Published online Mar 21, 2017. doi: 10.3748/wjg.v23.i11.1980
Peer-review started: October 27, 2016
First decision: November 9, 2016
Revised: December 16, 2016
Accepted: February 8, 2017
Article in press: February 8, 2017
Published online: March 21, 2017
Processing time: 144 Days and 7.7 Hours
To evaluate effect of treatment failure on cagA and vacA genotypes in Helicobacter pylori (H. pylori) isolates from Colombia.
One hundred and seventy-six participants infected with H. pylori from Colombia were treated during 14 d with the triple-standard therapy. Six weeks later, eradication was evaluated by 13C-Urea breath test. Patients with treatment failure were subjected to endoscopy control; biopsies obtained were used for histopathology and culture. DNA from H. pylori isolates was amplified using primers specific for cagA and vacA genes. The phylogenetic relationships among isolates obtained before and after treatment were established by conglomerate analysis based on random amplified polymorphic DNA (RAPD) fingerprinting.
Treatment effectiveness was at 74.6%. Of the participants with treatment failure, 25 accepted subjected to a second endoscopy. Prevalence of post-treatment infection was 64% (16/25) and 40% (10/25) by histology and culture, respectively. Upon comparing the cagA and vacA genotypes found before and after therapy, multiple cagA genotypes (cagA-positive and cagA-negative) were found before treatment; in contrast, cagA-negative genotypes decreased after treatment. vacA s1m1 genotype was highly prevalent in patients before and after therapy. The 3’cagA region was successfully amplified in 95.5% (21/22) of the isolates obtained before and in 81.8% (18/22) of the isolates obtained after treatment. In the isolates obtained from patients with treatment failure, it was found that 72.7% (16/22) presented alterations in the number of EPIYA motifs, compared to isolates found before treatment.
Unsuccessful treatment limits colonization by low-virulence strains resulting in partial and selective eradication in mixed infections, and acts on the cagA-positive strains inducing genetic rearrangements in cagA variable region that produces a loss or gain of EPIYA repetitions.
Core tip: This study evaluated the effect of treatment failure on cagA and vacA genotypes in Helicobacter pylori (H. pylori) isolates. It was found, that unsuccessful treatment of H. pylori limits the colonization by low-virulence strains, resulting in partial and selective eradication in mixed infections. Also, acts on the cagA-positive strains inducing genetic rearrangements (deletion or acquisition of EPIYA motifs) that could alter the adherence of CagA protein to the epithelial cell membrane, the level of tyrosine phosphorylation and CagA multimerization, impacting its effects on cellular signaling. Finally, in some cases, may lead to the divergence of H. pylori cagA-positive sub-clones.
- Citation: Bustamante-Rengifo JA, Matta AJ, Pazos AJ, Bravo LE. Effect of treatment failure on the CagA EPIYA motif in Helicobacter pylori strains from Colombian subjects. World J Gastroenterol 2017; 23(11): 1980-1989
- URL: https://www.wjgnet.com/1007-9327/full/v23/i11/1980.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i11.1980
Helicobacter pylori (H. pylori) colonize the gastric mucosa of over 50% of the population in the world[1-3]. This bacteria is generally acquired during childhood and persists throughout life[4]. Although most H. pylori-positive persons are asymptomatic, only a small set of infected individuals will progress to severe gastrointestinal diseases such as non- Hodgkin’s lymphoma of the stomach or distal gastric adenocarcinoma[1,2,4]. The clinical outcome of the infection is influenced by immune mechanisms in the host, environmental factors[3,5-7], and genetic heterogeneity of the strain[8]. To date several genes in the genome of H. pylori have been identified and associated with disease[9], however, the two genes best understood in terms of structure and function are cagA and vacA genes.
The cagA gene is an important constituent of cag pathogenicity island, present in 50%-60% of the Western H. pylori strains[8,10], and in more than 90% of the strains isolated in East Asia, encodes a bacterial oncoprotein (CagA)[2,11,12], which is directly translocated within the epithelial cells via a type IV secretion system; CagA undergoes tyrosine phosphorylation by host cell kinases within repeated sequences of five amino acids (glutamic acid-proline-isoleucine-tyrosine-alanine), called EPIYA motifs[1,11,13]. These motifs show variation in the number of repetitions present in the carboxyl-terminus region of the protein, and based on the sequences surrounding them, they are defined as EPIYA-A, -B, -C, and -D[1]. EPIYA-A and EPIYA-B motifs are typically present in the CagA proteins of all cagA-positive isolates, followed by one to three EPIYA-C motifs, or by an EPIYA-D motif in Western and East Asian-type isolates, respectively[12,13]. Phosphorylated CagA interacts with the SHP-2 phosphatase and the Crk protein[14], resulting in reorganization of the cytoskeleton, cell elongation (hummingbird phenotype), and abnormal proliferation[11]. Hence, the sequence polymorphisms and duplications shown by CagA protein in their C-terminal region can modify the risk of disease by H. pylori[13]. Due to this, characterization of the number and type of EPIYA motifs in clinical isolates provides an additional value to the detection of the cag island[15].
Unlike cag PAI, all the H. pylori strains carry the vacA gene[2,10], but this is only expressed in about half of all the strains[16,17], it encodes an vacuolating toxin known as VacA that exerts multiple effects in the epithelial cells, resulting in cell damage, and inhibits activation and proliferation of T cells[2]. vacA is a polymorphic gene that exhibits two major regions of sequence diversity: signal (s) and median (m) region. There are two types of signal sequence of vacA (s1 or s2) and two types of median region (m1 or m2) of vacA[8,17,18]. The combination of s and m alleles results in different degrees of cytotoxicity and influences the pathogenicity of bacteria[10,11]. The vacA s1m1 and s1m2 strains produce large and moderate amounts of vacuolating toxin, respectively, and are strongly associated with gastric adenocarcinoma and peptic ulcer[10,19], while the vacA s2m2 strains are virtually not toxic and rarely associated to disease[20].
This extraordinary genetic diversity is generated through a high rate of point mutations, slipped-strand mispairing, and frequent intra-genomic and inter-genomic recombination[2,4,19], it is creating a non-linear system for diversification[19]. Thus, H. pylori probably uses its genetic plasticity to adapt physiologically to changing conditions in its host through of the selection of clonal variants of the same strain with changes in the surface molecules or variations in factors of interaction with the cell (e.g., loss all or part of cag-PAI, change number of EPIYA repetitions of CagA protein), contributing to maintaining of host-pathogen equilibrium that promotes persistence.
From this point view, the eradication of H. pylori has been proposed as a promising measure in the prevention of gastric lesions associated with infection. Nevertheless, an ideal treatment is not yet available[21]; in practice, 20%-30% of the therapies fail[22]. In most cases, this failure is attributed to the acquired resistance of strains to antibiotics[23]. Other factors poorly understood may also influence treatment failure[24,25]. Previous studies suggest that eradication rates are associated with the genetic characteristics of H. pylori[8], thus the cagA-positive/vacA s1 genotypes show a higher sensitivity than cagA-negative/vacA s2 genotypes to eradication treatment[24,26]. These findings are consistent with the observations of Correa et al[22], who observed that unsuccessful treatment is associated to increased prevalence of less virulent genotypes. Therefore, for the purpose of this study, we will shift the focus of how the H. pylori genotype influences the outcome of eradication therapy to who the treatment failure modifies the genotype of the infecting strains. We evaluate here the effect of the triple-standard therapy on cagA and vacA genotypes in H. pylori strains from Colombian subjects with unsuccessful treatment.
In 2009, 206 adults were voluntarily recruited with symptoms of dyspepsia (91 male, 115 female, mean age of 40.5 ± 0.8 years), in Túquerres Colombia, a population with high prevalence of H. pylori and preneoplastic lesions[3,27]. The participants underwent to upper gastrointestinal tract endoscopy. Antrum and gastric body biopsies were obtained for histopathological evaluation and H. pylori culture. Informed consent was obtained from all participants, and the study was approved by the Human Ethics Committee at Universidad del Valle (CIREH), certificate of approval No l073-07.
Expert pathologists in gastric mucosa biopsies performed the histopathological diagnosis, according Sydney’s classification system[28]. The categories used were non-atrophic gastritis (NAG), multifocal atrophic gastritis without intestinal metaplasia (MAG), intestinal metaplasia (IM), and dysplasia (DYS). The Steiner silver stain allowed to evaluate the presence of H. pylori.
The 176 patients positive for H. pylori through histology were treated with Clarithromycin 500 mg, amoxicillin 1000 mg, and omeprazole 20 mg (Genfar laboratories, Bogotá, Cundinamarca, Colombia) twice daily for 14 d. Resolution of the infection was evaluated through a 13C-Urea breath test (UBT), six weeks later. Participants with treatment failure were subjected to a control endoscopy, the gastric mucosa fragments obtained were again used for histopathological diagnosis and culture.
H. pylori were cultured from biopsies of antrum and body gastric obtained before (first endoscopy) and after (second endoscopy) anti-H.pylori treatment in patients with treatment failure. Chromosomal DNA was extracted from lysis of pure H. pylori cultures following a protocol of digestion with Proteinase K and later steps of precipitation with ethanol as previously described[29]. The vacA and cagA status was evaluated using the primers described by van Doorn et al[30].
Primers cagA2530S and cagA3000AS previously described by Panayotopoulou et al[13], allowed to characterize the number of EPIYA repetitions present on H. pylori isolates obtained before and after treatment, resulting in the generation of several fragments separated equidistantly by 100 bp. The PCR amplicons ranged in the range of 390 bp (2 repetitions), 490 bp (3 repetitions), 570 bp (4 repetitions), and 670 bp (5 repetitions).
To study the DNA sequence diversity among H. pylori strains obtained at baseline and post-treatment, two random primers were used: 1254 and 1281[31]. The RAPD-PCR conditions employed were previously described[29]. Each strain was amplified by duplicate under the same conditions.
For categorical variables, the McNemar test on paired data was used to determine the significance of the differences in the proportions of cagA and vacA genotypes observed before and after treatment in patients with unsuccessful treatment. The clustering analysis and its association with anatomic location of the isolates, exposure to anti-H. pylori treatment, and histopathological diagnosis were evaluated using the χ2 test. All data were analyzed with the statistical software (SPSS version 15). A value of P < 0.05 was considered statistically significant.
Of the 206 patients initially recruited, 176 (85.4%) and 149 (72.3%) participants were H. pylori-positive by histopathology and culture, respectively. The isolates obtained were characterized by virulence factors and antibacterial susceptibility. Subsequently, 176 participants were treated with 14-d standard therapy. Six weeks after, it was possible to contact 174 participants to conduct the [13C]-Urea breath test, nine of these participants were excluded because they were pregnant or had changed their home address. With the 165 participants in which was possible to conduct post-treatment control, it was found that 11 cases were ambiguous and 31 participants were UBT-positive. The eradication rate was 74.6% (123/165). Of the participants with treatment failure, only 25 accepted to undergo a second endoscopy. Prevalence post-treatment infection was at 64% (16/25) and 40% (10/25) by histopathology and culture, respectively. Once again, the strains obtained were characterized by virulence markers.
Of sixteen subjects with treatment failure diagnosed by histopathology, thirteen (81.3%) presented chronic NAG and three (18.7%) presented IM. When comparing the histopathological diagnosis of each of the 10 patients with treatment failure in which it was possible to isolate H. pylori, with histopathological diagnosis before treatment, it was found that diagnosis did not coincide in only one case (SV512) (data not shown).
Of the 149 isolates obtained before treatment, 136 (91.3%) isolates showed in vitro sensitivity to the two antibiotics used in standard triple therapy, whereas 4% and 2.7% of isolates were resistant to amoxicillin and clarithromycin, respectively. The remaining isolates presented double resistance (data not shown). After of the treatment, it was observed that the isolates obtained from the 10 patients with treatment failure, only one of them (SV415) had previously shown isolates resistant to clarithromycin (MIC > 4.0 mg/L), while the remaining participants presented prior to treatment, isolates sensitive to amoxicillin and clarithromycin according to antibiotic susceptibility testing.
The cagA and vacA status of H. pylori strains obtained from antrum and gastric body, before and after treatment in the 10 patients with treatment failure, are shown in Table 1. In 90% (9/10) of the patients before treatment, the infection with multiple H. pylori strains within and among anatomical sites was observed. In contrast, in only 60% (6/10) of the patients after the treatment presented mixed colonization within and among anatomical locations. When comparing the cagA and vacA genotypes found in each patient before and after therapy, only one case was found (SV444) in which these were identical and a specific site within the stomach was confirmed, the remaining nine patients presented differences. Multiple cagA (cagA-positive and cagA-negative) genotypes were found in these nine participants before therapy, but in four of them, just cagA-positive genotype was found after intervention, differences that were significant (P = 0.000). The vacA s1m1 genotype was highly prevalent, present in isolates of eight patients before and after treatment; the remaining patients (SV377 and SV480) presented a vacA s2m2 genotype prior to treatment that then varied with administration of anti-H. pylori treatment toward a vacA s1m1 genotype, nevertheless, these findings were not significant (P = 0.125) (Tables 1 and 2). None of patients harbored isolates with vacA s1m2 or s2m1 genotypes. In general, for most subjects, virulent strains were present before and after therapy, nevertheless, the antibiotic treatment limited colonization by multiple strains among and within the anatomic sites evaluated for each patient.
| Patient (n = 10) | Anatomic location | Anti-H. pylori treatment | |||
| Genotype | EPIYA motifs (bp) | ||||
| Before | After | Before | After | ||
| SV314 | Antrum lesser curvature | cagA+/vacA s1m11 | cagA+/vacA s1m1 | 495 | 200 |
| Antrum greater curvature | cagA+/vacA s1m11 | cagA+/vacA s1m1 | 495/200 | 200 | |
| SV318 | Antrum lesser curvature | cagA+/vacA s1m11 | cagA+/vacA s1m1 | 495 | 595/495 |
| Antrum greater curvature | cagA+/vacA s1m11 | No change | 200 | 595/495 | |
| Body greater curvature | cagA+/vacA s1m11 | CagA+/vacA s1m1 | 495/200 | NA | |
| SV377 | Antrum lesser curvature | cagA+/vacA s2m22 | cagA+/vacA s1m1 | 695/595/495/200 | 200 |
| Antrum greater curvature | cagA+/vacA s2m22 | cagA+/vacA s1m1 | NA | 200 | |
| Body greater curvature | cagA+/vacA s2m22 | Undefined | 200 | 495 | |
| SV415 | Antrum lesser curvature | cagA+/vacA s1m11 | cagA+/vacA s1m1 | 495 | 495/200 |
| Body greater curvature | cagA+/vacA s1m11 | cagA+/vacA s1m1 | 495/200 | 495 | |
| SV444 | Body greater curvature | cagA+/vacA s1m1 | No change | 595 | 595 |
| SV471 | Antrum greater curvature | cagA+/vacA s1m11 | cagA+/vacA s1m1 | 495 | 595 |
| Body greater curvature | cagA+/vacA s1m11 | cagA+/vacA s1m1 | 495/200 | 595 | |
| SV480 | Antrum lesser curvature | cagA+/vacA s2m22 | cagA+/vacA s1m11 | 595/495 | 595/200 |
| Body greater curvature | cagA+/vacA s2m22 | cagA+/vacA s1/m1 | 200 | 595 | |
| SV509 | Antrum lesser curvature | cagA+/vacA s1m11 | cagA+/vacA s1m1 | 200 | NA |
| Antrum greater curvature | cagA+/vacA s1m11 | cagA+/vacA s1m1 | 200 | 495 | |
| SV512 | Antrum greater curvature | cagA+/vacA s1m11 | cagA+/ vacAs1m1 | 495 | NA |
| Body greater curvature | cagA+/vacA s1m11 | No change | 495/200 | 595 | |
| SV526 | Antrum lesser curvature | cagA+/vacA s1m1 | cagA+/vacA s1m11 | 595 | NA |
| Antrum greater curvature | cagA+/vacA s1m11 | cagA+/vacA s1m1 | 595/200 | 595 | |
| Body greater curvature | cagA+/vacA s1m11 | cagA+/vacA s1m1 | 595/200 | 595 | |
| Genotype | Before treatment (n = 22) | After treatment (n = 22) | P vaule |
| cagA | |||
| Positive | 2 (9.1) | 17 (77.3) | 0.000 |
| Mixed | 20 (90.9) | 5 (22.7) | |
| vacA | |||
| s1m1 | 17 (22.7) | 21 (95.5) | 0.125 |
| s2m2 | 5 (77.3) | 1 (4.5) |
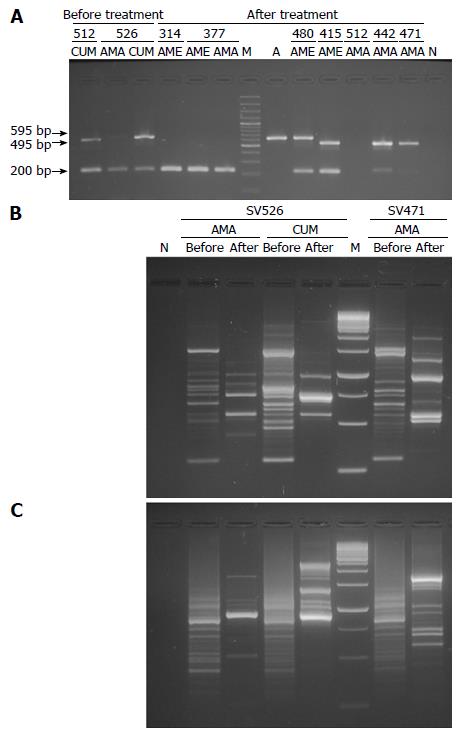
The 3’ end of cagA was successfully detected in 95.5% (21/22) of the isolates obtained before treatment and in 81.8% (18/22) of the isolates obtained after treatment in the 10 patients with therapeutic failure, which allowed corroborated the existence of cag locus and besides predicted the number of EPIYA repetitions (Table 1). The size of most PCR amplicons varied between 495 to 695 bp. However, reproducible bands with unexpected molecular weights of 200 bp were observed (Figure 1A and Table 1). A single-band was found in 59% (26/44) of the isolates obtained among antrum and body samples, before and after treatment, while 29.5% (13/44) of the isolates presented a double-band with different molecular weight, which confirms the presence of multiple cagA-positive strains in these subjects. All negative isolates for EPIYA-PCR (n = 5) were confirmed by cag empty-site, in two cases the test positivity (SV377 antrum greater curvature pre-treatment and SV526 antrum lesser curvature post-treatment) did not permit to establish the cag status by effect of multiple colonization. In the three remaining cases the amplification was not obtained. Finally, no statistical differences were observed when comparing the proportions of amplification of the EPIYA-PCR vs cagA-PCR in detecting of the locus cag in strains obtained before and after treatment (P > 0.05).
A reproducible RAPD pattern was observed using primers 1281 and 1254. These could discriminate 44 fingerprints. With primer 1281, well-defined profiles of one to 15 fragments of 74 to 1341 bp were observed (Figure 1B). It was evaluated whether exposure to anti-H. pylori treatment, anatomic location of the isolates within the stomach, and the histopathological result was associated to the RAPD conglomerates. Isolates obtained before treatment in patients with treatment failure (12/22; 54.5%) were included in conglomerate I, while isolates obtained after treatment (10/22; 45.5%) were included in conglomerate II (P = 0.042). In contrast, no significant differences were found in the segregation of the isolates by anatomic site and histopathological diagnosis (P = 0.414 and P = 0.339, respectively).
In the case of primer 1254, well-resolved fingerprints were also observed, the number and size oscillated from three to 30 and 87 to 1187 bp, respectively (Figure 1C). The dendrogram obtained includes anatomic location of the isolates, exposure to anti-H.pylori treatment, and histopathological result (Figure 2). The conglomerate analyses showed three principal conglomerates in a parsimonious arrangement. In no conglomerate, the distribution of the strains by anatomic site and exposure to treatment showed significant differences (P = 0.700 and P = 0.851, respectively). In contrast, significant differences were observed in the segregation of the histopathological diagnosis associated to each isolate (P = 0.006). Conglomerate I included 19/36 (52.8%) isolates associated to chronic NAG, and conglomerate II grouped 7/8 (87.5%) isolates associated to IM.
Analyzing the number of EPIYA motifs in H. pylori isolates obtained from patients with treatment failure with respect to isolates found before treatment in each of them according to the anatomic location. It was found an alteration in the number of EPIYA repeats in 72.7% (16/22) of the isolates due to the action of the antibiotics used. In six of these (SV377 BGC, SV471 AGC, SV509 AGC, SV512 BGC, SV318 ALC and AGC) the variation consisted in the gain of repeats, but in the last two isolates a divergence of cagA-positive subclones was observed. In contrast, in two isolates (SV314 ALC and SV480 ALC) the change consisted in the loss of EPIYA repetitions. While in SV377 ALC, SV415 ALC, and BGC isolates, the change evidenced after antibiotic pressure was the product of the replacement of the initial isolate with another isolate from a different region of the stomach of the same patient. Only in one isolate (SV480 BGC), the change in the number of EPIYA repetitions post-treatment was attributed to the re-infection of this patient with new H. pylori strain based on differences in genotyping of isolates and RAPD profiles. In the remaining four isolates (SV314 AGC, SV471 BGC, SV526 AGC and BCG), one of the CagA species initially present was lost, reflecting the co-infection by cagA-positive strains, and the subsequent selection of the resistant strain after treatment. Finally, one isolate (SV444 BGC) did not evidence any alteration, and in the five remaining isolates, the alteration could not be observed due to problems of amplification with the EPIYA-PCR. In all cases, the RAPD fingerprints generated by both primers supported the findings.
To remove H. pylori from the gastric mucosa, standard 14-d triple-drug including PPI-clarithromycin and amoxicillin is one of the most effective first-line therapy and best tolerated for patients[24]. Although high success rates have been obtained in clinical trials[32], the effectiveness of triple-therapy has decreased over time, achieved with its implementation eradication rates of up to 70%, less than 80% rate expected, and below what should be expected for an infectious disease[33], which is consistent with the 74.6% eradication rate found. This is worrisome because every failure to eradicate the infection can result in the emerging resistance of the microorganism to antibiotics employed, being unknown other possible implications on the genome of the strains. Thus, it is important to evaluate the effect of treatment failure on cagA and vacA genes in H. pylori isolates from Colombia.
As previously documented, the most important factors for treatment failure are pre-existing antibiotic resistance[33] and lack of compliance to treatment[34]. However, in this study, antimicrobial susceptibility testing performed on isolates obtained prior to treatment showed low rates of resistance to clarithromycin (2.7%) and amoxicillin (4%)[29], besides the compliance was strictly monitored during the treatment, without clinically important adverse sequelae during the treatment administration. Therefore, it seems that both factors are unlikely explanations for the results of our study. Thus, other factors such as increased free and prolonged use of PPIs[16], genetic polymorphisms of the CYP2C19[35], cigarette smoking, increased acidity[25], high bacterial load[36], and genotype of infecting strain[22,24] could be associated with treatment failure.
In this study, we found that 90% of the patients with treatment failure, before treatment carried multiple strains of H. pylori within and among the anatomic sites evaluated; after treatment, only 60% of the patients showed multiple colonization. One hypothesis is that presence of multiple H. pylori strains in an individual probably represents a stable association during the establishment of infection in the long-time[37]. When comparing cagA and vacA genotypes found in each patient, within the intragastric locations evaluated before and after therapy, it was observed that in several patients, virulent genotypes (cagA-positive; vacA s1m1) predominated before and after treatment; on the contrary, the low-virulence genotypes found before treatment were almost undetectable after therapy. These preliminary findings contrast with that reported by Correa et al[22] who suggest that failure to remove this bacteria can lead in some individuals to the survival of low-virulence strains (cagA-negative; vacA s2m2) because these strains seem to be more resistant to treatment.
The difference between these findings can be explained from the use in the present research of multiple biopsies (antrum and body) by patient to establish the genotypes, minimizing the effect of sampling error, without overestimating multiple colonization. Additionally, the results obtained could be reflecting a possible association between virulent genotypes and antibiotic resistance, which has been previously described in several studies[16,37,38]. In fact, the cagA-positive strains have a higher replication rate than the equivalent cagA-negative strains[39], thus, increasing the possibility of acquiring mutations that can be evolutionarily advantageous, supporting the hypothesis of treatment failure in these patients by acquisition of mutations that possibly modified the site of the antibiotic action (secondary antibiotic resistance). However, the presence of point mutations associated with resistance in the H. pylori isolates was not confirmed by sequencing, being a limitation of the study.
Additionally, the characterization of the number of EPIYA repetitions present between isolates obtained before and after treatment in patients with treatment failure, showed that antibiotic pressure in some strains induces genomic rearrangements inside the 3’ end of cagA gene that generate a non-directed alteration (gain or loss) of EPIYA repeats, agreeing with findings reported previously in in-vitro experiment[29]. It is hypothesized that these genetic changes could be directed through intergenomic and intragenomic recombination, processes that are improved through a second-order selection[13,19,40], allowing bacteria to confront variable and stressful environments within its current host[41].
In this case, the ability of H. pylori to edit particular immunostimulatory genetic regions (3’ end of cagA gene), it could lead to: (1) the synthesis of a non-phosphorylatable form of CagA by loss of all EPIYA motifs, which compromises their ability to interact with SHP-2, minimizing the alteration of signal transduction pathways of the cell, supported by the bands obtained with molecular weight not expected (200 bp); or (2) the alteration of number of EPIYA motifs impacting the adherence of CagA protein to the epithelial cell membrane[42]; level of tyrosine phosphorylation, the induction of IL-8 secretion[43], and multimerization of CagA[44]. Also, in some cases lead to differentiation of cagA-positive sub-clones with distinct numbers of EPIYA repeats (e.g., SV318 ALC and AGC, SV314 ALC), each acting as possible source of genetic elements for other clones.
These clonal variants cooperate by quorum sensing and recombination to downregulate their interplay with the individual, and thus produce less damage[19]. However, some tissue damage and inflammation are unavoidable, because in these strains diverse CagA species are secreted, altering multiple signaling pathways and inducing various degrees of elongation in gastric epithelial cells[13]. This hypothesis may partly explain the findings of Mera et al[45] who in a randomized trial of 795 adults with preneoplastic gastric lesions for 3, 6, and 12 years, observed that patients with treatment failure at 12 years of follow-up had a modest decrease in their histopathological score compared with average histopathology at baseline.
In general, the RAPD profiles obtained were of great utility because they initially supported the close clonal association among the H. pylori strains present in present in different gastric localizations of the same patient; for this reason, in the clusters analysis for both primers no differences were found in the segregation of the strains by anatomical site. Subsequently, the profiles generated by primer 1254 clearly defined a pathological conglomerate for IM (P = 0.006), in agreement with the hypothesized by Kidd et al[46] who suggest that clonal grouping by RAPD patterns can be associated to disease. Likewise, Vega et al[16] demonstrated the usefulness of the DNA fingerprints to discriminate H. pylori isolates associated with peptic ulcer, nevertheless, in the present research this association was not evident with primer 1281.
Additionally, the DNA fingerprints generated by primer 1254 permitted evidencing the close clonal relationship among some isolates obtained before and after treatment (Recrudescence), not showing significant differences in segregation of isolates according to the exposure to treatment, agreeing with that expected and contrasting with the results obtained with primer 1281. In spite of these differences, the conglomerate analysis for primer 1281 also reflected the close relationship of some isolates obtained before and after treatment, although in lesser number. There was only one case (SV480 BGC) where the lack of proximity within the clusters formed together with the differences in genotyping for the basal isolate and post-treatment suggest reinfection of the patient with a new H. pylori strain.
Our study has some limitations. First, due to the absence of sequencing of bacterial genes encoding 16S rRNA and pbp-1A in isolates of patients with treatment failure, it was no possible to detect point mutations that explain the acquisition of secondary resistance to clarithromycin and amoxicillin, respectively. Second, although EPIYA PCR is a practical tool for amplify the cagA gene, and characterize the number and type of EPIYA motifs, in the bands with unexpected molecular weight (200 bp), the number of motifs is unknown because it was not sequenced. Third, the number of H. pylori isolates obtained from subjects with unsuccessful treatment in this study is small, so our findings must be confirmed in other studies, where it is also advisable to evaluate the effect of treatment failure on the signal and median region of the vacA gene that can equally present genetic variation through recombination. Future interventions may be designed based on this research.
In conclusion, the present research shows that although the anti-H. pylori treatment fails to eradicate the infection in some patients, it limits colonization by low-virulence genotypes leading to a partial and selective elimination in mixed infections, and it acts in some cases on cagA-positive strains inducing: (1) mutations that make it resistant to antibiotics; and (2) genetic rearrangements as deletion or acquisition of EPIYA motifs, decreasing or increasing the phosphorylation of CagA and its binding to SHP-2, respectively. All this leads to divergence of cagA-positive strains with implications for pathogenesis that are not yet well understood and need to be studied. Finally, it is possible that indiscriminate use of antibiotics to treat upper respiratory and intestinal infections also alter the genetic structure of H. pylori strains that coexist within the host.
We thank to Cali Cancer Registry (RPCC for the term in Spanish) for their support in logistics and handling of information.
Infection with Helicobacter pylori (H. pylori) is the greatest risk factor for development of gastric adenocarcinoma, especially in individuals infected with cagA-positive strains. In developing countries, the colonization with multiple strains of high (cagA-positive) and low virulence (cagA-negative) is common, these H. pylori clones can co-exist in dynamic equilibrium within of host, cooperating through quorum sensing and recombination, which can lead to subregulation of its interaction to induce lower damage even when some damage is inevitable. In any case, the eradication of H. pylori with antibiotics constitutes an important primary prevention strategy of gastric lesions and atrophy. Although major improvements have been made in the efficacy of treatment regimes, all of them result in failures to eradicate the infection. Few studies have focused on evaluating the effect of treatment failure on virulence factors of H. pylori.
Virulence-associate genotypes of H. pylori are important determinants of the clinical outcome of the infection. In many series, patients with severe gastritis, atrophic gastritis, peptic ulcer disease and distal gastric cancer are predominantly infected with the cagA-positive/vacA s1m1 strains, whereas the cagA-negative/vacA s2m2 strains are more frequent in patients with non-ulcer dyspepsia and mild gastritis. That virulence factors are linked with disease implies that they are a fixed characteristic, but this is not the case because change in genotypes promoted by environmental pressures (e.g., antibiotic treatment, high uptake salt and hyperchlorhydria) can occur through of intragenomic recombination (e.g., number motif EPIYA in CagA) or recombination with clonal variants of the same strain or with other strains in cases of mixed infection that can lead to partial or complete loss of cag PAI and changes in vacA genotype, reflects local selection of H. pylori particular phenotypes that appears to be essential for persistent colonization of host.
A previous study indicates that the unsuccessful treatment of H. pylori results in a increase of less virulent genotypes in Colombian patients, it suggesting that the cagA-negative/vacA s2m2 strains were less responsive to treatment or a possible loss of specific virulence factors (cag PAI) induced by antibiotic pressure. In contrast, in a recent study, where we evaluated the in vitro effect of antibiotics using in the standard triple therapy, we were found that antibiotic pressure does not induce loss of the cag pathogenicity island, but it can lead in most cases to genetic rearrangements within the 3’ region cagA, these findings are consistent with the results found in this new study, it showing that the failure of anti-H. pylori treatment limits colonization by low-virulence strains resulting in partial and selective eradication of H. pylori in mixed infections, and acts on the cagA-positive strains inducing genetic rearrangements within the cagA variable region that produces a non-directed alteration like loss or gain of EPIYA motifs, which as a set could alter the pathogenic process induced by H. pylori. These events should be considered after failure of first-line treatment.
These findings have important implications for the treatment of gastro-duodenal diseases caused by H. pylori, suggesting that the failure of anti-H.pylori treatment results in survival of more virulent genotypes in mixed infections, this alters the dynamic balance between clones of H. pylori and host, leading to H. pylori to rapidly adapt to new conditions in the stomach by genetic rearrangements that favor the acquisition or deletion of EPIYA motifs, which may lead to upregulation of interaction with the host. In support of this, it has been reported that infection with a single antral-colonizing strain could lead to duodenal ulceration, while, the colonization with two different strains in antrum and corpus leads to lower physiological alterations. These determinants are important considerations in deciding who should be treated.
CagA is recognized as a major etiologic determinant of H. pylori-associated gastric disease. This bacterial protein is translocate into the gastric epithelial cell cytoplasm via the type IV secretion system. Once injected, CagA localizes to the plasma membrane and undergoes tyrosine phosphorylation by multiple members of the Src family of kinases on specific tyrosine residues within repeating Glu-Pro-Ile-Tyr-Ala (EPIYA) motifs, encoded in the 3’variable region of cagA gene. These EPIYA motifs are defined as EPIYA-A, -B -C, and -D, according to the amino acid sequence that surrounds the EPIYA sequence. Phosphorylated CagA interacts with the SHP-2 phosphatase and the Crk protein resulting in reorganization of the cytoskeleton, cell elongation, and abnormal proliferation.
In this study, the authors investigated to identify effects of treatment failure on the CagA EPIYA motif in H. pylori isolates. This study was well written.
| 1. | Sicinschi LA, Correa P, Peek RM, Camargo MC, Piazuelo MB, Romero-Gallo J, Hobbs SS, Krishna U, Delgado A, Mera R. CagA C-terminal variations in Helicobacter pylori strains from Colombian patients with gastric precancerous lesions. Clin Microbiol Infect. 2010;16:369-378. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 76] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 2. | Suerbaum S, Josenhans C. Helicobacter pylori evolution and phenotypic diversification in a changing host. Nat Rev Microbiol. 2007;5:441-452. [PubMed] |
| 3. | Ek C, Whary MT, Ihrig M, Bravo LE, Correa P, Fox JG. Serologic evidence that ascaris and toxoplasma infections impact inflammatory responses to Helicobacter pylori in Colombians. Helicobacter. 2012;17:107-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 4. | Dorer MS, Talarico S, Salama NR. Helicobacter pylori’s unconventional role in health and disease. PLoS Pathog. 2009;5:e1000544. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 79] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 5. | Noto JM, Gaddy JA, Lee JY, Piazuelo MB, Friedman DB, Colvin DC, Romero-Gallo J, Suarez G, Loh J, Slaughter JC. Iron deficiency accelerates Helicobacter pylori-induced carcinogenesis in rodents and humans. J Clin Invest. 2013;123:479-492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 6. | Torres J, Correa P, Ferreccio C, Hernandez-Suarez G, Herrero R, Cavazza-Porro M, Dominguez R, Morgan D. Gastric cancer incidence and mortality is associated with altitude in the mountainous regions of Pacific Latin America. Cancer Causes Control. 2013;24:249-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 90] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 7. | de Sablet T, Piazuelo MB, Shaffer CL, Schneider BG, Asim M, Chaturvedi R, Bravo LE, Sicinschi LA, Delgado AG, Mera RM. Phylogeographic origin of Helicobacter pylori is a determinant of gastric cancer risk. Gut. 2011;60:1189-1195. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 114] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 8. | van Doorn LJ, Schneeberger PM, Nouhan N, Plaisier AP, Quint WG, de Boer WA. Importance of Helicobacter pylori cagA and vacA status for the efficacy of antibiotic treatment. Gut. 2000;46:321-326. [PubMed] |
| 9. | da Costa DM, Pereira Edos S, Rabenhorst SH. What exists beyond cagA and vacA? Helicobacter pylori genes in gastric diseases. World J Gastroenterol. 2015;21:10563-10572. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 36] [Cited by in RCA: 34] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 10. | Sicinschi LA, Correa P, Peek RM, Camargo MC, Delgado A, Piazuelo MB, Romero-Gallo J, Bravo LE, Schneider BG. Helicobacter pylori Genotyping and Sequencing Using Paraffin-Embedded Biopsies from Residents of Colombian Areas with Contrasting Gastric Cancer Risks. Helicobacter. 2008;13:135-145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 11. | Wen S, Moss SF. Helicobacter pylori virulence factors in gastric carcinogenesis. Cancer Lett. 2009;282:1-8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 12. | Acosta N, Quiroga A, Delgado P, Bravo MM, Jaramillo C. Helicobacter pylori CagA protein polymorphisms and their lack of association with pathogenesis. World J Gastroenterol. 2010;16:3936-3943. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 33] [Cited by in RCA: 39] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 13. | Panayotopoulou EG, Sgouras DN, Papadakos K, Kalliaropoulos A, Papatheodoridis G, Mentis AF, Archimandritis AJ. Strategy to characterize the number and type of repeating EPIYA phosphorylation motifs in the carboxyl terminus of CagA protein in Helicobacter pylori clinical isolates. J Clin Microbiol. 2007;45:488-495. [PubMed] |
| 14. | Higashi H, Tsutsumi R, Muto S, Sugiyama T, Azuma T, Asaka M, Hatakeyama M. SHP-2 tyrosine phosphatase as an intracellular target of Helicobacter pylori CagA protein. Science. 2002;295:683-686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 755] [Cited by in RCA: 790] [Article Influence: 32.9] [Reference Citation Analysis (0)] |
| 15. | Argent RH, Zhang Y, Atherton JC. Simple method for determination of the number of Helicobacter pylori CagA variable-region EPIYA tyrosine phosphorylation motifs by PCR. J Clin Microbiol. 2005;43:791-795. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 86] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 16. | Vega AE, Cortiñas TI, Puig ON, Silva HJ. Molecular characterization and susceptibility testing of Helicobacter pylori strains isolated in western Argentina. Int J Infect Dis. 2010;14 Suppl 3:e85-e92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 17. | Peek RM, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer. 2002;2:28-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1317] [Cited by in RCA: 1352] [Article Influence: 56.3] [Reference Citation Analysis (1)] |
| 18. | Polk DB, Peek RM. Helicobacter pylori: gastric cancer and beyond. Nat Rev Cancer. 2010;10:403-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 19. | Atherton JC, Blaser MJ. Coadaptation of Helicobacter pylori and humans: ancient history, modern implications. J Clin Invest. 2009;119:2475-2487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 20. | Yamaoka Y, Kato M, Asaka M. Geographic differences in gastric cancer incidence can be explained by differences between Helicobacter pylori strains. Intern Med. 2008;47:1077-1083. |
| 21. | Gisbert JP. Tratamientos de rescate ante el fracaso erradicador de Helicobacter pylori. Gastroenterología y Hepatología. 2011;34:89-99. [RCA] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 14] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 22. | Correa P, van Doorn LJ, Bravo JC, Ruiz B, Bravo LE, Realpe JL. Unsuccessful treatment results in survival of less virulent genotypes of Helicobacter pylori in Colombian patients. Am J Gastroenterol. 2000;95:564-566. [PubMed] |
| 23. | Graham DY. Antibiotic resistance in Helicobacter pylori: implications for therapy. Gastroenterology. 1998;115:1272-1277. [PubMed] |
| 24. | Zhao JJ, Wang JB, Yang L, Li Y. Influence of Helicobacter pylori genotype on triple eradication therapy. J Gastroenterol Hepatol. 2007;22:2251-2255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 25. | Camargo MC, Piazuelo MB, Mera RM, Fontham ET, Delgado AG, Yepez MC, Ceron C, Bravo LE, Bravo JC, Correa P. Effect of smoking on failure of H. pylori therapy and gastric histology in a high gastric cancer risk area of Colombia. Acta Gastroenterol Latinoam. 2007;37:238-245. [PubMed] |
| 26. | Russo F, Berloco P, Cuomo R, Caruso ML, Di Matteo G, Giorgio P, De Francesco V, Di Leo A, Ierardi E. Helicobacter pylori strains and histologically-related lesions affect the outcome of triple eradication therapy: a study from southern Italy. Alimentary Pharmacol Therap. 2003;17:421-428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 27. | Bravo LE, van Doom LJ, Realpe JL, Correa P. Virulence-associated genotypes of Helicobacter pylori: do they explain the African enigma? Am J Gastroenterol. 2002;97:2839-2842. [PubMed] |
| 28. | Dixon MF, Genta RM, Yardley JH, Correa P. Classification and grading of gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am J Surg Pathol. 1996;20:1161-1181. [PubMed] |
| 29. | Bustamante-Rengifo JA, Matta AJ, Pazos A, Bravo LE. In vitro effect of amoxicillin and clarithromycin on the 3’ region of cagA gene in Helicobacter pylori isolates. World J Gastroenterol. 2013;19:6044-6054. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 9] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 30. | van Doorn LJ, Figueiredo C, Rossau R, Jannes G, van Asbroeck M, Sousa JC, Carneiro F, Quint WGV. Typing of Helicobacter pylori vacA Gene and Detection of cagA Gene by PCR and Reverse Hybridization. J Clin Microbiol. 1998;36:1271-1276. [PubMed] |
| 31. | Akopyanz N, Bukanov NO, Westblom TU, Kresovich S, Berg DE. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 1992;20:5137-5142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 565] [Cited by in RCA: 565] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 32. | Fischbach LA, Goodman KJ, Feldman M, Aragaki C. Sources of variation of Helicobacter pylori treatment success in adults worldwide: a meta-analysis. Int J Epidemiol. 2002;31:128-139. [PubMed] |
| 33. | Malfertheiner P, Megraud F, O’Morain CA, Atherton J, Axon AT, Bazzoli F, Gensini GF, Gisbert JP, Graham DY, Rokkas T. Management of Helicobacter pylori infection--the Maastricht IV/ Florence Consensus Report. Gut. 2012;61:646-664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1719] [Cited by in RCA: 1614] [Article Influence: 115.3] [Reference Citation Analysis (7)] |
| 34. | Gisbert JP, Gonzalez L, Calvet X. Systematic review and meta-analysis: proton pump inhibitor vs. ranitidine bismuth citrate plus two antibiotics in Helicobacter pylori eradication. Helicobacter. 2005;10:157-171. [PubMed] |
| 35. | Padol S, Yuan Y, Thabane M, Padol IT, Hunt RH. The effect of CYP2C19 polymorphisms on H. pylori eradication rate in dual and triple first-line PPI therapies: a meta-analysis. Am J Gastroenterol. 2006;101:1467-1475. [PubMed] |
| 36. | Moshkowitz M, Konikoff FM, Peled Y, Santo M, Hallak A, Bujanover Y, Tiomny E, Gilat T. High Helicobacter pylori numbers are associated with low eradication rate after triple therapy. Gut. 1995;36:845-847. [PubMed] |
| 37. | Cellini L, Grande R, Di Campli E, Di Bartolomeo S, Capodicasa S, Marzio L. Analysis of genetic variability, antimicrobial susceptibility and virulence markers in Helicobacter pylori identified in Central Italy. Scand J Gastroenterol. 2006;41:280-287. [PubMed] |
| 38. | Elviss NC, Owen RJ, Breathnach A, Palmer C, Shetty N. Helicobacter pylori antibiotic-resistance patterns and risk factors in adult dyspeptic patients from ethnically diverse populations in central and south London during 2000. J Med Microbiol. 2005;54:567-574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 39. | Mégraud F, Corti R. Resistencia bacteriana del Helicobacter pylori en el mundo en el año 2009. Acta Gastroenterol Latinoam. 2009;39:282-290. |
| 40. | Tenaillon O, Taddei F, Radmian M, Matic I. Second-order selection in bacterial evolution: selection acting on mutation and recombination rates in the course of adaptation. Res Microbiol. 2001;152:11-16. [PubMed] |
| 41. | Monack DM, Mueller A, Falkow S. Persistent bacterial infections: the interface of the pathogen and the host immune system. Nat Rev Microbiol. 2004;2:747-765. [PubMed] |
| 42. | Higashi H, Yokoyama K, Fujii Y, Ren S, Yuasa H, Saadat I, Murata-Kamiya N, Azuma T, Hatakeyama M. EPIYA motif is a membrane-targeting signal of Helicobacter pylori virulence factor CagA in mammalian cells. J Biol Chem. 2005;280:23130-23137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 107] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 43. | Kim SY, Lee YC, Kim HK, Blaser MJ. Helicobacter pylori CagA transfection of gastric epithelial cells induces interleukin-8. Cell Microbiol. 2006;8:97-106. [PubMed] |
| 44. | Ren S, Higashi H, Lu H, Azuma T, Hatakeyama M. Structural basis and functional consequence of Helicobacter pylori CagA multimerization in cells. J Biol Chem. 2006;281:32344-32352. [PubMed] |
| 45. | Mera R, Fontham ET, Bravo LE, Bravo JC, Piazuelo MB, Camargo MC, Correa P. Long term follow up of patients treated for Helicobacter pylori infection. Gut. 2005;54:1536-1540. [PubMed] |
| 46. | Kidd M, Atherton JC, Lastovica AJ, Louw JA. Clustering of South African Helicobacter pylori isolates from peptic ulcer disease patients is demonstrated by repetitive extragenic palindromic-PCR fingerprinting. J Clin Microbiol. 2001;39:1833-1839. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Colombia
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
P- Reviewer: Abadi ATB, Ierardi E, Sugimoto M, Tongtawee T S- Editor: Qi Y L- Editor: A E- Editor: Zhang FF