Published online Aug 7, 2017. doi: 10.3748/wjg.v23.i29.5364
Peer-review started: February 1, 2017
First decision: March 3, 2014
Revised: March 14, 2014
Accepted: July 4, 2017
Article in press: July 4, 2017
Published online: August 7, 2017
Processing time: 194 Days and 18.4 Hours
To investigate whether single nucleotide polymorphisms in maf protein K (MAFK), which encodes the MAFK, lead to increased susceptibility to ulcerative colitis in the Japanese population.
This case control study examined the associations between MAFK single nucleotide polymorphisms (rs4268033 G>A, rs3735656 T>C and rs10226620 C>T) and ulcerative colitis susceptibility in 174 patients with ulcerative colitis (UC) cases, and 748 subjects without no lower abdominal symptoms, diarrhea or hematochezia (controls). In addition, as the second controls, we set 360 subjects, who have an irregular bowel movement without abnormal lower endoscopic findings (IBM controls).
The genotype frequency of rs4268033 AA and allelic frequency of the rs4268033A allele were significantly higher in the UC cases than in both controls (P = 0.0005 and < 0.0001, P = 0.015 and 0.0027 vs controls and IBM controls, respectively). Logistic regression analysis after adjustment for age and gender showed that the rs4268033 AA and rs3735656 CC genotypes were significantly associated with susceptibility to UC development (OR = 2.63, 95%CI: 1.61-4.30, P = 0.0001 and OR = 1.81; 95%CI: 1.12-2.94, P = 0.015, respectively). Similar findings were observed by the comparison with IBM controls. In addition, the rs4268033 AA genotype was significantly associated with all phenotypes of UC except early onset. There was no significant association between rs10226620 and ulcerative colitis.
Our results provide the first evidence that MAFK genetic polymorphisms are significantly associated with susceptibility to UC development. In particular, rs4268033 is closely associated with an increased risk for the development of UC.
Core tip: We investigated the association between maf protein K (MAFK), polymorphisms and ulcerative colitis in Japan. Both rs4268033 and rs3735656 minor allele homozygotes were significantly associated with the susceptibility to ulcerative colitis (UC) development. In addition, rs4268033 minor allele homozygote was significantly associated with all phenotypes of UC except the phenotype with younger age onset. Our results provided the first evidence that MAFK genetic polymorphisms were significantly associated with the susceptibility to UC development.
- Citation: Arisawa T, Nakamura M, Otsuka T, Jing W, Sakurai N, Takano H, Hayashi T, Ota M, Nomura T, Hayashi R, Shimasaki T, Tahara T, Shibata T. Genetic polymorphisms of MAFK, encoding a small Maf protein, are associated with susceptibility to ulcerative colitis in Japan. World J Gastroenterol 2017; 23(29): 5364-5370
- URL: https://www.wjgnet.com/1007-9327/full/v23/i29/5364.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i29.5364
The number of inflammatory bowel disease (IBD) patients in several Asian countries, including Japan and China, has recently rapidly increased with adaptation of a westernization life style and diet[1]. Ulcerative colitis (UC) is a representative IBD whose exact etiology is unclear, but environmental and genetic factors are implicated in its onset[2,3]. UC is a nonspecific inflammatory disease possibly involving the colonic mucosa spanning from the rectum to the cecum. It has been clarified that the degradation of inflammation and immune response related molecules prescribed genetically participate[4]. UC is a multifactorial, polygenic disease with probable genetic heterogeneity, and the associations between the polymorphisms of various genes and UC have been studied[5,6].
Reactive oxygen species (ROS) are involved in promoting inflammation in various diseases, including UC[7,8]. Nuclear factor-erythroid 2-related factor 2 (Nrf2) plays an important role in the removal of ROS[9,10]. Nrf2 cannot induce anti-oxidant enzymes such as heme oxygenase-1 (HO-1) and peroxiredoxin 1 alone, and shows transcriptional activity when hetero-dimerized with small musculoaponeurotic fibrosarcoma (Maf)[11]. Thus, it has been clarified that small Maf proteins are regulated transcription factors through heterodimer formation, with Nrf2 and BTB And CNC Homology 1 (Bach1)[12], although small Mafs do not contain an obvious transcriptional activation domain[13]. We previously reported a significant association between Nrf2 genetic polymorphisms and susceptibility to UC[14].
In the present study, we investigated the association between genetic polymorphisms of MAFK, one of the small Mafs, and UC susceptibility in a Japanese population.
The study was performed using a population comprising 226 patients with UC (UC cases) and 748 subjects without lower abdominal symptoms, diarrhea or hematochezia (controls). In addition, as the second controls, we prepared 360 subjects, who have an irregular bowel movement without abnormal lower endoscopic findings (IBM controls). UC was diagnosed according to standard clinical, endoscopic, radiological, and histological criteria[15]. Genomic DNA was isolated from peripheral blood using FlexiGene DNA Kit (QIAGEN GmbH, Hilden, Germany).
The Ethics Committees of Fujita Health University and Kanazawa Medical University approved the protocol, and written informed consent was obtained from all participating subjects.
According to their clinical courses, UC cases were classified into 2 types: continuous, or not continuous, disease (i.e., relapsing or only one episode)[16]. UC patients were also classified by endoscopic findings as total or not total colitis (left sided, distal colitis) according to the location and extension of the inflammatory lesions. In addition, the cases were classified into two groups according to the past highest UC disease activity index (UCDAI) during the course of the disease (≤ 8 or ≤ 9)[17].
There are two large linkage disequilibrium blocks within 20kbp of MAFK with a Hardy-Weinberg equilibrium (HWE) P value of above 0.05 and a minor allele frequency of above 0.05. We selected rs4268033 G>A and rs3735656 T>C (*910 T>C) as a Tag single nucleotide polymorphism (SNP) in each block and another SNP, rs10226620 C>T (*1506 C>T), located in the 3’-UTR where several microRNA bind, was also selected.
Polymorphisms were genotyped using the PCR-SSCP method as reported previously[14,18]. The PCR and SSCP conditions are shown in Table 1. All PCR reactions were carried out in a volume of 20 μL containing 0.1 μg of genomic DNA using Takara HS Taq (TAKARA Bio Inc., Japan). SSCP was carried out using a GenePhor DNA separation system with GeneGel Excel 12.5/24 (GE Health Care Bio-Sciences AB, Sweden) at 6 °Ctemperature, and then the denatured single strand DNA bands were detected using a DNA Silver Staining Kit (GE Health Care Bio-Sciences AB).
| Primer set | PCR condition | SSCP temperature |
| (rs4268033) | ||
| 5'-TAATCCCAACTCGCAGCATCTGTGT-3' | 96 °C 15 s, 60 °C 30 s, 72 °C 30 s | 6 °C |
| 5'-GGTCTGACTTAGCTGGGGAAAGTGC-3' | 35 cycle | |
| (rs3735656) | ||
| 5'-ATCTCAGCGGACACAGGCAGGA-3' | 96 °C 15 s, 54 °C 40 s, 72 °C 30 s | 6 °C |
| 5'-CTGCACTGACCACAGTTGGTGAGAA-3' | 35 cycle | |
| (rs10226620) | ||
| 5'-GTCCCTCCTGTGACTGGGGTCTCT-3' | 96 °C 15 s, 60 °C 30 s, 72 °C 30 s | 6 °C |
| 5'-AGGCACCACCTTGCAGGTCTTATGT-3' | 35 cycle |
HWE was assessed by χ2 statistics. The age data were expressed as mean ± SD. Mean ages between the cases and the controls was compared by Student’s t-test, and the male/female ratio was compared by Fisher’s exact test. The allele counts and the distribution of genotype were compared between the two groups by a 2 × 2 table using Fisher’s exact test. The odds ratios (OR) and 95%CI were calculated by logistic regression with adjustment for age and gender. A probability value of less than 0.05 was considered statistically significant in all analyses.
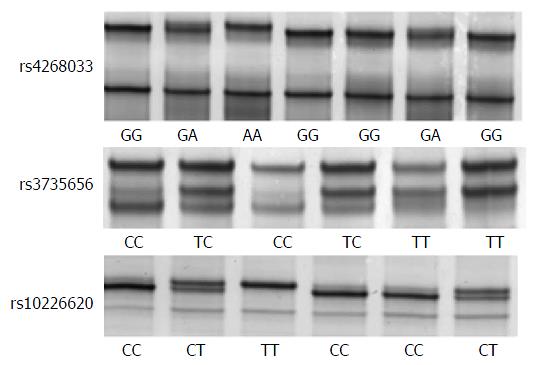
The characteristics of the subjects in this study are summarized in Table 2. The mean ages of both controls and IBM controls were significantly higher than that of the UC cases. The male/female ratio of IBM controls was significantly lower than those of the controls and UC cases. Single strand DNAs of all genotypes were clearly separated by SSCP (Figure 1). The distribution of genotypes in the controls was in Hardy-Weinberg equilibrium (rs4268033, rs3735656 and rs10226620: P = 0.25, P = 0.21 and P = 0.87, respectively). The ratio of the mutant homozygote (AA genotype) of rs4268033 was significantly higher in the UC cases compared to the controls and IBM controls (P = 0.0005 and < 0.0001, respectively). However, no significant differences in the distributions of rs3735656 and rs10226620 genotypes were observed. There were no significant differences in the distributions of genotypes between the controls and IBM controls.
| Controls | IBM controls | UC cases | |
| Number of sample | 748 | 360 | 226 |
| Mean age ± SD | 57.1 ± 17.0 | 58.5 ± 14.5 | 40.7 ± 14.2a |
| (age of onset) | (33.6 ± 13.5) | ||
| Male:female | 438:310 | 165:195b | 125:101 |
| rs4268033 G>A | |||
| GG | 366 | 184 | 101 |
| GA | 324 | 156 | 89 |
| AA | 58 | 20 | 36c |
| A allele freqency | 29.40% | 27.20% | 35.6%d |
| rs3735656 T>C | |||
| TT | 329 | 168 | 103 |
| TC | 346 | 162 | 90 |
| CC | 73 | 30 | 33 |
| C allele freqency | 32.90% | 30.80% | 34.50% |
| rs10226620 C>T | |||
| CC | 345 | 171 | 104 |
| CT | 328 | 159 | 94 |
| TT | 75 | 30 | 28 |
| T allele freqency | 32.00% | 30.40% | 33.20% |
When the controls was compared with UC cases, logistic regression analysis after adjustment for age and gender showed that the rs4268033 mutant homozygote (AA genotype) was strongly associated with susceptibility to UC (OR = 2.63, 95%CI: 1.61-4.30, P = 0.0001; Table 3), and the rs3735656 mutant homozygote (CC genotype) was also significantly associated with UC susceptibility (OR = 1.81, 95%CI: 1.12-2.94, P = 0.015). When IBM controls was compared with UC cases, similar findings were obtained (OR = 3.54, 95%CI: 1.82-6.88, P = 0.0002 and OR = 2.14; 95%CI: 1.16-3.95, P = 0.015, for rs4268033 and rs3735656, respectively). No significant association between rs10226620 and UC was seen. The influence of MAFK genotypes on the symptoms from irregular bowel movement did not seem to be large.
| Genotype (n) | AA vs others | Genotype (n) | CC vs others | Genotype (n) | TT vs others | ||||||||||
| GG | GA | AA | OR (95%CI); P value | TT | TC | CC | OR (95%CI); P value | CC | CT | TT | OR (95%CI); P value | ||||
| rs4268033 G>A | |||||||||||||||
| Controls (748) | 366 | 324 | 58 | Reference | - | ||||||||||
| IBM controls (358) | 184 | 156 | 18 | 0.682 (0.402-1.16); P = 0.16 | Reference | ||||||||||
| UC cases (226) | 101 | 89 | 36 | 2.63 (1.61-4.30); P = 0.0001 | 3.54 (1.82-6.88); P = 0.0002 | ||||||||||
| rs3735656 T>C | |||||||||||||||
| Controls (748) | 329 | 346 | 73 | Reference | - | ||||||||||
| IBM controls (358) | 168 | 162 | 28 | 0.789 (0.503-1.24); P = 0.30 | Reference | ||||||||||
| UC cases (174) | 103 | 90 | 33 | 1.81 (1.12-2.94); P = 0.015 | 2.14 (1.16-3.95); P = 0.015 | ||||||||||
| rs10226620 C>T | |||||||||||||||
| Controls (748) | 345 | 328 | 75 | Reference | - | ||||||||||
| IBM controls (358) | 171 | 159 | 28 | 0.764 (0.488-1.20); P = 0.24 | Reference | ||||||||||
| UC cases (174) | 104 | 94 | 28 | 1.37 (0.822 - 2.28); P = 0.23 | 1.57 (0.827 - 3.00); P = 0.17 | ||||||||||
The observed strong association between rs4268033 and UC susceptibility prompted us to investigate an association between this SNP and UC phenotypes. The rs4268033 genotype was only associated with UC cases with onset after 21 years of age. Regarding as clinical type, disease extension, the past history of hospitalization and the past maximum UCDAI score, rs4268033 was significantly associated with each phenotype of UC (Table 4).
| Genotype (n) | AA vs others | ||||
| GG | GA | AA | OR (95%CI) | P value | |
| Controls (748) | 366 | 324 | 58 | Reference | - |
| Age of onset | |||||
| 20 (31) | 16 | 12 | 3 | 2.06 (0.499-8.52) | 0.32 |
| 21 (176) | 76 | 71 | 29 | 2.51 (1.51-4.19) | 0.0004 |
| Unknown (19) | |||||
| Clinical type | |||||
| Not continuous (124) | 51 | 52 | 21 | 2.92 (1.63-5.25) | 0.0003 |
| Continuous (97) | 46 | 36 | 15 | 2.32 (1.20-4.48) | 0.012 |
| Unknown (5) | |||||
| Extension | |||||
| Not total colitis (115) | 50 | 46 | 19 | 2.72 (1.48-5.00) | 0.0013 |
| Total colitis (105) | 49 | 41 | 15 | 2.18 (1.14-4.16) | 0.019 |
| Unknown (6) | |||||
| Hospitalization | |||||
| None (139) | 59 | 59 | 21 | 2.34 (1.32-4.14) | 0.0035 |
| One time £ (76) | 36 | 27 | 13 | 2.91 (1.42-5.97) | 0.0034 |
| Unknown (11) | |||||
| Past max. UCDAI score | |||||
| 8 (135) | 56 | 58 | 21 | 2.41 (1.36-4.27) | 0.0025 |
| 9 (81) | 40 | 27 | 14 | 3.01 (1.49-6.05) | 0.002 |
| Unknown (10) | |||||
Three small Maf proteins have been identified to date: MafG, MafK and MafF were identified[19]. These three Maf proteins have high homology and form homodimers or heterodimers with each another and also heterodimers with a group of other b-Zip proteins[20]. Previous studies using knockout mice showed that MafG-/-/K-/- mice are die by the peri- or postnatal stage, whereas both MafF-/-/G-/- and MafF-/-/K-/- mice are viable and fertile[21-23]. We therefore selected the two small Mafs, MafG and MafK, because of their apparent importance in biology. In HapMap-JPT, there are large linkage blocks of SNPs around MAFK but not around MAFG. Furthermore, previous studies revealed that MafK-Bach1 controls the expression of a subset of oxidative stress-inducible genes, such as HO-1 and ferritins[24], whereas MafK-Nrf2 heterodimer activates their expression[25]. We therefore suspected that MAFK genetic variations might affect the development and process of inflammatory diseases, including UC.
Our results provide the first evidence that MAFK genetic polymorphisms are significantly associated with susceptibility to UC in the Japanese population. In these polymorphisms, the rs4268033 G>A minor allele homozygote is closely associated with an increased risk for the development of UC. The allele frequencies of rs4268033 and rs10226620 in our controls, which were in HWE, were the same as that in the Japanese population reported in the HapMap database (P = 0.73 and 0.39, respectively). However, the distribution of rs3735656 genotype in our controls was different from that in HapMap-JPT (P = 0.011). The distribution of rs3735656 in our controls is in HWE (P = 0.21) whereas it is not in HapMap-JPT (P = 0.025). The cause of this discrepancy is unknown, but we believe that rs3735656 is worthy of further examination.
There are few reports linking MAFK genetic variations and clinical disease susceptibility[26,27]. Nanashima et al[26] reported that the mutant allele of rs4720833, located in the same linkage block as rs4268033, is significantly associated with anti-tuberculosis drug induced hepatotoxicity susceptibility, whereas rs3808337, located in the same block as rs3735656, is not associated. This suggests that the rs4720833 mutant genotype might be associated with an increased risk of drug-induced injury via alteration of the toxicity of drug metabolites, although the detailed mechanisms remain unclear. Similarly, in our study, the rs4268033 mutant homozygote was strongly associated with UC susceptibility. In addition, this genotype was associated with all phenotypes of UC except age of onset. These findings suggest that rs4268033 may be associated with the development but not the progression of UC. The lack of association of early onset with UC may be due to the small number of cases and/or the younger controls.
It is not clear how rs4268033 participates in the development of UC. Small Maf proteins are transcription factors localized to the nucleus that dimerize with CNC family proteins, including Nrf2 and Bach1[13]. Small Maf-Bach1 heterodimers are removed from antioxidant-responsive element (ARE) by oxidative stress, and small Maf-Nrf2 heterodimers replace and bind to ARE, leading to the activation of antioxidant enzyme expression. Therefore, our data suggest that rs4268033 may act as a repressor for the expression of small Maf (dimerized with Nrf2) and/or as an activator for the expression of small Maf (dimerized with Bach1), resulting in association with UC susceptibility. Another possibility is that the mechanism involves the inflammatory response. Overexpression of MafK protein in T cells decreased T-cell proliferation and interleukin-2 (IL-2) secretion[28]. Therefore, IL-2 secretion, resulting in persistent inflammation, may be increased by the diminished expression of MafK protein in the rs4260833 mutant homozygote. However, the presence of enhancers or repressors in the genome region containing the linkage block with rs4268033 remains unknown. In addition, the downregulation of Nrf2 or upregulation of Bach1 in UC patients with the rs4268033 mutant homozygote should be verified by further studies.
There are some clinical limitations in our study. We recruited patients who visited our hospital for various reasons and therefore the mean age of the controls was relatively high. Age-matched subjects with no symptom are essential for the control group. In addition, the UC treatment regimens were not standardized, possibly affecting the clinical type and extent of inflammation observed. Furthermore, we had no data on the effects of MAFK gene polymorphisms on the expressions of MafK protein. Our study was a case-control studyand therefore further examination is necessary regarding this point. The major problem in this study is the relatively small sample size, especially the number of UC cases. A larger cohort will be required to clearly assess the association of genetic variation with disease susceptibility. Finally, the design of this study used only samples stored at a single center and were analyzed retrospectively.
In conclusion, the present study demonstrated that MAFK polymorphisms are significantly associated with susceptibility to UC. In particular, the rs4268033 minor homozygote is strongly associated with increased risk for the development of UC.
Both environmental and genetic factors participate in the onset of ulcerative colitis (UC). Therefore, genetic alteration of inflammation related molecules may affect the susceptibility to UC.
Reactive oxygen species (ROS) are involved in promoting inflammation in various diseases, including UC. Small Maf proteins are regulated transcription factors thorough heterodimer formation, such as nuclear factor-erythroid 2-related factor 2 which play an important role in a removal process of these ROS. Therefore, it is of interest whether genetic alteration of Maf protein K (MAFK), encoding small MAFK, may affect the susceptibility to UC.
The results provide the first evidence that MAFK genetic polymorphisms are significantly associated with the susceptibility to UC in the Japanese population.
MAFK single nucleotide polymorphisms should be added to the spectrum of genetic factors involved in UC in Japanese population.
This case control study study investigated the association between genetic polymorphisms of MafK and UC susceptibility. The authors found that rs4268033 AA and rs3735656 CC genotypes were significantly associated with the susceptibility to UC.
| 1. | Sakamoto N, Kono S, Wakai K, Fukuda Y, Satomi M, Shimoyama T, Inaba Y, Miyake Y, Sasaki S, Okamoto K. Dietary risk factors for inflammatory bowel disease: a multicenter case-control study in Japan. Inflamm Bowel Dis. 2005;11:154-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 277] [Cited by in RCA: 284] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 2. | Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347:417-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2693] [Cited by in RCA: 2770] [Article Influence: 115.4] [Reference Citation Analysis (3)] |
| 3. | Newman B, Siminovitch KA. Recent advances in the genetics of inflammatory bowel disease. Curr Opin Gastroenterol. 2005;21:401-407. [PubMed] |
| 4. | Head KA, Jurenka JS. Inflammatory bowel disease Part 1: ulcerative colitis--pathophysiology and conventional and alternative treatment options. Altern Med Rev. 2003;8:247-283. [PubMed] |
| 5. | Uniken Venema WT, Voskuil MD, Dijkstra G, Weersma RK, Festen EA. The genetic background of inflammatory bowel disease: from correlation to causality. J Pathol. 2017;241:146-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 91] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 6. | Ye BD, McGovern DP. Genetic variation in IBD: progress, clues to pathogenesis and possible clinical utility. Expert Rev Clin Immunol. 2016;12:1091-1107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 70] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 7. | Pérez S, Taléns-Visconti R, Rius-Pérez S, Finamor I, Sastre J. Redox signaling in the gastrointestinal tract. Free Radic Biol Med. 2017;104:75-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 212] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 8. | Pravda J. Radical induction theory of ulcerative colitis. World J Gastroenterol. 2005;11:2371-2384. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 142] [Cited by in RCA: 169] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 9. | Ishii T, Itoh K, Takahashi S, Sato H, Yanagawa T, Katoh Y, Bannai S, Yamamoto M. Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J Biol Chem. 2000;275:16023-16029. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1144] [Cited by in RCA: 1237] [Article Influence: 47.6] [Reference Citation Analysis (0)] |
| 10. | Loboda A, Damulewicz M, Pyza E, Jozkowicz A, Dulak J. Role of Nrf2/HO-1 system in development, oxidative stress response and diseases: an evolutionarily conserved mechanism. Cell Mol Life Sci. 2016;73:3221-3247. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1641] [Cited by in RCA: 2050] [Article Influence: 205.0] [Reference Citation Analysis (0)] |
| 11. | Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2966] [Cited by in RCA: 3286] [Article Influence: 113.3] [Reference Citation Analysis (0)] |
| 12. | Raval CM, Zhong JL, Mitchell SA, Tyrrell RM. The role of Bach1 in ultraviolet A-mediated human heme oxygenase 1 regulation in human skin fibroblasts. Free Radic Biol Med. 2012;52:227-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 13. | Fujiwara KT, Kataoka K, Nishizawa M. Two new members of the maf oncogene family, mafK and mafF, encode nuclear b-Zip proteins lacking putative trans-activator domain. Oncogene. 1993;8:2371-2380. [PubMed] |
| 14. | Arisawa T, Tahara T, Shibata T, Nagasaka M, Nakamura M, Kamiya Y, Fujita H, Yoshioka D, Okubo M, Sakata M. Nrf2 gene promoter polymorphism is associated with ulcerative colitis in a Japanese population. Hepatogastroenterology. 2008;55:394-397. [PubMed] |
| 15. | Podolsky DK. Inflammatory bowel disease (1). N Engl J Med. 1991;325:928-937. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 762] [Cited by in RCA: 780] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 16. | Langholz E, Munkholm P, Davidsen M, Binder V. Course of ulcerative colitis: analysis of changes in disease activity over years. Gastroenterology. 1994;107:3-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 483] [Cited by in RCA: 425] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 17. | Rizzello F, Gionchetti P, Venturi A, Amadini C, Romagnoli R, Campieri M. Review article: monitoring activity in ulcerative colitis. Aliment Pharmacol Ther. 2002;16 Suppl 4:3-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 18. | Arisawa T, Tahara T, Ozaki K, Matsue Y, Minato T, Yamada H, Nomura T, Hayashi R, Matsunaga K, Fukumura A. Association between common genetic variant of HRH2 and gastric cancer risk. Int J Oncol. 2012;41:497-503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 19. | Kataoka K, Igarashi K, Itoh K, Fujiwara KT, Noda M, Yamamoto M, Nishizawa M. Small Maf proteins heterodimerize with Fos and may act as competitive repressors of the NF-E2 transcription factor. Mol Cell Biol. 1995;15:2180-2190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 187] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 20. | Igarashi K, Kataoka K, Itoh K, Hayashi N, Nishizawa M, Yamamoto M. Regulation of transcription by dimerization of erythroid factor NF-E2 p45 with small Maf proteins. Nature. 1994;367:568-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 21. | Onodera K, Shavit JA, Motohashi H, Yamamoto M, Engel JD. Perinatal synthetic lethality and hematopoietic defects in compound mafG::mafK mutant mice. EMBO J. 2000;19:1335-1345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 69] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 22. | Motohashi H, Katsuoka F, Engel JD, Yamamoto M. Small Maf proteins serve as transcriptional cofactors for keratinocyte differentiation in the Keap1-Nrf2 regulatory pathway. Proc Natl Acad Sci USA. 2004;101:6379-6384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 290] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 23. | Katsuoka F, Motohashi H, Ishii T, Aburatani H, Engel JD, Yamamoto M. Genetic evidence that small maf proteins are essential for the activation of antioxidant response element-dependent genes. Mol Cell Biol. 2005;25:8044-8051. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 233] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 24. | Sun J, Hoshino H, Takaku K, Nakajima O, Muto A, Suzuki H, Tashiro S, Takahashi S, Shibahara S, Alam J. Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. EMBO J. 2002;21:5216-5224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 478] [Cited by in RCA: 542] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 25. | Zhang J, Ohta T, Maruyama A, Hosoya T, Nishikawa K, Maher JM, Shibahara S, Itoh K, Yamamoto M. BRG1 interacts with Nrf2 to selectively mediate HO-1 induction in response to oxidative stress. Mol Cell Biol. 2006;26:7942-7952. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 188] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 26. | Nanashima K, Mawatari T, Tahara N, Higuchi N, Nakaura A, Inamine T, Kondo S, Yanagihara K, Fukushima K, Suyama N. Genetic variants in antioxidant pathway: risk factors for hepatotoxicity in tuberculosis patients. Tuberculosis (Edinb). 2012;92:253-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 50] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 27. | Martínez-Hernández A, Gutierrez-Malacatt H, Carrillo-Sánchez K, Saldaña-Alvarez Y, Rojas-Ochoa A, Crespo-Solis E, Aguayo-González A, Rosas-López A, Ayala-Sanchez JM, Aquino-Ortega X. Small MAF genes variants and chronic myeloid leukemia. Eur J Haematol. 2014;92:35-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 28. | Yoh K, Sugawara T, Motohashi H, Takahama Y, Koyama A, Yamamoto M, Takahashi S. Transgenic over-expression of MafK suppresses T cell proliferation and function in vivo. Genes Cells. 2001;6:1055-1066. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Japan
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
P- Reviewer: Lakatos PL, Rong L, Swierczynski JT S- Editor: Qi Y L- Editor: A E- Editor: Huang Y