Published online Jan 7, 2017. doi: 10.3748/wjg.v23.i1.76
Peer-review started: September 6, 2016
First decision: September 28, 2016
Revised: October 9, 2016
Accepted: November 15, 2016
Article in press: November 16, 2016
Published online: January 7, 2017
Processing time: 121 Days and 16.7 Hours
To investigate the role of the miR-133a-UCP2 pathway in the pathogenesis of inflammatory bowel disease (IBD) and to explore the potential downstream mechanisms with respect to inflammation, oxidative stress and energy metabolism.
C57BL/6 mice were fed dextran sulfate sodium (DSS) liquid for 7 consecutive days, followed by the administration of saline to the DSS group, UCP2 siRNA to the UCP2 group and a miR-133a mimic to the miR-133a group on days 8 and 11. Body weight, stool consistency and rectal bleeding were recorded daily, and these composed the disease activity index (DAI) score for the assessment of disease severity. After cervical dislocation was performed on day 14, the length of the colon in each mouse was measured, and colonic tissue was collected for further study, which included the following: haematoxylin and eosin staining, UCP2 and miR-133a detection by immunohistochemical staining, western blot and quantitative real-time PCR, measurement of apoptosis by TUNEL assay, and the assessment of inflammation (TNF-α, IL-1β, IL-6 and MCP1), oxidative stress (H2O2 and MDA) and metabolic parameters (ATP) by ELISA and colorimetric methods.
An animal model of IBD was successfully established, as shown by an increased DAI score, shortened colon length and specific pathologic changes, along with significantly increased UCP2 and decreased miR-133a levels. Compared with the DSS group, the severity of IBD was alleviated in the UCP2 and the miR-133a groups after successful UCP2 knockdown and miR-133a overexpression. The extent of apoptosis, as well as the levels of TNF-α, IL-1β, MDA and ATP, were significantly increased in both the UCP2 and miR-133a groups compared with the DSS group.
The miR-133a-UCP2 pathway participates in IBD by altering downstream inflammation, oxidative stress and markers of energy metabolism, which provides novel clues and potential therapeutic targets for IBD.
Core tip: The pathogenesis of inflammatory bowel disease (IBD) is unclear, but increasing evidence supports the involvement of epigenetic regulation such as the formation of miRNA-mRNA pairs. In this study, we investigated the role of the miR-133a-UCP2 pathway in the pathogenesis of IBD in a well-established mouse model. We found that the severity of IBD was alleviated after the UCP2 and miR-133a levels were antagonized and that the underlying mechanism may involve changes in inflammation, oxidative stress and energy metabolism. Our data provide novel clues and potential therapeutic targets for IBD.
- Citation: Jin X, Chen D, Zheng RH, Zhang H, Chen YP, Xiang Z. miRNA-133a-UCP2 pathway regulates inflammatory bowel disease progress by influencing inflammation, oxidative stress and energy metabolism. World J Gastroenterol 2017; 23(1): 76-86
- URL: https://www.wjgnet.com/1007-9327/full/v23/i1/76.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i1.76
Inflammatory bowel disease (IBD) is characterized by chronic, unspecific inflammation of the gastrointestinal tract and may be classified into two major subtypes: Crohn’s disease (CD) and ulcerative colitis (UC)[1]. Although the prevalence of IBD is high in western countries, a rapid rise in IBD has been observed in Asia[2], where its prevalence has reached approximately 11.6 × 105 for UC and 1.4 × 105 for CD in China[3]. Although a combination of genetic, environmental, infectious and immunologic factors is considered to be involved in the pathogenesis of IBD, the underlying mechanism is still unclear[4]. Moreover, the effectiveness of routine therapy is discouraging, and aetiology-directed therapy is rare[5]. Therefore, an in-depth investigation of IBD would help reveal the pathogenesis of IBD and provide a novel theoretical base for innovative treatments.
MicroRNAs (miRNAs) are a family of non-coding RNAs that are 19-25 nucleotides (nt) long. They are processed from double-stranded hairpin precursors, which are 70-100 nt in length, by the RNaseIII family member Dicer. Dicer is endogenously expressed in the cytoplasm as part of the RNA-induced silencing complex[6]. miRNAs recognize the 3’ untranslated region of target mRNAs with imperfect complementarity, which leads to translational repression in mammals and mRNA cleavage in plants[7]. With the development of high-throughput methods such as microarray and deep sequencing, miRNA profiling has been widely applied to determine molecular biomarkers in tumours[8] and other diseases such as nonalcoholic fatty liver disease (NAFLD)[9,10]. Additionally, miRNA-mRNA pathways have been revealed to participate in various diseases[11]. These features make miRNAs attractive research targets.
The effect and underlying mechanism of miRNAs in the pathogenesis and progression of IBD have become popular research topics. With the use of microarray screening and bioinformatics approaches, researchers have revealed specific tissue[12] and serum[13] miRNA profiles of IBD, which have provided researchers with novel diagnostic biomarkers. Further functional studies have reported the regulation of IBD inflammation by miR-146a[14] and miR-132[15] through the hedgehog and cholinergic pathways. Moreover, the involvement of the miR-224-p21 pathway was found to participate in the early pathogenesis of IBD[16]. However, compared with the large amount of miRNA data from microarray analyses, few miRNA-mRNA pathways have been reported in IBD, which is why this topic is worth more intensive study in the future.
We previously summarized the effect of uncoupling protein (UCP), which belongs to a specific mitochondrial inner membrane protein family with the capacity to uncouple oxidative phosphorylation in NAFLD[17]; we also revealed the effect of liver-specific UCP in conditions of oxidative stress[18]. Since oxidative stress has been demonstrated to be strongly associated with IBD[19] and previous studies have identified the association between the genetic polymorphism 866G/A of UCP2[20] and IBD, we proposed an important role for UCP2 (the most common and widely studied UCP) in IBD. More importantly, previous studies confirmed the direct regulation of UCP2 by miR-133a[21,22], which highlights the possible effect of the miR-133a-UCP2 pathway in IBD and why this pathway has become the research focus of this study.
This study was performed in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The animal protocol was approved by the institutional review board of the First Affiliated Hospital of Zhejiang University.
A total of 50 female C57BL/6 mice aged 8-12 wk were purchased from Shilek Lab Animal (Shanghai, China). All mice received food and water ad libitum and were maintained on a 12/12-h light/dark cycle at 25 °C for 7 d as part of adaptive feeding. Thereafter, the first 10 mice were randomly divided into two groups to establish an animal model of IBD. Under a common basic diet, the control (n = 5) group received normal liquid, while the DSS (n = 5) group received 3% Dextran Sulfate Sodium (DSS) liquid (MP Biomedicals, Shanghai, China) instead of normal liquid for 7 consecutive days. The body weight, stool consistency and rectal bleeding of each mouse were recorded daily; these constituted the DAI (range from 0 to 12), which was used to assess the clinical severity of colitis, as previously described[23]. After 7 d of feeding, all mice were sacrificed by cervical dislocation, after which the colonic tissue was collected for haematoxylin and eosin (HE) staining and for the detection of the UCP2 and miR-133a levels.
After the animal model of IBD was verified, the remaining mice were further randomly divided into four groups as follows: control (n = 10), DSS (n = 10), UCP2 (n = 10) and miR-133a (n = 10), and the diet of mice in the UCP2 and the miR-133a groups was the same as that of mice in the DSS group. After 7 consecutive days of feeding, an intravenous injection of 100-200 μL saline was given to the control and DSS groups; similarly, UCP2 siRNA was given to mice in the UCP2 group and miR-133a mimics were given to mice in the miR-133a group on days 8 and 11 with the final dose of 33 μg according to the manufacturer’s instruction, respectively. There was no anesthesia before intravenous injection. All mice were sacrificed by cervical dislocation on day 14, after which the colonic tissue was collected for HE staining and for the detection of inflammation, oxidative indicators and energy-related markers.
Specific siRNA against the UCP2 gene and a scrambled siRNA, which was used as a negative control, were synthesized at and purchased from GenePharma (Shanghai, China). The mouse miR-133aRNA mimic was also designed at and purchased from GenePharma. The sequence of the negative control (5’-3’) was sense-UUCUCCGA ACGUGUCACGUTT and antisense-ACGUGACACGUUCGGAGAATT; the sequence of the miR133a mimic (5’-3’) was sense-UUUGGUCCCCUUCAACCAG CUG and antisense-GCUGGUUGAAGGGGACCAAAUU; the sequence of the UCP2 siRNA(5’-3’) was sense-GGAAAGGGACUUCUCCCAATT and antisense-U UGGGAGAAGUCCCUUUCCTT. The administration of the siRNA and RNA mimic was performed through intravenous injection using an in vivo transfection reagent (Auckland, New Zealand) according to the method of Entranster.
The mouse colonic tissue was routinely homogenized and harvested for the detection of TNF-α, IL-1β, IL-6 and MCP1 by ELISA according to the manufacturer’s instructions (Neobioscience, Shenzhen, China). The level of apoptosis in the colonic tissue was also routinely detected by TUNEL staining according to the manufacturer’s instructions (Roche, Shanghai, China), as previously reported[24]. Specifically, the apoptotic cells were dyed and were observed under an Olympus microscope. Typically, 10 visual fields were selected and 100 cells within each field were counted, where the apoptosis index = (apoptosis cell/total cell) × 100%.
The tissues were routinely assessed for the oxidative stress-related markers H2O2 and MDA by a colorimetric method using an OD value of 405 nm at 37 °C and by a thiobarbituric acid method using an OD value of 532 nm at 95 °C, respectively. The level of the major energy product ATP was also routinely measured using a colorimetric method and an OD value of 636 nm at 37 °C. All these experiments were performed according to the manufacturer’s instructions (Nanjing Jiancheng Bioengineering Institute, China).
For immunohistochemical staining, the colon tissues collected from the mice were sequentially fixed in 10% formaldehyde solution for at least 24 h, processed and paraffin embedded, cut into 4-μm-thick sections, and mounted onto glass slides. Each slide was deparaffinized and rehydrated in xylene and graded alcohol solutions, respectively, which was followed by antigen retrieval in Citrate Antigen Retrieval solution at 126 °C for 40 min. The slides were then washed in PBS and blocked with goat serum for 10 to 30 min and incubated overnight at 4 °C with diluted anti-UCP2 antibody. After three washes in PBS, the slides were incubated with goat-anti-rabbit IgG for 20 min, which was followed by reaction with diaminobenzidine and counterstaining with haematoxylin. The slides were finally mounted in neutral gum for semiquantitative analysis by microscopy. Negative control slides were incubated with PBS instead of the primary antibody.
For a more accurate analysis, the UCP2 protein level was further quantified by Western blot with a primary mouse polyclonal antibody raised against UCP2 (Abcam, ab77363) and an ECL chemiluminescence kit (Santa Cruz, United States). Normalization was performed by blotting the same samples with a mouse anti-β-actin antibody. For miR-133a quantitative analysis, 2 μg of retrieved total RNA was reverse transcribed using stem-loop antisense primer mix and AMV transcriptase (TaKaRa, Dalian, China). Real-time PCR was routinely performed in an MX3000p real time PCR system (Stratagene, United States). U6 snRNA and GAPDH were amplified as normalization controls, and the relative amount of miR-133a compared with U6 and the relative amount of UCP2 compared with GAPDH were calculated using the equation 2-ΔCT, where ΔCT= CTmiRNA/UCP2-CTu6/GAPDH. The detailed primer sequences are shown in Supplementary Figure 1.
Statistical analyses were performed using SPSS version 16 (Chicago, IL, United States). Data are presented as the mean ± standard deviation when normally distributed or as the median if the distribution was skewed. Differences between two groups were analysed using Student’s t-test or the Mann-Whitney U test; differences among four groups were analysed with One-Way Anova.
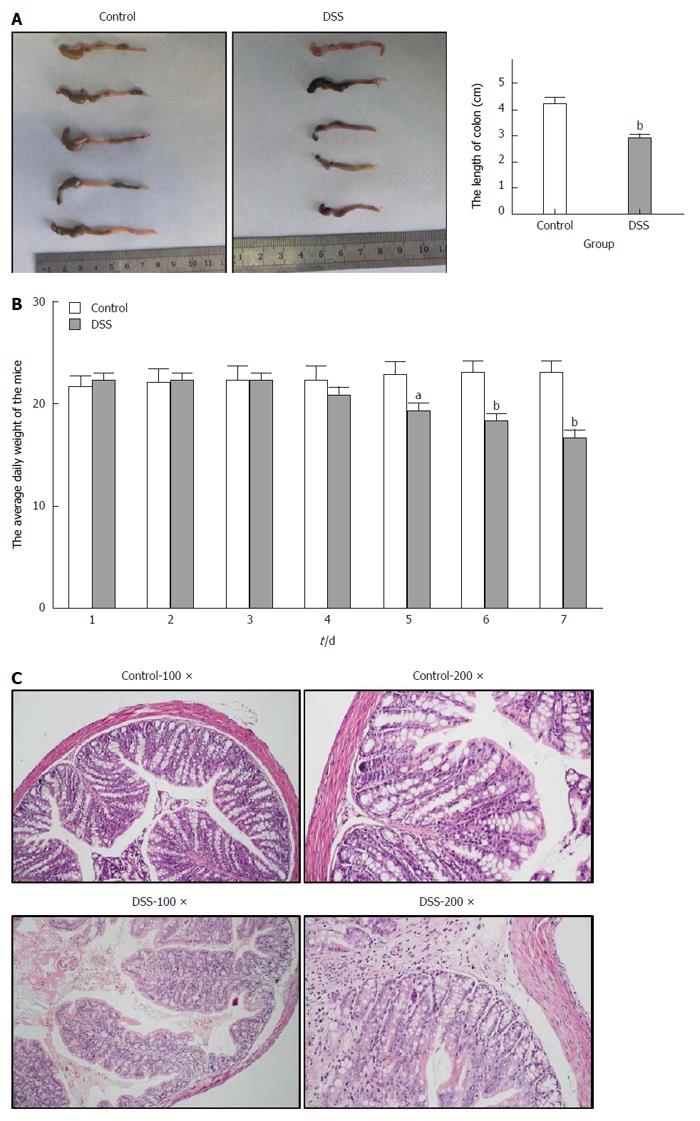
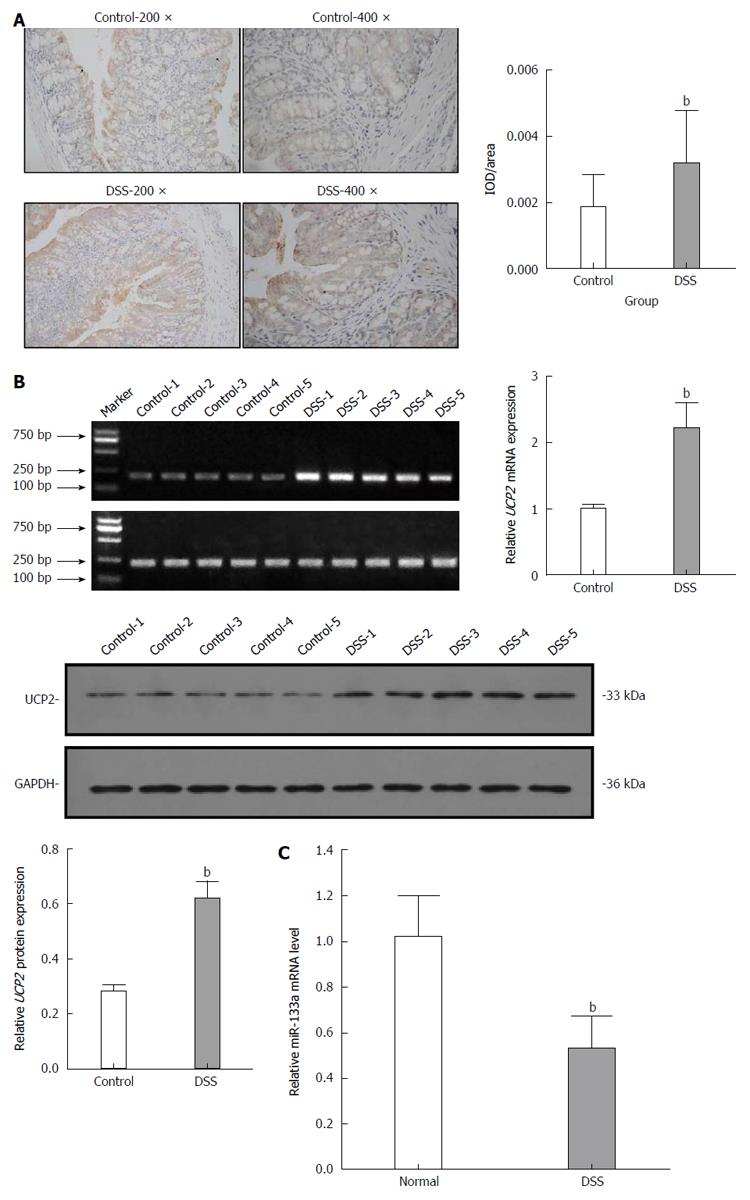
After the C57BL/6 mice were fed 3% DSS for 7 consecutive days, a representative IBD mouse model was successfully established (Figure 1). Generally, after the first 3 d of DSS feeding, the water and food intake and body weight of mice in the DSS group were slightly increased, while on the 4th day, which was the turning point, the above-mentioned markers had steadily decreased. In addition, the total colon length was also significantly shortened in the DSS group compared with the control group. Finally, HE staining of the colon tissue showed intact colonic mucosa and regularly arranged colonic glands in the control group, whereas destroyed colonic glands, mucosal ulceration and inflammatory cell infiltration were observed in the DSS group. The average DAI for the DSS group was 11.67 on day 7. As shown in Figure 2, we also identified significantly increased UCP2 mRNA and protein levels through qRT-PCR and by a combination of western blot and immunohistochemical staining of colon tissue from mice in the DSS group. Furthermore, a significantly decreased miR-133a level was also detected in the colon tissue of mice in the DSS group by quantitative real-time (qRT)-PCR.
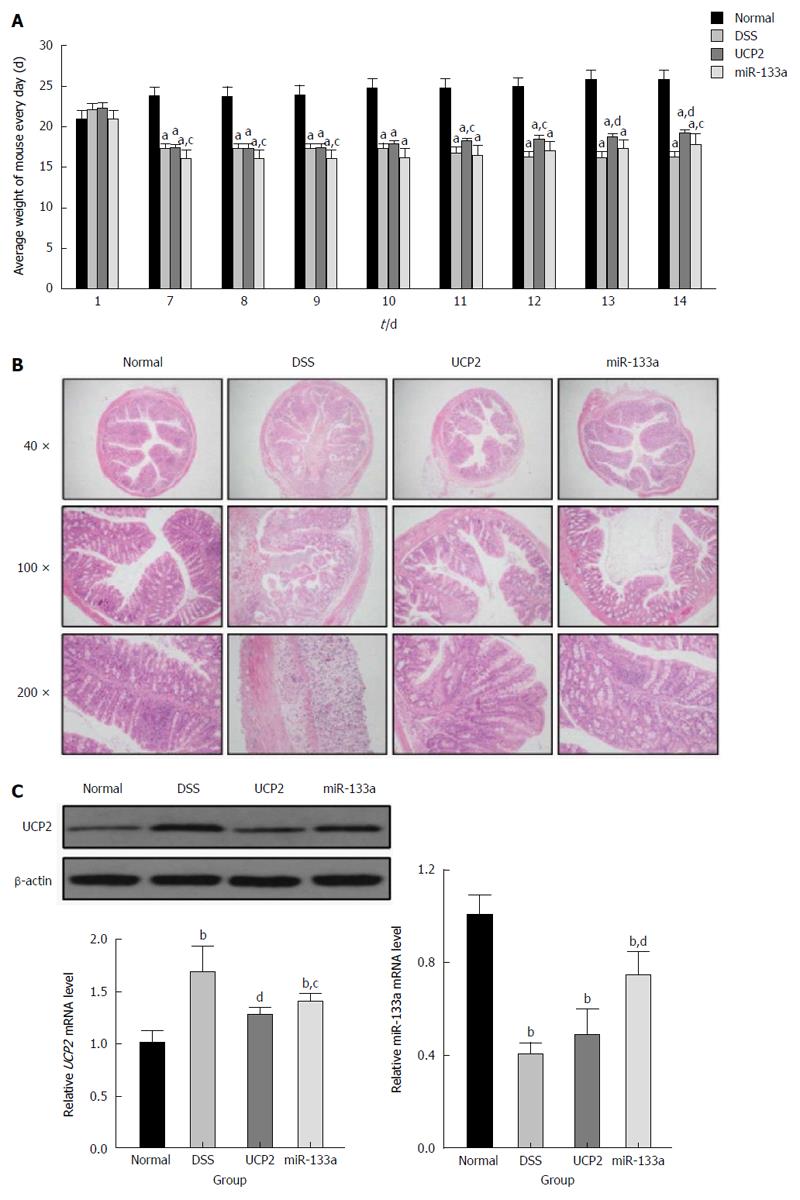
Compared with the DSS group, the severity of IBD was alleviated in the UCP2 and miR-133a groups after successful UCP2 down regulation and miR-133a over expression. Specifically, on day 7, the mice in the DSS, UCP2 and miR-133a groups all experienced significant weight loss, diarrhoea and rectal bleeding. However, as shown in Figure 3A, compared with the continuing weight loss and rectal bleeding in the DSS group, rectal bleeding ceased in the mice in the UCP2 and miR-133a groups on days 10 and 14, respectively. Mice in these two groups also experienced similar significant weight gain by day 14, although they did not recover to the level of the control group. A decrease in the DAI score was achieved (8, 6.8 and 7.2 for the DSS, UCP2 and miR-133a groups, respectively) on day 14, although the differences were not statistically significant. Moreover, compared with the DSS group, the colon tissue from mice in the UCP2 and miR-133a groups demonstrated intact tissue structure, well-organized glands as well as rare mucosal oedema, congestion and inflammatory cell infiltration. In addition, H&E staining revealed that disease alleviation was achieved in the miR-133a group, but the degree of alleviation was less than that in the UCP2 group (Figure 3B). Taken together, the targeting of both UCP2 and miR-133a was able to reduce the severity of IBD, but the targeting of UCP2 was more effective. More intriguingly, compared with the DSS group, the UCP2 level was decreased in the miR-133a group, in which the level of miR133a was up regulated, which indirectly supports the regulatory role of miR-133a on UCP2 (Figure 3C).
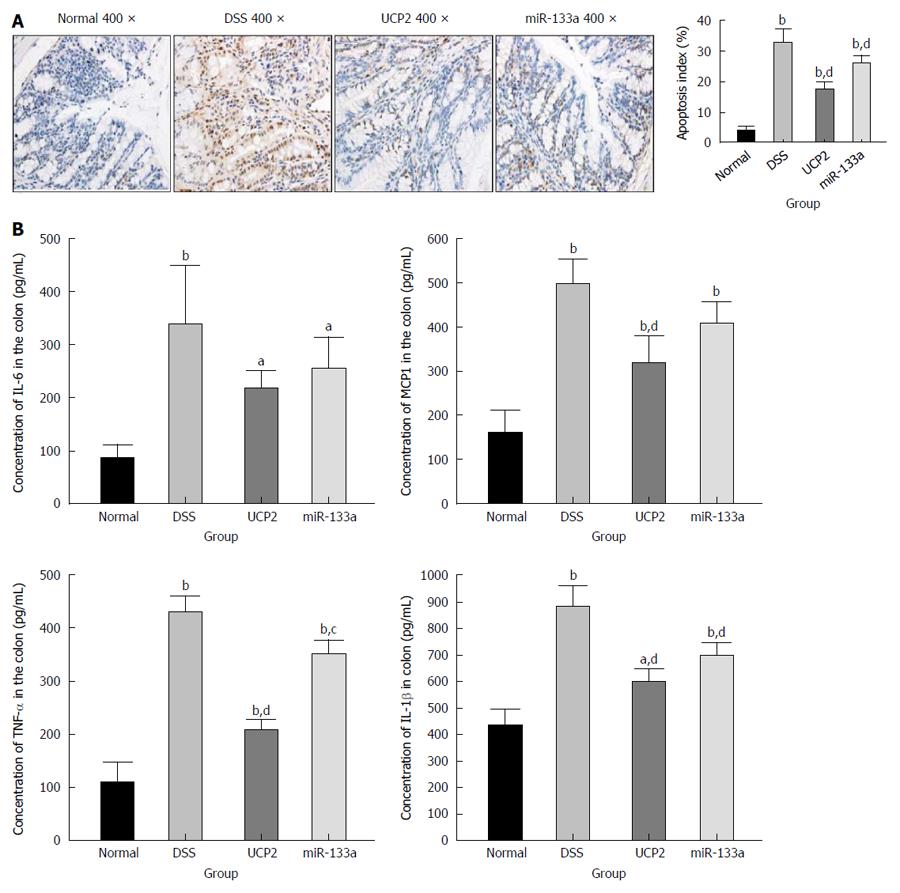
To explore the potential underlying mechanism of IBD alleviation, we further investigated the changes in apoptosis and other common inflammation-associated markers. We found a significantly increased level of apoptosis in the DSS group compared with the control group, as shown by TUNEL staining. Further UCP2 down regulation and miR-133a over expression could significantly inhibit apoptosis, as indicated by the notable decrease in the brown colour of the cells in Figure 4A. Furthermore, as shown in Figure 4B, the levels of inflammation-associated markers TNF-α, IL-1β, IL-6 and MCP1 were significantly increased in the DSS group compared with the control group. Additionally, although all four inflammation-associated markers showed a tendency to decrease, only the differences in the levels of TNF-α, IL-1β, and MCP1 in the UCP2 group and the levels of TNF-α and IL-1β in the miR-133a group reached statistical significance compared with the corresponding levels in the DSS group. Moreover, compared with the DSS group, the degree of declination of all markers was larger in the UCP2 group than in the miR-133a group, which indicates a greater therapeutic effect of UCP2 over miR-133a.
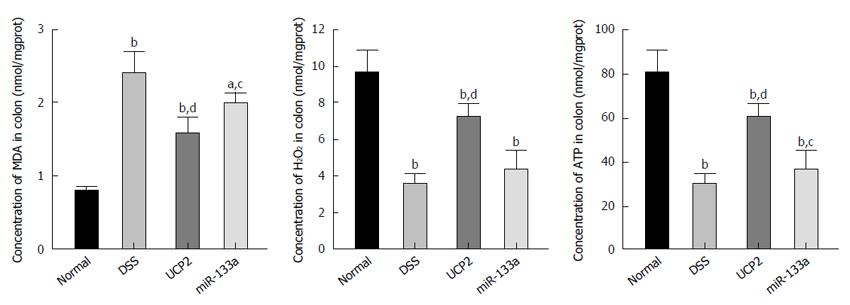
Oxidative stress-related markers include MDA and H2O2, while ATP is a vital representative of energy metabolism. In this study, we identified a significantly increased MDA level and decreased H2O2 and ATP levels in the DSS group compared with the control group. Nevertheless, after UCP2 down regulation or miR-133a over expression, the H2O2 and ATP levels were significantly increased, while the MDA level was significantly decreased compared with the DSS group. This may be attributed to the diminished uncoupling of oxidative phosphorylation as a result of the decreased UCP2 level (Figure 5).
Recently, IBD has become a global health problem as its prevalence has gradually increased, which has placed a heavy burden on the economy and has increased the suffering of patients. However, the unclear aetiology of IBD has made effective therapy a huge challenge. Therefore, the exploration of the potential underlying mechanism of IBD is of great clinical importance. The effect of miRNA, which is an important type of non-coding RNA, in IBD was summarized in 2016[25], while novel serum miRNA signatures used in the diagnosis of IBD were also proposed in mice[26] and humans[27]. Additionally, an interesting study established an association between miRNA polymorphisms and the risk of IBD[28]. All these studies support the importance of miRNA in IBD.
It is well acknowledged that UCPs play a pivotal role in various diseases due to their ability to influence energy metabolism, peroxidation and inflammation through the uncoupling of oxidative phosphorylation. A previous study found increased UCP2 expression in colon cancer and identified an association between the level of UCP2 and cancer metastasis[29]. Currently, the effects of specific miRNAs in the pathogenesis of IBD have not been frequently reported, and the role of miRNA-mRNA pathways in IBD has been reported even less frequently. After a literature review, we only found the following: the miR-346-TNF-α pathway influences Vitamin D metabolism[30], the miR-193a-3p-PepT1 pathway reduces intestinal inflammation[31], and the miR-132/miR-223-FOXO3a pathway increases the production of inflammatory cytokines[32]. Intriguingly, the effect of miR-133a in colonic epithelial cells and in experimental colitis was reported[33]. Considering the important role of UCP2 and the direct regulation of UCP2 by miR-133a[21,22,34], it is theoretically reasonable to explore the role of the miR-133a-UCP2 pathway in the pathogenesis of IBD.
In this study, we firstly reported increased UCP2 and decreased miR-133a levels in a successfully established animal model of IBD, which indicates the potential role of miR-133a and UCP2 in IBD. This result was in accordance with the result of a previous study, which supports the protective role of UCP2 in DSS-induced colitis[35]. Secondly, we found an inverse association between UCP2 and miR-133a, and more importantly, miR133a over expression could significantly decrease the UCP2 level. This finding indirectly verified the regulation of UCP2 by miR-133a at the post transcriptional level. Thirdly, with the use of RNA interference and gene over expression, we found that antagonizing both the UCP2 and miR-133a level could alleviate the severity of IBD.
To investigate the potential underlying mechanism and downstream targets of the miR-133a-UCP2 pathway, we selected typical inflammation-, oxidative- and energy metabolism-associated markers. Briefly, TNF-α, IL-1β, IL-6 and MCP1 are classical markers of inflammation with broad applications. TNF-α overproduction has been associated with mucosal damage in IBD, while anti-TNF-α therapy with infliximab is considered the representative biological therapy for IBD[36]. IL-1β and IL-6 belong to the interleukin family, and the immune response they mediate has been tightly linked with IBD progression[37]. MCP1 functions as a monocyte chemoattractant, and its A2518G polymorphism is related to IBD risk[38]. With respect to oxidative stress, MDA is an important product of lipid peroxidation, and its diagnostic value in CD has been reported[39]. H2O2 is mainly produced during the process of oxidative phosphorylation and is regarded as a universal mediator of the inflammatory process in the colon[40]. Finally, ATP is a basic energy form used in various pathophysiological processes, and its impaired production due to mitochondrial dysfunction has been identified in the intestines of patients with IBD[41]. Taken together, the results showed that increased apoptosis, inflammation, oxidative stress and ATP depletion were involved as downstream effectors of the miR-133a-UCP2 pathway.
This study has several limitations that should be acknowledged. First, whether the miR-133a-UCP2 pathway in an animal model could be generalized to humans requires further study. Second, it is better to use UCP2 knockout mice rather than RNAi to obtain more convincing data of the effects of UCP2 in IBD. Third, since novel studies have advocated the “miRNA sponge” effect of circRNA[42], the existence of circRNA as a regulator of miR-133a warrants more in depth studies. Fourth, although it is well known that increased UCP2 can enhance the uncoupling of oxidative phosphorylation, which results in a decrease in ATP and an increase in H2O2, the detailed mechanism of the miR-133a-UCP2 pathway on other inflammatory- and oxidative stress-associated markers is still unclear and requires further investigation. Finally, as distinct representatives of oxidative stress, H2O2 and MDA demonstrated inverse changes in their levels in the IBD animal model, which calls for further studies of the underlying mechanisms.
Overall, our study revealed the role of the miR-133a-UCP2 pathway in IBD and the potential downstream effectors, including inflammation-, oxidative stress- and energy metabolism-associated markers, which provide new clues in the pathogenesis of IBD and potential targets for disease therapy.
Inflammatory bowel disease (IBD) has been considered as a chronic and relapsing inflammatory disease that could affect any part of the intestine, with major two subtypes as Crohn’s disease and ulcerative colitis. The prevalence of IBD is increasing in developing and developed countries, making it a global health care problem and an interesting research area.
The mechanism of IBD remains vague; the involvement of genetic predisposition, immune response and environmental factors has been advocated. Currently, the effect of microRNA (miRNA) and uncoupling protein was intensively investigated for their respective effect in epigenomic regulation and uncoupling energy metabolism.
However, the specific miRNA-UCP pathway in the pathogenesis of IBD has not been reported hitherto. In this study, the authors investigated the role of the miR-133a-UCP2 pathway in the pathogenesis of IBD and to explore the potential downstream mechanisms with respect to inflammation, oxidative stress and energy metabolism.
The miR-133a-UCP2 pathway participates in IBD by altering downstream inflammation, oxidative stress and markers of energy metabolism, which provides novel clues and potential therapeutic targets for IBD.
miRNA belongs to a family of non-coding RNAs that are 19-25 nucleotides (nt) long. They are processed from double-stranded hairpin precursors, which are 70-100 nt in length, by the RNaseIII family member Dicer.
The present study shows an investigation of the miR-133a-UCP2 pathway in the pathogenesis of IBD. For this used an adequate methodology and experimental conditions. With the found results in this study the author concluded that miR-133a-UCP2 pathway participates of IBD influencing the inflammatory process and this pathway could be a potential therapeutic targets for IBD. The manuscript is clear and well written.
| 1. | Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361:2066-2078. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1967] [Cited by in RCA: 2254] [Article Influence: 132.6] [Reference Citation Analysis (10)] |
| 2. | Engel MA, Khalil M, Neurath MF. Highlights in inflammatory bowel disease--from bench to bedside. Clin Chem Lab Med. 2012;50:1229-1235. [PubMed] |
| 3. | Ouyang Q, Hu PJ, Qian JM, Zheng JJ, Hu RW. Consensus on the management of inflammatory bowel disease in China in 2007. J Dig Dis. 2008;9:52-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 4. | Zhang YZ, Li YY. Inflammatory bowel disease: pathogenesis. World J Gastroenterol. 2014;20:91-99. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 751] [Cited by in RCA: 1133] [Article Influence: 94.4] [Reference Citation Analysis (32)] |
| 5. | Mao R, Hu PJ. The Future of IBD Therapy: Where Are We and Where Should We Go Next? Dig Dis. 2016;34:175-179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 35] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 6. | Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858-862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2408] [Cited by in RCA: 2426] [Article Influence: 97.0] [Reference Citation Analysis (1)] |
| 7. | Ambros V. The functions of animal microRNAs. Nature. 2004;431:350-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7919] [Cited by in RCA: 8669] [Article Influence: 394.0] [Reference Citation Analysis (3)] |
| 8. | Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857-866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5705] [Cited by in RCA: 6084] [Article Influence: 304.2] [Reference Citation Analysis (0)] |
| 9. | Jin X, Chen YP, Kong M, Zheng L, Yang YD, Li YM. Transition from hepatic steatosis to steatohepatitis: unique microRNA patterns and potential downstream functions and pathways. J Gastroenterol Hepatol. 2012;27:331-340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 37] [Article Influence: 2.6] [Reference Citation Analysis (1)] |
| 10. | Jin X, Ye YF, Chen SH, Yu CH, Liu J, Li YM. MicroRNA expression pattern in different stages of nonalcoholic fatty liver disease. Dig Liver Dis. 2009;41:289-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 78] [Article Influence: 4.6] [Reference Citation Analysis (1)] |
| 11. | Moreno-Moya JM, Vilella F, Simón C. MicroRNA: key gene expression regulators. Fertil Steril. 2014;101:1516-1523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 121] [Article Influence: 9.3] [Reference Citation Analysis (3)] |
| 12. | Clark PM, Dawany N, Dampier W, Byers SW, Pestell RG, Tozeren A. Bioinformatics analysis reveals transcriptome and microRNA signatures and drug repositioning targets for IBD and other autoimmune diseases. Inflamm Bowel Dis. 2012;18:2315-2333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 46] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 13. | Paraskevi A, Theodoropoulos G, Papaconstantinou I, Mantzaris G, Nikiteas N, Gazouli M. Circulating MicroRNA in inflammatory bowel disease. J Crohns Colitis. 2012;6:900-904. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 207] [Article Influence: 14.8] [Reference Citation Analysis (2)] |
| 14. | Ghorpade DS, Sinha AY, Holla S, Singh V, Balaji KN. NOD2-nitric oxide-responsive microRNA-146a activates Sonic hedgehog signaling to orchestrate inflammatory responses in murine model of inflammatory bowel disease. J Biol Chem. 2013;288:33037-33048. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 80] [Article Influence: 6.2] [Reference Citation Analysis (1)] |
| 15. | Maharshak N, Shenhar-Tsarfaty S, Aroyo N, Orpaz N, Guberman I, Canaani J, Halpern Z, Dotan I, Berliner S, Soreq H. MicroRNA-132 modulates cholinergic signaling and inflammation in human inflammatory bowel disease. Inflamm Bowel Dis. 2013;19:1346-1353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 65] [Article Influence: 5.0] [Reference Citation Analysis (1)] |
| 16. | Olaru AV, Yamanaka S, Vazquez C, Mori Y, Cheng Y, Abraham JM, Bayless TM, Harpaz N, Selaru FM, Meltzer SJ. MicroRNA-224 negatively regulates p21 expression during late neoplastic progression in inflammatory bowel disease. Inflamm Bowel Dis. 2013;19:471-480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 53] [Article Influence: 4.1] [Reference Citation Analysis (1)] |
| 17. | Jin X, Xiang Z, Chen YP, Ma KF, Ye YF, Li YM. Uncoupling protein and nonalcoholic fatty liver disease. Chin Med J (Engl). 2013;126:3151-3155. [PubMed] |
| 18. | Jin X, Yang YD, Chen K, Lv ZY, Zheng L, Liu YP, Chen SH, Yu CH, Jiang XY, Zhang CY. HDMCP uncouples yeast mitochondrial respiration and alleviates steatosis in L02 and hepG2 cells by decreasing ATP and H2O2 levels: a novel mechanism for NAFLD. J Hepatol. 2009;50:1019-1028. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 33] [Article Influence: 1.9] [Reference Citation Analysis (1)] |
| 19. | Zhu H, Li YR. Oxidative stress and redox signaling mechanisms of inflammatory bowel disease: updated experimental and clinical evidence. Exp Biol Med (Maywood). 2012;237:474-480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 266] [Cited by in RCA: 348] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 20. | Yu X, Wieczorek S, Franke A, Yin H, Pierer M, Sina C, Karlsen TH, Boberg KM, Bergquist A, Kunz M. Association of UCP2 -866 G/A polymorphism with chronic inflammatory diseases. Genes Immun. 2009;10:601-605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 73] [Article Influence: 4.3] [Reference Citation Analysis (2)] |
| 21. | Bandyopadhyay S, Lane T, Venugopal R, Parthasarathy PT, Cho Y, Galam L, Lockey R, Kolliputi N. MicroRNA-133a-1 regulates inflammasome activation through uncoupling protein-2. Biochem Biophys Res Commun. 2013;439:407-412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 67] [Article Influence: 5.2] [Reference Citation Analysis (2)] |
| 22. | Chen X, Wang K, Chen J, Guo J, Yin Y, Cai X, Guo X, Wang G, Yang R, Zhu L. In vitro evidence suggests that miR-133a-mediated regulation of uncoupling protein 2 (UCP2) is an indispensable step in myogenic differentiation. J Biol Chem. 2009;284:5362-5369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 85] [Article Influence: 4.7] [Reference Citation Analysis (2)] |
| 23. | Fitzpatrick LR, Wang J, Le T. In vitro and in vivo effects of gliotoxin, a fungal metabolite: efficacy against dextran sodium sulfate-induced colitis in rats. Dig Dis Sci. 2000;45:2327-2336. [PubMed] |
| 24. | Yamada S, Koyama T, Noguchi H, Ueda Y, Kitsuyama R, Shimizu H, Tanimoto A, Wang KY, Nawata A, Nakayama T. Marine hydroquinone zonarol prevents inflammation and apoptosis in dextran sulfate sodium-induced mice ulcerative colitis. PLoS One. 2014;9:e113509. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 44] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 25. | Soubières AA, Poullis A. Emerging Biomarkers for the Diagnosis and Monitoring of Inflammatory Bowel Diseases. Inflamm Bowel Dis. 2016;22:2016-2022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 35] [Article Influence: 3.5] [Reference Citation Analysis (1)] |
| 26. | Viennois E, Zhao Y, Zhang M, Han M, Merlin D. O-016 YI Longitudinal Study of Circulating miRNA Biomarkers in Inflammatory Bowel Disease. Inflamm Bowel Dis. 2016;22 Suppl 1:S6. [PubMed] [DOI] [Full Text] |
| 27. | Hübenthal M, Hemmrich-Stanisak G, Degenhardt F, Szymczak S, Du Z, Elsharawy A, Keller A, Schreiber S, Franke A. Sparse Modeling Reveals miRNA Signatures for Diagnostics of Inflammatory Bowel Disease. PLoS One. 2015;10:e0140155. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 32] [Article Influence: 2.9] [Reference Citation Analysis (1)] |
| 28. | Zhu M, Li D, Jin M, Li M. Association between microRNA polymorphisms and the risk of inflammatory bowel disease. Mol Med Rep. 2016;13:5297-5308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 29. | Kuai XY, Ji ZY, Zhang HJ. Mitochondrial uncoupling protein 2 expression in colon cancer and its clinical significance. World J Gastroenterol. 2010;16:5773-5778. [PubMed] |
| 30. | Chen Y, Du J, Zhang Z, Liu T, Shi Y, Ge X, Li YC. MicroRNA-346 mediates tumor necrosis factor α-induced downregulation of gut epithelial vitamin D receptor in inflammatory bowel diseases. Inflamm Bowel Dis. 2014;20:1910-1918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 88] [Article Influence: 7.3] [Reference Citation Analysis (1)] |
| 31. | Dai X, Chen X, Chen Q, Shi L, Liang H, Zhou Z, Liu Q, Pang W, Hou D, Wang C. MicroRNA-193a-3p Reduces Intestinal Inflammation in Response to Microbiota via Down-regulation of Colonic PepT1. J Biol Chem. 2015;290:16099-16115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 72] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 32. | Kim HY, Kwon HY, Ha Thi HT, Lee HJ, Kim GI, Hahm KB, Hong S. MicroRNA-132 and microRNA-223 control positive feedback circuit by regulating FOXO3a in inflammatory bowel disease. J Gastroenterol Hepatol. 2016;31:1727-1735. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 48] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 33. | Law IK, Pothoulakis C. MicroRNA-133α regulates neurotensin-associated colonic inflammation in colonic epithelial cells and experimental colitis. RNA Dis. 2015;2:pii: e472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 34. | Yuan Y, Yao YF, Hu SN, Gao J, Zhang LL. MiR-133a Is Functionally Involved in Doxorubicin-Resistance in Breast Cancer Cells MCF-7 via Its Regulation of the Expression of Uncoupling Protein 2. PLoS One. 2015;10:e0129843. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 35. | Zhang H, Kuai XY, Yu P, Lin L, Shi R. Protective role of uncoupling protein-2 against dextran sodium sulfate-induced colitis. J Gastroenterol Hepatol. 2012;27:603-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 15] [Article Influence: 1.1] [Reference Citation Analysis (3)] |
| 36. | Komaki Y, Komaki F, Sakuraba A, Cohen R. Approach to Optimize Anti-TNF-α Therapy in Patients With IBD. Curr Treat Options Gastroenterol. 2016;14:83-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 1.5] [Reference Citation Analysis (1)] |
| 37. | Magyari L, Kovesdi E, Sarlos P, Javorhazy A, Sumegi K, Melegh B. Interleukin and interleukin receptor gene polymorphisms in inflammatory bowel diseases susceptibility. World J Gastroenterol. 2014;20:3208-3222. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 28] [Cited by in RCA: 30] [Article Influence: 2.5] [Reference Citation Analysis (2)] |
| 38. | Li YW, Yang CQ, Xiao YL, Li J, Xie CX, Zhang SH, Yu Q, Wang HL, Lu WM, Chen MH. The -A2518G polymorphism in the MCP-1 gene and inflammatory bowel disease risk: A meta-analysis. J Dig Dis. 2015;16:177-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 39. | Boehm D, Krzystek-Korpacka M, Neubauer K, Matusiewicz M, Paradowski L, Gamian A. Lipid peroxidation markers in Crohn’s disease: the associations and diagnostic value. Clin Chem Lab Med. 2012;50:1359-1366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 27] [Article Influence: 1.9] [Reference Citation Analysis (1)] |
| 40. | Te Velde AA, Pronk I, de Kort F, Stokkers PC. Glutathione peroxidase 2 and aquaporin 8 as new markers for colonic inflammation in experimental colitis and inflammatory bowel diseases: an important role for H2O2? Eur J Gastroenterol Hepatol. 2008;20:555-560. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 76] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 41. | Novak EA, Mollen KP. Mitochondrial dysfunction in inflammatory bowel disease. Front Cell Dev Biol. 2015;3:62. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 187] [Article Influence: 17.0] [Reference Citation Analysis (1)] |
| 42. | Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science. 2013;340:440-441. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 326] [Cited by in RCA: 424] [Article Influence: 32.6] [Reference Citation Analysis (0)] |
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
P- Reviewer: Coelho T, Lakatos PL, Witaicenis A S- Editor: Yu J L- Editor: A E- Editor: Wang CH