a Caucasian population (http://www.ncbi.nlm.nih.gov/snp).
| Gene | SNP | Genotype | HCC group | HBV/HCV group | Healthy group |
| ERCC1 | rs3212986 | GG | 126 (67.7) | 82 (49.1) | 91 (47.4) |
| GT | 44 (23.7) | 78 (46.7) | 81 (42.2) | ||
| TT | 16 (8.6) | 7 (4.2) | 20 (10.4) | ||
| XRCC3 | rs1799794 | AA | 128 (66.7) | 108 (65.1) | 137 (71.7) |
| AG | 52 (27.1) | 55 (33.1) | 49 (25.7) | ||
| GG | 12 (6.3) | 3 (1.8) | 5 (2.6) | ||
| GST-P1 | rs1138272 | CC | 172 (92.5) | 139 (83.2) | 182 (94.8) |
| CT | 13 (7.0) | 27 (16.2) | 10 (5.2) | ||
| TT | 1 (0.5) | 1 (0.6) | 0 (0.0) | ||
| CYP17A1 | rs743572 | TT | 72 (38.1) | 39 (23.4) | 54 (28.4) |
| TC | 84 (44.4) | 88 (52.7) | 91 (47.9) | ||
| CC | 33 (17.5) | 40 (24.0) | 45 (23.7) | ||
| ABCB1 | rs1128503 | CC | 54 (28.1) | 52 (31.1) | 58 (30.2) |
| CT | 92 (47.9) | 93 (55.7) | 95 (49.5) | ||
| TT | 46 (24.0) | 22 (13.2) | 39 (20.3) |
| Gene | SNP | Allelic change | HCC vs HBV/HCV | HCC vs Healthy | ||||
| Mod | OR (95%CI)1 | P value | Mod | OR (95%CI)2 | P value | |||
| ERCC1 | rs3212986 | G>T | Dom | 0.46 (0.30-0.71) | 0.0005b | Dom | 0.43 (0.28-0.65) | < 0.0001b |
| XRCC3 | rs1799794 | A>G | Rec | 3.70 (1.02-13.39) | 0.0461a | Rec | 2.44 (0.84-7.08) | 0.1003 |
| GST-P1 | rs1138272 | C>T | Dom | 0.41 (0.21-0.81) | 0.0097b | Add | 1.58 (0.70-3.57) | 0.2699 |
| CYP17A1 | rs743572 | T>C | Dom | 0.50 (0.31-0.79) | 0.0032b | Add | 0.73 (0.55-0.97) | 0.0310a |
| ABCB1 | rs1128503 | C>T | Rec | 2.06 (1.18-3.61) | 0.0111a | Dom | 1.22 (0.75-1.98) | 0.4194 |
Specifically, ERCC1 rs3212986 (OR = 0.46, P = 0.0005), GST-P1 rs1138272 (OR = 0.41, P = 0.0097), and CYP17A1 rs743572 (OR = 0.50, P = 0.0032) markers were observed more frequently in the HBV/HCV group than in the HCC group, and they were significantly associated with reduced liver cancer risk, according to the dominant model. In contrast, the frequency of XRCC3 rs1799794 (OR = 3.70; P = 0.0461) and ABCB1 rs1128503 (OR = 2.06; P = 0.0111) markers were higher in the HCC group than in the HBV/HCV group, and they were associated with increased HCC risk, according to the recessive model (Table 3).
Given the molecular and clinicopathological differences between HBV- and HCV- related hepatocarcinogenesis[18], a similar analysis was also carried out in subgroups, divided according to viral status (Table 4). In all cases, the ORs calculated for the HBV- and HCV-positive subgroups were concordant with those obtained for the entire combined group. The association was significant in both sub-groups for CYP17A1 rs743572 (OR = 0.45, P = 0.0315 and OR = 0.51, P = 0.0384 in HBV-positive and HCV-positive subgroups, respectively). In contrast, ABCB1 rs1128503 (OR = 4.18, P = 0.0048, HBV-positive subgroup) and ERCC1 rs3212986 (OR = 0.33, P = 0.0003, HCV-positive subgroup) showed significant associations in only one sub-group.
| Gene | SNP | Allelic change | Mod | HCC vs HBV/HCV | |||
| HBV-positive | HCV-positive | ||||||
| OR (95%CI)1 | P value | OR (95%CI)1 | P value | ||||
| ERCC1 | rs3212986 | G>T | Dom | 0.59 (0.30-1.15) | 0.1231 | 0.33 (0.18-0.60) | 0.0003b |
| XRCC3 | rs1799794 | A>G | Rec | 2.23 (0.39-12.82) | 0.3695 | 6.73 (0.82-55.03) | 0.0753 |
| GST-P1 | rs1138272 | C>T | Dom | 0.40 (0.14-1.15) | 0.0881 | 0.41 (0.16-1.02) | 0.0558 |
| CYP17A1 | rs743572 | T>C | Dom | 0.45 (0.22-0.93) | 0.0315a | 0.51 (0.27-0.97) | 0.0384a |
| ABCB1 | rs1128503 | C>T | Rec | 4.18 (1.55-11.29) | 0.0048b | 1.49 (0.71-3.11) | 0.2893 |
HCC vs healthy group: We also conducted a case case/control analysis of the genotype frequency distribution in patients with HCC compared to individuals without HCC or virus infections to evaluate the extent of the predictive value of the selected five markers (Table 3). Two markers were significantly associated with HCC risk. Specifically, ERCC1 rs3212986 (OR = 0.43, P < 0.0001) and CYP17A1 rs743572 (OR = 0.73, P = 0.0310) were associated with reduced HCC risk, according to the dominant and additive models, respectively.
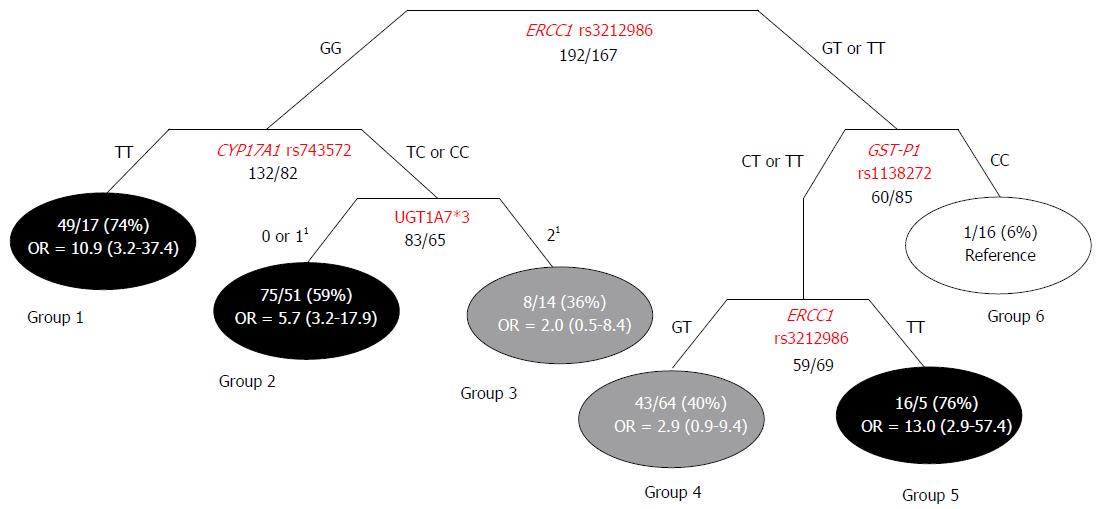
Classification and regression tree analysis: The Classification and Regression Tree (CART) method allowed us to identify 6 subgroups of subjects according to genotype features, each with a different probability of HCC occurrence. Three of the terminal nodes exhibited high-probability (59%-76%), two exhibited intermediate-probability (36% and 40%), and one exhibited low-probability (6%) of developing HCC (Figure 1). The ORs and 95%CIs were calculated for each high/intermediate-probability group, with respect to the reference low-probability group. The group with the highest percentage of HCC cases (group 5) was associated with an approximately 13-fold higher HCC risk compared to the reference low-probability group. The first node of the tree represented the ERCC1 rs3212986 variant, which significantly interacted with CYP17A1 rs743572, GST-P1 rs1138272, and the UGT1A7*3 marker in the generation of the tree (Figure 1).
HCC is a global healthcare problem. It is one of the most common malignancies worldwide, with an increasing incidence in Western developed countries. It represents the second cause of death attributable to cancer[1,2]. The genetic origin of HCC has been a focus of research in the past few years. Some studies found that genetic variants could be predictive of the clinical course of HBV or HCV and an individual’s predisposition to developing liver cancer[4-8]. However, those studies were performed mainly in Asian populations; thus, few data are available on Caucasian populations. The ethnicity of the population being studied is a crucial factor in case-control studies, because polymorphism frequency varies greatly with geographical origin. This association could give rise to ethnicity-specific phenotypic effects[4,14]. The present study revealed some novel genetic markers that could be used as early diagnosis indicators for liver cancer susceptibility in Caucasians. The availability of these markers could improve HCC risk stratification algorithms and surveillance programs.
The main result of this work was the identification of five polymorphisms (ERCC1 rs3212986, GST-P1 rs1138272, CYP17A1 rs743572, XRCC3 rs1799794, and ABCB1 rs1128503) that could identify sub-populations of patients among high-risk Caucasian individuals (HBV/HCV-positive), which had differential predispositions for HCC development. The ABCB1 rs1128503 and ERCC1 rs3212986 variants displayed different effects, based on the viral status of the patient (HBV- or HCV-positive serology). This result was consistent with the molecular and clinical-pathological differences between HBV- and HCV-related hepatocarcinogenesis[18]. Two polymorphisms (ERCC1 rs3212986, and CYP17A1 rs743572) could also predict HCC risk among the healthy Caucasian population. A CART analysis provided evidence of gene-gene interactions between ERCC1 rs3212986, CYP17A1 rs743572, GST-P1 rs1138272, and the previously described UGT1A7*3 marker. These interactions defined subgroups of patients with genetic combinations that could further increase the risk of HCC.
The ERCC1 rs3212986 (8092G>T) polymorphism was demonstrated to be a strong protective factor against liver cancer development in both high-risk (HCV/HBV-positive) and healthy individuals. Furthermore, in an analysis of subgroups with different viral statuses, a particularly significant effect was detected in the HCV-positive subgroup.
ERCC1 is one of several key rate-limiting enzymes in the nucleotide excision repair (NER) pathway. The NER mechanism of the DNA repair system maintains genomic integrity by removing inter-strand DNA crosslinks, and thus, it influences the cell’s sensitivity to DNA-adducting carcinogens[19]. A NER-related reduction in DNA repair capacity has been associated with some types of tumors (i.e., lung, head and neck, prostate, glioma, ovarian cancers). Accordingly, ERCC1 gene variants were shown to influence susceptibility to cancer[20,21]. However, few studies have investigated the implication of the NER biological pathway and ERCC1 polymorphisms, including rs3212986[20,21], in hepatocarcinogenesis, and no data are available in Caucasian populations. The ERCC1 rs3212986 variant, located in the 3’ untranslated region (3’UTR) of the gene, is thought to increase transcript stability, although further biochemical studies are required to clarify the effect of this polymorphism on gene transcription and protein translation[22,23]. Based on our data, we inferred that the rs3212986 variant allele was associated with elevated ERCC1 expression and/or activity, which could lead to enhanced protection against the accumulation of mutagenic DNA lesions, and consequently, against hepatocarcinogenesis. However, we could not exclude that this marker might also be closely linked to other functionally relevant genetic variants[21] that might alter ERCC activity and/or HCC onset.
The XRCC3 rs1799794 (4541A>G) marker was associated with elevated HCC susceptibility in a high-risk (HCV/HBV-positive) population. XRCC3 is another DNA repair protein that participates in the DNA double-strand break/recombination repair machinery. XRCC3 was shown to be involved in the pathogenesis of many cancers, including HCC[24,25]. Reduced XRCC3 activity was associated with significantly elevated levels of bulky DNA adducts[26]. Furthermore, epidemiological evidence, derived mainly from studies in an Asian population, have shown that XRCC3 contributed to the repair of DNA damage induced by environmental toxins, such as aflatoxin B1, and that this effect depended on viral status and ethnicity[25,27]. The XRCC3 rs1799794 variant, located in a regulatory region in the 5’UTR of the gene, was predicted to have a potential effect on protein expression levels[28]. The results of the present study suggested that this polymorphism could reduce the ability to repair DNA adducts, which would contribute to the accumulation of mutagenic lesions, and thus, increase the risk of HCC onset. However, the exact molecular mechanism of the rs1799794 variant is currently unknown, and we could not exclude a linkage with another functional marker[29].
The GST-P1 rs1138272 (341C>T) polymorphism was found to have a protective effect on liver cancer development in a high-risk HCV/HBV-positive population. GST-P1 belongs to the GST multigene family. It is a phase II enzyme involved in the inactivation of electrophilic xenobiotics by conjugation with glutathione[30], which facilitates the excretion of molecules that are potentially toxic to the liver[31]. This enzyme also contributes to protecting liver cells from damage related to reactive oxygen species generated, for instance, by chronic alcohol and tobacco abuse[32]. Several studies have clearly demonstrated that silencing GST-P1 expression by promoter methylation was positively correlated with the incidence of HCC[31] and with virus-related hepatocarcinogenesis[33]. In contrast, in meta-analyses, GST-P1 genetic polymorphisms (i.e., Ile105Val, rs1695) were not associated with HCC risk[34,35] although the few data available were insufficient for drawing any definite conclusions. The less investigated GST-P1 rs1138272 polymorphism is a missense variant (Val114Ala) that lies in the electron binding, active domain of the protein. This variant was reported to affect substrate specificity by altering its ability to distinguish between planar and non-planar substrates[36,37]. Our results suggested that this functional change in GST-P1 activity may result in a lower risk of HCC development; further clinical experimental studies are required to clarify the role of the rs1138272 polymorphism in the response of GST-P1 to environmental xenobiotics and oxidative stress, particularly in a virus-infected liver.
The CYP17A1 rs743572 (-34T>C) polymorphism was found to be a strong protective factor against HCC development in both the high-risk HCV/HBV-positive group and healthy individuals. CYP17A1 is an enzyme that plays a critical role in the biosynthesis of steroid hormones by catalyzing both 17a-hydroxylase and 17, 20-lyase activities[38]. Some data derived from animal models and human epidemiologic studies have demonstrated that estrogens play an essential role in hepatocarcinogenesis; however, the exact mechanism underlying the effect of estrogen levels on HCC development is currently controversial[39]. It was initially suggested that the CYP17A1 rs743572 variant, located in the 5’UTR of the gene, served as a new Sp1-type (CCACC box) binding site, which provided an additional promoter, and thus, it could increase estrogen biosynthesis[40]. However, subsequent studies could not replicate the phenotypic impact of this variant on CYP17A1 mRNA expression or estrogen levels[41]. Moreover, the involvement of the rs743572 marker on HCC development was not well defined[42,43]. From the present data, it could be speculated that the rs743572 variant might contribute to changing CYP17A1 activity, and its dependent steroid levels, to provide a protective effect against liver cancer development. Estrogen is known to participate in various biological functions. For example, an increase in sex hormone levels was suggested to suppress the inflammatory processes mediated by pro-inflammatory cytokines[44,45]. Additional work will be required to clarify the functional significance of the rs743572 marker and the molecular mechanism of CYP17A1 in hepatocellular carcinogenesis.
Finally, ABCB1 rs1128503 (1236C>T) was associated with elevated HCC susceptibility in a high-risk (HCV/HBV-positive) population. Furthermore, an analysis of subgroups with different viral statuses showed that a particularly significant effect was detected in the HBV-positive subgroup. The ABCB1 gene encodes MDR-1, also known as P-glycoprotein (P-gp). MDR-1 belongs to the ATP-binding cassette (ABC) transporter superfamily[46,47]. This membrane efflux pump is widely expressed in various organs, including the liver, and it is physiologically involved in the protection of normal tissue from environmental and endogenous toxins[46-48]. MDR1 is implicated in hepatic clearance of oxidative and other genotoxic products generated by chronic inflammation induced by HCC risk factors. Thus, it contributes to preventing the initiation of hepatocarcinogenesis[49,50]. Many studies have described MDR1 over-expression in various tumor tissues, including liver cancer[51]; thus, this membrane carrier and its functional genetic variants are good candidates for influencing liver susceptibility to HCC. ABCB1 rs1128503 is a synonymous variant (Gly412Gly) of a residue located in the cytoplasmic domain of the transporter. It is reported to affect mRNA stability[52], but the exact phenotypic impact of this variant on protein expression/activity remains to be defined[53,54]. Several studies, performed in Asian populations, have investigated associations between ABCB1 polymorphisms (including rs1128503) and the risk of developing HCC. A recent meta-analysis[55] showed that polymorphic alleles of the ABCB1 gene significantly increased HCC risk. In analyses of subgroups with different types of variations, both synonymous and non-synonymous polymorphisms were associated with elevated liver cancer susceptibility. In particular, synonymous variants (i.e., rs1128503) were suggested to be correlated with changes in transcriptional and translational processes. In addition, genetic markers located in the cytoplasmic domain of the transporter, like rs1128503, were shown to have the greatest predictive power for HCC risk. These data, although obtained in Asian cohorts, were consistent with the result of the present study, which provided the first evidence of a role of the ABCB1 rs1128503 marker in HCC risk in a Caucasian population.
Due to the multifactorial nature of hepatocarcinogenesis, an exploratory CART analysis was applied to investigate interactions between the genetic factors identified as predictors of liver cancer susceptibility in the high-risk population (HBV/HCV-infected group). The polymorphism in the gene of the DNA repair system (i.e., ERCC1 rs3212986) was demonstrated to significantly interact with variants of genes that encoded phase I (CYP17A1 rs74357) and II (GST-P1 rs1138272, UGT1A7*3) metabolic enzymes; these interactions were important in defining individual HCC risk. The CART analysis generated a distinctive clustering of subjects, according to different probabilities of developing liver cancer. These data pointed out that previously acknowledged predictive markers, such as the UGT1A*3 polymorphism[14], should be considered in association with novel identified genetic variants for properly defining the complex trait of HCC susceptibility. In particular, ERCC1 rs3212986 was shown to be the most relevant polymorphism for HCC risk stratification, because it appeared twice in the generated tree. This marker significantly interacted with three additional genetic variants (CYP17A1 rs74357, GST-P1 rs1138272, and UGT1A7*3) in defining individual susceptibility. The analysis of gene-gene interactions and the network shape uncovered specific genetic-context effects that could not be detected in evaluations of single markers. These findings highlighted the importance of combining genetic markers to obtain a better representation of the biological cooperation among multiple pathways in hepatocarcinogenesis.
The present study had some limitations. First, our case-cohort comprised a highly specific group of patients with HCC (patients that received OLT). These patients had definite clinical features that could limit the generalizability of the present findings. The validity of the present results was also limited to Caucasian individuals; the results remain to be verified in other populations, like Asians or Africans. Second, our work did not evaluate other confounding etiological factors, such as smoking and alcohol intake. However, the use of a control group matched to HCC cases for HBV/HCV viral status permitted us to control for this crucial confounding variable. Third, the exact functional and phenotypic properties of some of the genetic markers that we identified as predictors of HCC risk have not been well established. Moreover, we could not exclude any linkages with other functionally relevant genetic variants. Finally, considering the retrospective and exploratory nature of our case-control analysis, the results obtained should be interpreted with caution. Our findings will require independent validation in future prospective, large-scale epidemiological investigations that are performed in well-defined, homogeneous populations.
The validation of the present findings could have important clinical implications for improving HCC management. The novel identified genetic markers and their gene-gene interactions, in combination with other well-known serum biomarkers (e.g., alphafetoprotein) and diagnostic imaging (e.g., ultrasonography)[9], could contribute to improving the HCC risk stratification algorithms and surveillance programs, particularly for patients at risk. Those improvements might facilitate early cancer detection and might lead to potentially curative therapies. Furthermore, an enhanced understanding of the genetic factors associated with the evolution of HCV infection could be essential for the optimized use of the recently developed, but highly expensive, anti-HCV DAAs[12,13]. Thus, improved screening could assist the health care system in containing costs and ensuring the most clinically appropriate treatments.
The present study identified five variants in genes that encode DNA repair system proteins (i.e., ERCC1, XRCC3), phase I (i.e., CYP17A1) and II (i.e., GST-P1) enzymes, and a membrane transporter (i.e., MDR-1). Our data suggested that these genes might play a role in predicting HCC susceptibility in a high-risk (HBV-HCV infected) population and in a healthy Caucasian population. The CART analysis pointed out the potential value of studying of gene-gene interactions for the stratification of individuals at different risks of developing HCC. Although validation is required, these preliminary results might have important clinical implications in the management of HCC prevention and patient care.
The authors wish to thank Dr. Alessandro Fornasier, who developed the genetic data management software used in the present research.
Hepatocellular carcinoma (HCC) represents a global health problem, due to its high incidence and mortality rates. Although the etiologies of this neoplasia are well-defined, the majority of patients are diagnosed at advanced-stage disease, which excludes them from receiving a potentially curative therapy. Hence, they urgently need the definition of new biomarkers of HCC susceptibility that can permit early cancer detection, particularly for individuals in a high-risk population [hepatitis B (HBV)/hepatitis C (HCV)-positive]. Moreover, in developed countries (i.e., United States and Europe), HCV-related HCC can account for most liver cancers; therefore, increasing our knowledge of the genetic contribution to the clinical course of viral infections could be essential in optimizing the use of the newly developed, but highly expensive, direct acting antiviral treatments.
Accumulating data derived from candidate-gene investigations and genome-wide association studies have highlighted the possible role of genetic variants as significant predictors of HCC susceptibility and as determinants of the clinical course of HBV or HCV infections. However, the high heterogeneity among published works has prevented drawing definite conclusions. Furthermore, the majority of case-control investigations were performed in Asian populations; thus, replication studies with different ethnicities are warranted. Consequently, there is a critical need to intensify research efforts in identifying effective molecular biomarkers predictive of individual HCC risk in a homogeneous, well-defined cohort study, with particular attention to the (currently) poorly investigated Caucasian population.
This work uncovered five markers in genes that encode key proteins involved in pathways underlying hepatocarcinogenesis (ERCC1-rs3212986, GSTP1-rs1138272, CYP17A1-rs743572, XRCC3- rs1799794, and ABCB1-rs112850). These markers were predictive of individual HCC susceptibility in a high-risk Caucasian population (HBV/HCV-positive). Two variants also exhibited serology-specific effects when a subgroup analysis was performed in patients stratified according to HBV or HCV positive serology. That analysis showed that ABCB1 rs1128503 specifically affected the HBV-positive subgroup and ERCC1 rs3212986 specifically affected the HCV-positive subgroup. Among the five markers identified, ERCC1 rs3212986 and CYP17A1 rs743572 retained predictive value for HCC risk among healthy individuals. In addition, they found gene-gene interactions between ERCC1 rs3212986, CYP17A1 rs743572, GST-P1 rs1138272, and the previously described UGT1A7*3 marker, which were included in the definition of the complex trait of HCC susceptibility.
The definition of genetic traits that confer susceptibility to liver carcinogenesis is of potential clinical interest. This information can facilitate improvements in preventive, diagnostic, and therapeutic strategies for HCC management. If validated, these markers and their interactions should be considered for improving HCC risk stratification algorithms and surveillance programs. Their inclusion in screening could facilitate early cancer detection and might lead to potentially curative therapies. For patients with HCV infections, the biomarkers we discovered could also contribute to optimizing the use of recently developed, but highly expensive, anti-viral therapies. These findings address a great challenge in HCC management and might help enhance care for patients with HCC.
Polymorphism: a variation in the DNA sequence that occurs in a population at a frequency of 1% or higher; CART: classification and regression tree analysis; this is a statistical method, based on recursive partitioning, that is applied to elucidate potential high-order gene-gene and gene-environmental relationships among discriminating factors that contribute to a specific end-point; ERCC1: a key rate-limiting enzyme that acts in the nucleotide excision repair pathway. This pathway represents a DNA repair mechanism designed to maintain genomic integrity by removing inter-strand DNA crosslinks; XRCC3: a DNA repair protein that participates in DNA double-strand break/recombination repair machinery; GST-P1: a phase II enzyme involved in the inactivation of electrophilic xenobiotics; it acts by conjugating with glutathione, which facilitates excretion of xenobiotics from the body; CYP17A1: an enzyme that plays a critical role in the in the biosynthesis of steroid hormones; MDR-1 (encoded by ABCB1): A widely expressed membrane efflux pump, physiologically involved in the protection of normal tissue from environmental and endogenous toxins.
The aim of this study was to find novel genetic markers that could contribute in predicting HCC susceptibility in Caucasians. The authors identified five genetic markers in key pathways linked to hepatocarcinogenesis (ERCC1-rs3212986, GSTP1-rs1138272, CYP17A1-rs743572, XRCC3- rs1799794, ABCB1-rs112850) that could predict the individual HCC susceptibility, especially in high-risk (HBV/HCV-infected) Caucasians population. The article provides the novel informations that could contribute to improving HCC surveillance, allowing early cancer diagnosis and possible curative therapy.
| 1. | Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18694] [Cited by in RCA: 21466] [Article Influence: 1951.5] [Reference Citation Analysis (6)] |
| 2. | McGlynn KA, Petrick JL, London WT. Global epidemiology of hepatocellular carcinoma: an emphasis on demographic and regional variability. Clin Liver Dis. 2015;19:223-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 491] [Cited by in RCA: 678] [Article Influence: 61.6] [Reference Citation Analysis (0)] |
| 3. | Galun D, Basaric D, Zuvela M, Bulajic P, Bogdanovic A, Bidzic N, Milicevic M. Hepatocellular carcinoma: From clinical practice to evidence-based treatment protocols. World J Hepatol. 2015;7:2274-2291. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 4. | Jin F, Xiong WJ, Jing JC, Feng Z, Qu LS, Shen XZ. Evaluation of the association studies of single nucleotide polymorphisms and hepatocellular carcinoma: a systematic review. J Cancer Res Clin Oncol. 2011;137:1095-1104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 48] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 5. | Miki D, Ochi H, Hayes CN, Aikata H, Chayama K. Hepatocellular carcinoma: towards personalized medicine. Cancer Sci. 2012;103:846-850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 6. | Nahon P, Zucman-Rossi J. Single nucleotide polymorphisms and risk of hepatocellular carcinoma in cirrhosis. J Hepatol. 2012;57:663-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 136] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 7. | Matsuura K, Isogawa M, Tanaka Y. Host genetic variants influencing the clinical course of hepatitis B virus infection. J Med Virol. 2016;88:371-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 43] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 8. | Matsuura K, Tanaka Y. Host genetic variants influencing the clinical course of hepatitis C virus infection. J Med Virol. 2016;88:185-195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 25] [Article Influence: 2.5] [Reference Citation Analysis (1)] |
| 9. | Marquardt JU, Nguyen-Tat M, Galle PR, Wörns MA. Surveillance of Hepatocellular Carcinoma and Diagnostic Algorithms in Patients with Liver Cirrhosis. Visc Med. 2016;32:110-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 10. | Mazzanti R, Arena U, Tassi R. Hepatocellular carcinoma: Where are we? World J Exp Med. 2016;6:21-36. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 94] [Cited by in RCA: 94] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 11. | Wang CH, Wey KC, Mo LR, Chang KK, Lin RC, Kuo JJ. Current trends and recent advances in diagnosis, therapy, and prevention of hepatocellular carcinoma. Asian Pac J Cancer Prev. 2015;16:3595-3604. [PubMed] |
| 12. | Luhnen M, Waffenschmidt S, Gerber-Grote A, Hanke G. Health Economic Evaluations of Sofosbuvir for Treatment of Chronic Hepatitis C: a Systematic Review. Appl Health Econ Health Policy. 2016;14:527-543. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 13. | Roderburg C, Tacke F, Trautwein C. Antiviral Therapy in Patients with Viral Hepatitis and Hepatocellular Carcinoma: Indications and Prognosis. Visc Med. 2016;32:121-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 14. | De Mattia E, Cecchin E, Polesel J, Lupo F, Tiribelli C, Crovatto M, Buonadonna A, Toffoli G. UGT1A polymorphisms as genetic biomarkers for hepatocellular carcinoma risk in Caucasian population. Liver Int. 2017;37:1345-1353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 15. | Cecchin E, Russo A, Corona G, Campagnutta E, Martella L, Boiocchi M, Toffoli G. UGT1A1*28 polymorphism in ovarian cancer patients. Oncol Rep. 2004;12:457-462. [PubMed] |
| 16. | Cecchin E, Russo A, Campagnutta E, Martella L, Toffoli G. Lack of association of CYP1 B1*3 polymorphism and ovarian cancer in a Caucasian population. Int J Biol Markers. 2004;19:160-163. [PubMed] |
| 17. | Toffoli G, Rossi D, Gaidano G, Cecchin E, Boiocchi M, Carbone A. Methylenetetrahydrofolate reductase genotype in diffuse large B-cell lymphomas with and without hypermethylation of the DNA repair gene O6-methylguanine DNA methyltransferase. Int J Biol Markers. 2003;18:218-221. [PubMed] |
| 18. | Sukowati CH, El-Khobar KE, Ie SI, Anfuso B, Muljono DH, Tiribelli C. Significance of hepatitis virus infection in the oncogenic initiation of hepatocellular carcinoma. World J Gastroenterol. 2016;22:1497-1512. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 49] [Cited by in RCA: 51] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 19. | Li C, Wang LE, Wei Q. DNA repair phenotype and cancer susceptibility--a mini review. Int J Cancer. 2009;124:999-1007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 71] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 20. | Li Y, Ou C, Shu H, Zhao H, Zhu B. The ERCC1-4533/8092, TNF-α 238/308 polymorphisms and the risk of hepatocellular carcinoma in Guangxi Zhuang populations of China: Case-control study. Medicine (Baltimore). 2016;95:e5217. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 21. | Wang B, Xu Q, Yang HW, Sun LP, Yuan Y. The association of six polymorphisms of five genes involved in three steps of nucleotide excision repair pathways with hepatocellular cancer risk. Oncotarget. 2016;7:20357-20367. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 31] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 22. | Rulli E, Marabese M, Piva S, Bonomi L, Caiola E, Ganzinelli M. DNA repair gene polymorphisms in non-small-cell lung cancer patients treated with first-line platinum-containing chemotherapy. Tumori. 2016;102:367-375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 23. | Zhu J, Hua RX, Jiang J, Zhao LQ, Sun X, Luan J, Lang Y, Sun Y, Shang K, Peng S. Association studies of ERCC1 polymorphisms with lung cancer susceptibility: a systematic review and meta-analysis. PLoS One. 2014;9:e97616. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 24. | Ji RB, Qian YS, Hu AR, Hu YR. DNA repair gene XRCC3 T241M polymorphism and susceptibility to hepatocellular carcinoma in a Chinese population: a meta-analysis. Genet Mol Res. 2015;14:15988-15996. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 25. | Zeng X, Liu S, Yu H, Ji L, Li L, Huang J, Bai H, Qiu X. DNA repair capacity, DNA-strand break repair gene polymorphisms, and the incidence of hepatocellular carcinoma in southwestern Guangxi of China. DNA Cell Biol. 2012;31:1384-1391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 26. | Matullo G, Palli D, Peluso M, Guarrera S, Carturan S, Celentano E, Krogh V, Munnia A, Tumino R, Polidoro S. XRCC1, XRCC3, XPD gene polymorphisms, smoking and (32)P-DNA adducts in a sample of healthy subjects. Carcinogenesis. 2001;22:1437-1445. [PubMed] |
| 27. | Yao JG, Huang XY, Long XD. Interaction of DNA repair gene polymorphisms and aflatoxin B1 in the risk of hepatocellular carcinoma. Int J Clin Exp Pathol. 2014;7:6231-6244. [PubMed] |
| 28. | Fachal L, Gómez-Caamaño A, Peleteiro P, Carballo A, Calvo-Crespo P, Sánchez-García M, Lobato-Busto R, Carracedo A, Vega A. Association of a XRCC3 polymorphism and rectum mean dose with the risk of acute radio-induced gastrointestinal toxicity in prostate cancer patients. Radiother Oncol. 2012;105:321-328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 29. | Smolkova B, Dusinska M, Hemminki K. NBN and XRCC3 genetic variants in childhood acute lymphoblastic leukaemia. Cancer Epidemiol. 2014;38:563-568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 30. | Watson MA, Stewart RK, Smith GB, Massey TE, Bell DA. Human glutathione S-transferase P1 polymorphisms: relationship to lung tissue enzyme activity and population frequency distribution. Carcinogenesis. 1998;19:275-280. [PubMed] |
| 31. | Tian M, Zhao B, Zhang J, Martin FL, Huang Q, Liu L, Shen H. Association of environmental benzo[a]pyrene exposure and DNA methylation alterations in hepatocellular carcinoma: A Chinese case-control study. Sci Total Environ. 2016;541:1243-1252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 32. | Mansoori AA, Jain SK. Molecular Links between Alcohol and Tobacco Induced DNA Damage, Gene Polymorphisms and Patho-physiological Consequences: A Systematic Review of Hepatic Carcinogenesis. Asian Pac J Cancer Prev. 2015;16:4803-4812. [PubMed] |
| 33. | Kiran M, Chawla YK, Kaur J. Methylation profiling of tumor suppressor genes and oncogenes in hepatitis virus-related hepatocellular carcinoma in northern India. Cancer Genet Cytogenet. 2009;195:112-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 34. | Chen J, Ma L, Peng NF, Wang SJ, Li LQ. A meta-analysis of the relationship between glutathione S-transferases gene polymorphism and hepatocellular carcinoma in Asian population. Mol Biol Rep. 2012;39:10383-10393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 35. | White DL, Li D, Nurgalieva Z, El-Serag HB. Genetic variants of glutathione S-transferase as possible risk factors for hepatocellular carcinoma: a HuGE systematic review and meta-analysis. Am J Epidemiol. 2008;167:377-389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 97] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 36. | Ali-Osman F, Akande O, Antoun G, Mao JX, Buolamwini J. Molecular cloning, characterization, and expression in Escherichia coli of full-length cDNAs of three human glutathione S-transferase Pi gene variants. Evidence for differential catalytic activity of the encoded proteins. J Biol Chem. 1997;272:10004-10012. [PubMed] |
| 37. | Ji X, Blaszczyk J, Xiao B, O’Donnell R, Hu X, Herzog C, Singh SV, Zimniak P. Structure and function of residue 104 and water molecules in the xenobiotic substrate-binding site in human glutathione S-transferase P1-1. Biochemistry. 1999;38:10231-10238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 32] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 38. | Goldstone JV, Sundaramoorthy M, Zhao B, Waterman MR, Stegeman JJ, Lamb DC. Genetic and structural analyses of cytochrome P450 hydroxylases in sex hormone biosynthesis: Sequential origin and subsequent coevolution. Mol Phylogenet Evol. 2016;94:676-687. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 39] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 39. | Baldissera VD, Alves AF, Almeida S, Porawski M, Giovenardi M. Hepatocellular carcinoma and estrogen receptors: Polymorphisms and isoforms relations and implications. Med Hypotheses. 2016;86:67-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 40. | Carey AH, Waterworth D, Patel K, White D, Little J, Novelli P, Franks S, Williamson R. Polycystic ovaries and premature male pattern baldness are associated with one allele of the steroid metabolism gene CYP17. Hum Mol Genet. 1994;3:1873-1876. [PubMed] |
| 41. | Xu J, Lin X, Zhu H, Zhang Z, Yang B. Genetic variation of the CYP17 and susceptibility to endometrial cancer: a meta-analysis. Mol Biol Rep. 2013;40:5085-5091. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 42. | Rossi L, Leveri M, Gritti C, De Silvestri A, Zavaglia C, Sonzogni L, Silvestri L, Civardi E, Mondelli MU, Silini EM. Genetic polymorphisms of steroid hormone metabolizing enzymes and risk of liver cancer in hepatitis C-infected patients. J Hepatol. 2003;39:564-570. [PubMed] |
| 43. | Yuan X, Zhou G, Zhai Y, Xie W, Cui Y, Cao J, Zhi L, Zhang H, Yang H, Zhang X. Lack of association between the functional polymorphisms in the estrogen-metabolizing genes and risk for hepatocellular carcinoma. Cancer Epidemiol Biomarkers Prev. 2008;17:3621-3627. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 44. | Hartwell HJ, Petrosky KY, Fox JG, Horseman ND, Rogers AB. Prolactin prevents hepatocellular carcinoma by restricting innate immune activation of c-Myc in mice. Proc Natl Acad Sci USA. 2014;111:11455-11460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 78] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 45. | Montella M, D’Arena G, Crispo A, Capunzo M, Nocerino F, Grimaldi M, Barbieri A, D’Ursi AM, Tecce MF, Amore A. Role of Sex Hormones in the Development and Progression of Hepatitis B Virus-Associated Hepatocellular Carcinoma. Int J Endocrinol. 2015;2015:854530. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 47] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 46. | Choudhuri S, Klaassen CD. Structure, function, expression, genomic organization, and single nucleotide polymorphisms of human ABCB1 (MDR1), ABCC (MRP), and ABCG2 (BCRP) efflux transporters. Int J Toxicol. 2006;25:231-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 271] [Cited by in RCA: 279] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 47. | Zhou SF. Structure, function and regulation of P-glycoprotein and its clinical relevance in drug disposition. Xenobiotica. 2008;38:802-832. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 387] [Cited by in RCA: 393] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 48. | Leslie EM, Deeley RG, Cole SP. Multidrug resistance proteins: role of P-glycoprotein, MRP1, MRP2, and BCRP (ABCG2) in tissue defense. Toxicol Appl Pharmacol. 2005;204:216-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 996] [Cited by in RCA: 1054] [Article Influence: 50.2] [Reference Citation Analysis (0)] |
| 49. | Aleksandrova K, Boeing H, Nöthlings U, Jenab M, Fedirko V, Kaaks R, Lukanova A, Trichopoulou A, Trichopoulos D, Boffetta P. Inflammatory and metabolic biomarkers and risk of liver and biliary tract cancer. Hepatology. 2014;60:858-871. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 150] [Cited by in RCA: 172] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 50. | Sun B, Karin M. Obesity, inflammation, and liver cancer. J Hepatol. 2012;56:704-713. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 348] [Cited by in RCA: 405] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 51. | Baldissera VD, de Mattos AA, Coral GP, de Araujo FB, Marroni CA, de Mello Brandão AB, Ott Fontes PR, Schmidt Cerski CT, Hartmann AA, Kretzmann Filho NA. Evaluation of the C3435T polymorphism in the MDR1 gene in patients with hepatocellular carcinoma. Ann Hepatol. 2012;11:899-906. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 369] [Cited by in RCA: 395] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 52. | Frittitta L, Ercolino T, Bozzali M, Argiolas A, Graci S, Santagati MG, Spampinato D, Di Paola R, Cisternino C, Tassi V. A cluster of three single nucleotide polymorphisms in the 3’-untranslated region of human glycoprotein PC-1 gene stabilizes PC-1 mRNA and is associated with increased PC-1 protein content and insulin resistance-related abnormalities. Diabetes. 2001;50:1952-1955. [PubMed] |
| 53. | Fung KL, Gottesman MM. A synonymous polymorphism in a common MDR1 (ABCB1) haplotype shapes protein function. Biochim Biophys Acta. 2009;1794:860-871. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 256] [Cited by in RCA: 262] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 54. | Haufroid V. Genetic polymorphisms of ATP-binding cassette transporters ABCB1 and ABCC2 and their impact on drug disposition. Curr Drug Targets. 2011;12:631-646. [PubMed] |
| 55. | Wang ZC, Liu LZ, Liu XY, Hu JJ, Wu YN, Shi JY, Yang LX, Duan M, Wang XY, Zhou J. Genetic polymorphisms of the multidrug resistance 1 gene MDR1 and the risk of hepatocellular carcinoma. Tumour Biol. 2015;36:7007-7015. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Italy
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
P- Reviewer: Kasprzak A S- Editor: Qi Y L- Editor: A E- Editor: Li D