Published online Nov 14, 2016. doi: 10.3748/wjg.v22.i42.9346
Peer-review started: April 6, 2016
First decision: May 30, 2016
Revised: August 3, 2016
Accepted: September 14, 2016
Article in press: September 14, 2016
Published online: November 14, 2016
Processing time: 226 Days and 5.8 Hours
To evaluate the pathogenic role of toll-like receptor (TLR) gene polymorphisms in patients with non-alcoholic fatty liver disease (NAFLD).
Two hundred and fifty subjects (NAFLD = 200, healthy volunteers = 50) underwent polymerase chain reaction and restriction fragment length polymorphism to assess one polymorphism in the toll-like receptor 2 (TLR2) gene (A753G), two polymorphisms in the TLR4 gene (TLR4 Asp299Gly and Thr399Ile allele), and two polymorphisms in the cluster of differentiation 14 (CD14) (C-159T and C-550T) gene, a co-receptor of TLR4. Association of TLR gene polymorphisms with NAFLD and its severity was evaluated by genetic models of association.
On both multiplicative and recessive models of gene polymorphism association, there was significant association of CD14 C (-159) T polymorphism with NAFLD; patients with TT genotype had a 2.6 fold increased risk of developing NAFLD in comparison to CC genotype. There was no association of TLR2 Arg753Gln, TLR4 Asp299Gly, Thr399Ile, and CD14 C (-550) T polymorphisms with NAFLD. None of the TLR gene polymorphisms had an association with histological severity of NAFLD.
Patients with CD14 C (-159) T gene polymorphism, a co-receptor of TLR4, have an increased risk of NAFLD development.
Core tip: Our study demonstrated that non-alcoholic fatty liver disease (NAFLD) patients with TT genotype of C (-159) T polymorphism in cluster of differentiation 14 promoter gene have a higher risk of NAFLD development. However, this polymorphism did not affect liver disease severity. We found no association of toll-like receptor (TLR) 2 ARG753, TLR4 (Asp299Gly), TLR4 (Thr399Ile), and CD 14 C/T 550 polymorphisms with the risk of NAFLD development.
- Citation: Kapil S, Duseja A, Sharma BK, Singla B, Chakraborti A, Das A, Ray P, Dhiman RK, Chawla Y. Genetic polymorphism in CD14 gene, a co-receptor of TLR4 associated with non-alcoholic fatty liver disease. World J Gastroenterol 2016; 22(42): 9346-9355
- URL: https://www.wjgnet.com/1007-9327/full/v22/i42/9346.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i42.9346
Non-alcoholic fatty liver disease (NAFLD) is a spectrum condition, ranging from simple steatosis to its progressive form of non-alcoholic steatohepatitis (NASH), and has emerged as an important cause of the otherwise unexplained increase in hepatic transaminases, cryptogenic cirrhosis, and cryptogenic hepatocellular carcinoma (HCC)[1-3]. Familial studies and inter-ethnic variation in susceptibility to the disease suggest that genetic factors are important in the occurrence and determining the risk of progressive NAFLD[4]. Studies suggests a strong association of NAFLD with patatin-like phospholipase domain containing 3 gene polymorphism[5-7], as well as inconclusive association with apolipoprotein C-III[8,9] and human hemochromatosis gene mutations[10].
Emerging data suggest the role of small intestinal bacterial overgrowth (SIBO) and gut-derived endotoxins in the pathogenesis of NAFLD via inducing the release of proinflammatory cytokines, including tumor necrosis factor-α (TNF-α) from hepatic Kupffer cells.
Toll-like receptors (TLRs) are the most important family of pattern recognition receptors[11,12]; they are the sensors for recognizing bacterial and viral components, such as lipopolysaccharides, bacterial DNA, and peptidoglycan. Of the various TLRs, toll-like receptor 2 (TLR2), TLR4, the co-receptor cluster of differentiation 14 (CD14), and TLR9 have been well studied in the pathogenesis of NAFLD. In addition to animal studies, two human studies[13,14] have also suggested a higher risk of developing NASH in patients with NAFLD that possess CD14 gene polymorphism.
The aim of our study was to evaluate the role of TLR polymorphisms in the causation and severity of NAFLD.
This was a case-controlled study where 250 subjects [NAFLD (n = 200, males = 122, mean age 38.27± 10.3 years) and healthy volunteers (HVs) (n = 50, males = 38, mean age 36.56 ± 4.2 years] were enrolled after informed consent. The study had the approval of the Institute’s Ethics Committee. All subjects were enrolled prospectively, with the exception of 28 patients with biopsy-proven NAFLD; their records were retrieved from the existing database and were called again for consent and fresh sampling for TLR polymorphisms. Patients with NAFLD were recruited as per the standard inclusion criteria. Fifty age and gender matched HVs were recruited, as per the guidelines from the Indian Council of Medical Research, and they all had normal liver function tests, normal fasting plasma glucose, normal lipid profile, and no evidence of fatty liver on ultrasound.
Inclusion criteria for NAFLD: (1) age greater than 13 years; (2) non-alcoholic individuals, defined as either total abstainers or individuals who consumed less than 20 g of alcohol per day. History of alcohol consumption was confirmed by two family members of the patient; (3) raised serum transaminases more than one and a half times the upper limit of normal for at least 3 mo; (4) ultrasound showing features of steatosis; (5) negative viral markers (HBsAg/Anti HCV), negative autoimmune markers [antinuclear antibody (ANA), anti-smooth muscle antibody, anti-liver kidney microsomal antibody, and anti-mitochondrial antibody (AMA)]; (6) normal ceruloplasmin/negative Kayser-Fleischer rings; (7) normal iron work up [serum iron, total iron binding capacity (TIBC), ferritin, and transferrin saturation]; and (8) liver biopsy consistent with NAFLD (60 cases where liver biopsy was performed).
Exclusion criteria for NAFLD: (1) pregnant females; (2) patients with history of drug intake likely to cause NAFLD (e.g., corticosteroids, methotrexate, and tamoxifen); (3) jejunoileal bypass or extensive small bowel resection; (4) total parenteral nutrition at the time of liver biopsy; and (5) clinical, imaging, or liver biopsy features of liver cirrhosis.
Height was determined with a measuring tape to the nearest cm. Subjects were requested to stand upright without shoes with their heels tight against the wall and eyes directed forward. Weight was measured in kilograms (kg) with a traditional spring balance, which was kept on a firm horizontal surface and the scale checked every day. Body Mass Index (BMI) was calculated using the formula: weight (kg)/height (m2). Waist circumference was taken as the average of two measurements taken after inspiration and after expiration at the midpoint between the lowest rib and the iliac crest. Hip circumference was taken at the level of greater trochanter and waist-to-hip ratio was defined as the ratio of the waist and hip circumference.
Patients were classified as being lean, overweight, class I obese, class II obese, or centrally obese as per the Asian Pacific criteria (lean: BMI = 18-23 kg/m2; overweight: > 23 < BMI < 25 kg/m2; class I obesity: BMI ≥ 25-30 kg/m2; class II obesity: BMI > 30 kg/m2; central obesity: waist circumference > 90 cm in males and > 80 cm in females)[15,16].
All patients with NAFLD underwent detailed baseline investigations, with selective investigations performed with HVs. In patients undergoing liver biopsy (32 = prospective, with 28 retrieved from the existing database), laboratory parameters were measured before the procedure. Serum bilirubin, aspartate aminotransferases, alanine aminotransferases (ALT) (Diagnosticum Rt., Budapest, Hungary), alkaline phosphatase (Reckon Diagnostics, Baroda, India), albumin, globulin (Far Diagnostics, Verona, Italy), total cholesterol, triglycerides (TG), high-density lipoprotein (HDL) cholesterol, and low-density lipoprotein (LDL) cholesterol (Roche Diagnostics, Indianapolis, United States) were determined as per the standard methodology. A fasting plasma glucose of > 126 mg/dL on more than one occasion or a random plasma glucose of > 200 mg/dL in a symptomatic patient was defined as diabetes mellitus. Fasting plasma glucose of > 110 and < 126 mg/dL was defined as impaired fasting glucose and 2 h post-prandial plasma glucose between 140 and 200 mg/dL as impaired glucose tolerance. Lipid profile was determined in all patients and serum cholesterol > 200 mg/dL, HDL < 40 mg/dL in males and < 50 mg/dL in females, LDL > 130 mg/dL, and serum TG > 150 mg/dL was taken as abnormal. Serum iron and TIBC were measured by the colorimetric method and serum ferritin was measured using an enzyme immunoassay kit (Orgentec Diagnostika GmbH, Germany). Virological markers, such as HBV (HBsAg, HBeAg), and HCV (anti-HCV), and auto immune markers including ANA anti-smooth muscle antibodies, anti-liver-kidney microsomes, and AMA were determined using enzyme-linked immunosorbent assay (ELISA).
All patients with NAFLD and CVH, as well as the HVs, were subjected to an abdominal ultrasound to detect and grade the degree of hepatic steatosis.
Metabolic syndrome was defined by the presence of ≥ 3 out of 5 modified adult treatment panel III criteria, including modified abnormal waist as per the Asia Pacific criteria, FPG > 110 mg/dL or known diabetes, hypertension (blood pressure ≥ 130/85 mmHg or on antihypertensive drugs), serum TG > 150 mg/dL, and HDL < 40 mg/dL in males or < 50 mg/dL in females[17].
Sixty patients with NAFLD (32 recruited prospectively and 28 retrieved from database) were histologically assessed for degree of hepatic steatosis and fibrosis, and then divided into NASH, borderline NASH and no-NASH, as per the NAFLD activity score (NAS) given by the Nonalcoholic Steatohepatitis Clinical Research Network[18].
Polymorphisms in the genes encoding for receptors (TLR2, TLR4, and CD14) were detected by polymerase chain reaction (PCR) and restriction fragment length polymorphism (RFLP) to assess one polymorphism in the TLR2 gene (Arg753Gln), two in the TLR4 gene (TLR4 Asp299Gly and Thr399Ile allele), and two polymorphisms in the CD14 gene (C-159T and C-550T).
PBMC and genomic DNA isolation: PBMC were isolated from whole blood samples using Ficoll-Hypaque differential density gradient centrifugation procedures. Genomic DNA was extracted from cells in the buffy coat using QIAamp DNA mini kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s instructions. The extracted DNA was used for detecting polymorphisms in different genes via PCR and RFLP.
PCR: Genomic DNA was amplified at specific regions containing the polymorphic sites using specific primer pairs flanking the respective polymorphic sites[19,20]. A list of the PCR’s conducted primer sequences, reaction conditions, and product sizes are given in Table 1.
| Polymorphism site | Primer sequence (5'-3') | PCR conditions | Product size (bp) |
| TLR4 (Asp299gly) | (1) 95 °C for 10 min | 249 | |
| (2) 35 cycles of : | |||
| F: GATTAGCATACTTAGACTACTACCTCCATG | 94 °C for 1 min | ||
| R: GATCAACTTCTGAAAAAGCATTCCCAC | 61.5 °C for 1 min | ||
| 72 °C for 1 min | |||
| (3) 72 °C for 7 min | |||
| TLR4 (Thr399Ile) | (1) 95 °C for 10 min | 406 | |
| (2) 35 cycles of : | |||
| F: GGTTGCTGTTCTCAAAGTGATTTTGGGAGAA | 94 °C for 1 min | ||
| R: CCTGAAGACTGGAGAGTGAGTTAAATGCT | 64.8 °C for 1 min | ||
| 72 °C for 1 min | |||
| (3) 72 °C for 10 min | |||
| TLR2 (Arg753Gln) | (1) 95 °C for 10 min | 289 | |
| (2) 35 cycles of : | |||
| F: 5’-CCTTCAAGTTGTGTCTTCATAAG-3’ | 94 °C for 1 min | ||
| R: 5’-GGCCACTCCAGGTAGGTCTT-3’ | 58.6 °C for 1 min | ||
| 72 °C for 1 min | |||
| (3) 72 °C for 10 min | |||
| CD14 (-550C/T) | (1) 95 °C for 10 min | 368 | |
| (2) 32 cycles of : | |||
| F: GGAAGGGGGAATTTTTCTTTAGGC | 94 °C for 1 min | ||
| R: -GGCAGTGTCCTGATGACTCA | 59.8 °C for 1 min | ||
| 72 °C for 1 min | |||
| (3) 72 °C for 7 min | |||
| CD14 (-159C/T) | (1) 95 °C for 10 min | 296 | |
| (2) 35 cycles of : | |||
| F: ATCATCCTTTTCCCACACC | 94 °C for 40 s | ||
| R: AACTCTTCGGCTGCCTCT | 61 °C for 40 s | ||
| 72 °C for 40 s | |||
| (3) 72 °C for 10 min |
RFLP: PCR products were digested with appropriate restriction endonucleases to differentiate different genotypes. A list of restriction enzymes, incubation temperatures and times, and RFLP patterns of different genotypes are given in Table 2.
| Polymorphic site | Restriction enzyme | Incubation temperature and duration | Genotype and restriction fragment pattern (bp) |
| (units used) | |||
| TLR4 (Asp299gly) | NcoI | 37 °C for 16 h | AA: 249 |
| AG: 249, 223, 26 | |||
| GG: 223, 26 | |||
| TLR4 (Thr399Ile) | HinfI | 37 °C for 8 h | CC: 406 |
| CT: 406, 377, 29 | |||
| TT: 377, 29 | |||
| TLR2 (Arg753Gln) | AciI | 37 °C for 16 h | GG: 252 and 37 |
| GA: 252, 37, 289 | |||
| AA:289 | |||
| CD14 (-550C/T) | HaeIII | 37 °C for 12 h | CC: 23, 236, 109 |
| CT: 23, 236, 109, 259 | |||
| TT: 109, 259 | |||
| CD14 (-159C/T) | HaeIII | 37 °C for 16 h | CC: 141, 154 |
| CT: 141, 154, 296 | |||
| TT: 296 |
PCR products for all genotypes of all genes were validated commercially by DNA sequencing.
Serum levels of various cytokines viz., adiponectin, TNF-α, and interleukin-1β were estimated by (ELISA, Ray Biotech, Norcross GA.). Homeostasis Model Assessment for Insulin Resistance (HOMA-IR) was calculated as the product of fasting insulin (μU/mL) (Roche Diagnostics GmbH, Mannheim, Germany) and fasting plasma glucose (mmol/L) divided by 22.5. An absolute value of HOMA-IR > 1.64 was taken as abnormal[21].
Data were analyzed for comparison between the two groups using SPSS version 15 for Windows (SPSS Inc. Chicago IL United States). Skewed clinical and biochemical data were expressed as a median (interquartile range), whereas normally-distributed variables were expressed as mean ± SD. For continuous data, groups were tested for normal distribution using the Kolmogorov-Smirnov test (K-S test). For skewed data, the Mann-Whitney U test was applied for comparison between the two groups. For categorical data, the Chi-square test or Fischer exact test was applied. Genotypic association and the odds ratio with 95%CI were estimated by binary or multinomial logistic regression analysis. Haplotype analysis was carried out for TLR4 and CD14 gene using SHEsis software. In all cases, a P value less than 0.05 was considered significant.
Models and measures of association of gene polymorphisms: Recessive and dominant models were applied to determine the association of these polymorphisms with the recessive and dominant alleles; a recessive model indicates that two copies of allele A are required for a γ-fold increase in disease risk, while a dominant model indicates that either one or two copies of allele A are required for a γ-fold increase in disease risk[22,23]. Patients with and without significant polymorphisms were compared to assess the difference between the two groups.
The demographic, anthropometric, and biochemical characteristics of the two groups (NAFLD and HVs) are summarized in Table 3.
| Parameters | NAFLD (n = 200) | HVs (n = 50) | P value |
| Mean age (yr) | 38.27 ± 10.3 | 36.56 ± 4.2 | 0.218 |
| Gender | 122 M/78 F | 38 M/12 F | |
| Mean BMI (kg/m2) | 27.16 ± 4.7 | 22.1 ± 1.2 | 0.0002 |
| Lean | 35 (17.5) | 46 (92) | 0.0001 |
| Overweight | 39 (19.5) | 3 (6) | 0.02 |
| Class I obesity | 87 (43.5) | 1 (2) | 0.0001 |
| Class II obesity | 39 (19.5) | 0 (0) | 0.003 |
| Waist (cm) | 91.20 ± 9.5 | 78.12 ± 4.6 | 0.0001 |
| Hip (cm) | 91.32 (89.90-93.94) | 89.18 (86.18-93.98) | 0.3 |
| Waist/hip ratio | 0.99 (0.98-1.01) | 0.93 (0.89-0.97) | 0.016 |
| Central obesity | 135 (67.5) | 0 (0) | 0.0001 |
| Mean AST (IU/L) | 60.27 (46.0-78.7) | 22.9 (18.75-29.50) | 0.0001 |
| Mean ALT (IU/L) | 88.04 (68.9-118.9) | 24 (17-29.5) | 0.0001 |
| Mean fasting sugar (mg/dL) | 94.95 (87-105.8) | 83.50 (75-89.25) | 0.0001 |
| Diabetes mellitus | 29 (14.5) | 0 (0) | 0.005 |
| Mean HDL (mg/dL) | 43 (38-48.9) | 52 (47.5-56) | 0.0001 |
| Mean Triglyceride (mg/dL) | 156 (126-200.2) | 102 (93-127.5) | 0.0001 |
| Hypertension | 48 (24) | 0 (0) | 0.0001 |
Patients with NAFLD had higher BMI, higher overall and central obesity, elevated liver enzymes, and a higher prevalence of diabetes mellitus and dyslipidemia in comparison to HVs (Table 3). In the NAFLD group, 78 patients (39%) displayed the presence of metabolic syndrome.
TLR2 Arg753Gln polymorphism: A PCR product size of 289 bp was subjected to restriction digestion with ACI 1 restriction enzyme at 37 °C to get the respective band size of 289 bp, 252 bp, and 37 bp, depending upon the deletion of restriction sites by mutant allele. GG was found to be the predominant genotype (89.5%), followed by GA (7.5%) and AA (3%) in NAFLD patients and GG (90%), GA (10%) and AA (0%) in HVs. There was no difference in GG [179/200 (89.5%) vs 45/50 (90%), P = 0.91] GA [15 (7.5%) vs 5 (10%), P = 0.56] and AA [6/200 (3%) vs 0/50 (0%), P = 0.21] genotypes in either study group. No association of TLR2 Arg753Gln polymorphism with NAFLD was found on multiplicative, dominant, or recessive models of analysis.
TLR4 Asp299Gly polymorphism: TLR4 Asp299Gly polymorphism in exon 4 is an A/G polymorphism that creates a recognition site for restriction enzyme NcoI. Amplification with primer pairs yielded a 249 bp fragment. Respective band sizes of 249 bp, 223 bp, and 26 bp, depending upon the restriction sites created by mutant allele, were obtained. Digestion with enzyme gave a 249 bp fragment in the presence of AA genotype, three bands of 249 bp, 223 bp, and 26 bp in the presence of AG genotype, and two bands of 223 bp and 26 bp fragments in presence of GG genotype. The genotype frequency of AA, AG, and GG were 79%, 17%, and 4%, respectively, in the NAFLD group and 82%, 18%, and 0%, respectively, in the HV group. Distribution was in accordance with Hardy-Weinberg equilibrium. The AA, AG, and GG genotype were not statistically different in NAFLD compared to HVs (P = 0.15). No association of TLR4 Asp299Gly polymorphism with NAFLD was found on multiplicative, dominant, or recessive models of analysis.
TLR4 Thr399Ile polymorphism: Our findings demonstrated a PCR product size of 406bp for TLR4 Thr399Ile gene. After restriction digestion of PCR product with HinfI restriction enzyme at 37 °C, respective band sizes of 406 bp, 377 bp, and 29 bp were obtained depending upon the creation of restriction sites by mutant allele. CC was found to be the predominant genotype (83%), followed by CT (12.5%) and TT (4.5%) in NAFLD patients vs CC (84%), and by CT (16%) and TT (0%) in HVs.
Among TLR4 Thr399Ile polymorphisms, the frequency of CC [166/200 (83%) vs 42/50 (84%), P = 0.86], CT [25 (12.5%) vs 8 (16%), P = 0.51], and TT [(9 (4.5%) vs 0 (0%), P = 0.12] genotypes were not different among patients with NAFLD and HVs. There was no difference in T allele frequency between the NAFLD or HV groups (10.75% vs 8%, P = 0.41), and no association of TLR4 Thr399Ile polymorphism with NAFLD was found on multiplicative, dominant, or recessive models of analysis.
Haplotype analysis for TLR4 Asp299Gly and TLR4 Thr399Ile polymorphism: A haplotype comprised of a combination of alleles present on the same chromosome. Haplotype analysis revealed that haplotypes AC, AT, GC, and GT were not associated with an increased risk of NAFLD.
CD14 C (-550) T polymorphism: CD14 C (-550) T is a C/T polymorphism that creates a recognition site for restriction enzyme Hae111. Amplification with primer pairs yielded a 368 bp fragment. Respective band sizes of 259, 236, 109, and 23 bp were obtained depending upon the deletion of restriction sites by mutant allele. Digestion with enzyme gave 23, 236, and 109 fragments in the presence of CC genotype, four bands of 23, 236, 109, and 259 in the presence of CT genotype, and two bands of 109 bp and 259 bp fragments in the presence of TT genotype. The genotype frequency of CC, CT, and TT was 60%, 32.5%, and 7.5%, respectively, in the NAFLD group and 68%, 28%, and 4%, respectively, in the HV group (Table 3). Distribution was in accordance with Hardy-Weinberg equilibrium. The TT genotype was not significantly overrepresented in NAFLD (P = 0.322) compared to HVs.
Among CD14 C (-550) T polymorphisms, the frequency of CC [120/200 (60%) vs 34/50 (68%)], CT [65 (32.5%) vs 14 (28%), OR = 1.3 (0.6-2.6), P = 0.43], and TT [(15 (7.5%) vs 2 (4%), OR = 2.1 (0.4-9.7), P = 0.322] genotypes were not different between patients with NAFLD and HVs. There was no difference in T allele frequency between the NAFLD or HV groups (23.75% vs 18%, P = 0.21) and no association of CD14 C (-550) T polymorphism with NAFLD was found on multiplicative, dominant, or recessive models of analysis (Table 4).
| CD14 C (-550) T | CC | CT | TT |
| genotypes | |||
| Cases (n = 200) | 120 (60) | 65 (32.5) | 15 (7.5) |
| Control (n = 50) | 34 (68) | 14 (28) | 2 (4) |
| OR | 1 | 1.3 (0.6-2.6) | 2.1 (0.4-9.7) |
| P = 0.43 | P = 0.322 | ||
| Genetic models | |||
| Multiplicative model | Allele C | Allele T | |
| Cases | 305 (76.25) | 95 (23.75) | P = 0.21 |
| Control | 82 (82) | 18 (18) | OR = 1.41 |
| CI: 0.81-2.48 | |||
| Dominant model | CC | CT + TT | |
| Cases | 120 (60) | 80 (40) | P = 0.29 |
| Control | 34 (68) | 16 (32) | OR = 0.7 |
| CI: 0.3-1.3 | |||
| Recessive model | TT | CC + CT | |
| P = 0.37 | |||
| OR = 1.9 | |||
| CI: 0.4-8.8 | |||
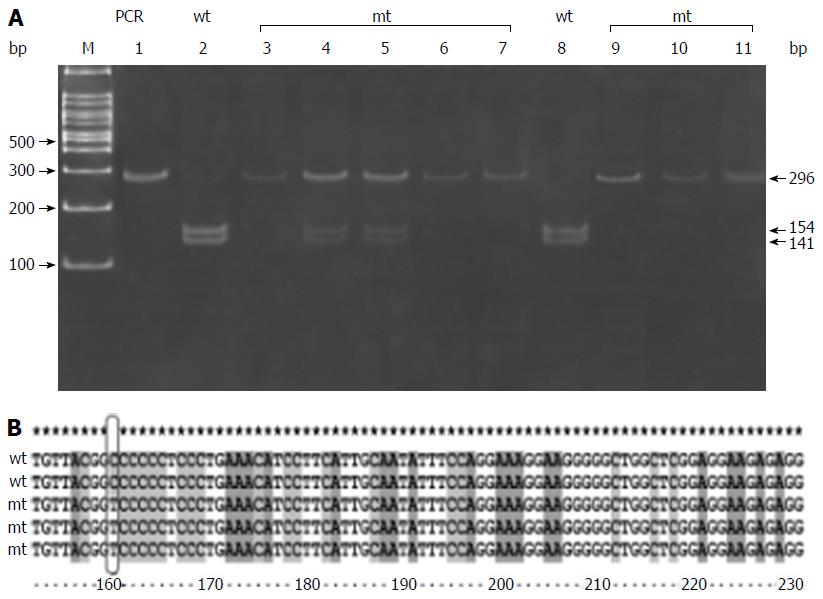
CD14 C (-159) T polymorphism is a C/T polymorphism that creates a recognition site for restriction enzyme Hae111. Amplification with primer pairs yielded a 296 bp fragment. Respective band sizes 259, 236, 109, and 23 bp were obtained depending upon the deletion of restriction sites by mutant allele. Digestion with enzyme gave a 296 bp fragment in the presence of CC genotype, three bands of 296, 154, and 141 in the presence of CT genotype, and two bands of 154 bp and 141 bp fragments in the presence of TT genotype (Figure 1A and Table 5).
| CD14 C (-159) T | CC | CT | TT |
| Genotypes | |||
| Cases (n = 200) | 36 (18) | 70 (35) | 94 (47) |
| Control (n = 50) | 10 (20) | 30 (60) | 10 (20) |
| OR | 1 | 0.6 (0.2-1.4) | 2.6 (1-6.7) |
| P = 0.29 | P = 0.043 | ||
| Genetic models | |||
| Multiplicative model | Allele C | Allele T | |
| Cases | 142 (35.50) | 258 (64.50) | P = 0.007 |
| Control | 50 (50) | 50 (50) | OR = 0.8 |
| CI: 1.1-2.8 | |||
| Dominant model | CC | CT + TT | |
| Cases | 36 (18) | 164 (82) | P = 0.74 |
| Control | 10 (20) | 16 (32) | OR = 0.7 |
| CI: 0.4-1.9 | |||
| Recessive model | TT | CC + CT | |
| Cases | 94 (47) | 106 (53) | P = 0.0005 |
| Control | 10 (20) | 40 (80) | OR = 3.5 |
| CI: 1.6-7.4 -8.8 | |||
There was no difference in the CC [36 (18%) vs 10 (20%), P = 0.74] genotype between patients with NAFLD and HVs. Even though there was difference in the CT [70/200 (35%) vs 30 (60%), P = 0.001] genotypes between patients with NAFLD and HVs, the CT genotype was not associated with an increased risk of NAFLD (Table 2). Distribution was in accordance with Hardy-Weinberg equilibrium. The TT genotype [94/200 (47%) vs 10/50 (20%), P = 0.0005] and T allele frequency (64% vs 50%, P = 0.007) were significantly higher among patients with NAFLD than HVs, with significant association of C (-159) T polymorphism with NAFLD on multiplicative (P = 0.007) and recessive models (P = 0.0005). The risk of developing NAFLD with the TT genotype was 2.6 fold higher than in CC genotypes (Table 5).
The DNA sequencing data for CD14 C (-159) T polymorphism confirmed our PCR-RFLP findings, wherein we found that C allele is replaced by T allele (Figure 1B). The results of PCR-RFLP of all other studied polymorphisms were comprised of 5 samples of each genotype in patients with NAFLD, which was also confirmed by DNA sequencing (5 homozygous wild, 5 heterozygous, and 5 homozygous variant) (data not shown).
Haplotype analysis for CD14 C (-159) T and CD14 C (-550) T polymorphisms: Haplotype analysis revealed that haplotype TC had a significantly higher (P = 0.0002) frequency in patients with NAFLD in comparison to HVs, with an odds ratio of 2.3; 95%CI: 1.4-3.7. Analysis of individual haplotypes in the CD14 gene as a method of determining the risk of developing NAFLD revealed that the TC haplotype was more frequently seen in NAFLD. None of the other haplotypes showed any association with the risk of developing NAFLD (Table 6).
| Haplotypes | NAFLD (frequency) | HVs (frequency) | P value | OR (95%CI) |
| (Fisher’s exact test) | ||||
| CC | 23.4% | 50% | 1.58E-007 | 0.3 (0.19-0.48) |
| TC | 52.3% | 32% | 0.0002 | 2.3 (1.4-3.7) |
| TT | 12.2% | 18% | 0.12 | 0.6 (0.3-1.1) |
| CT | 12.1% | 0% | 0.0002 | Undefined |
Role of CD14 C (-159) T polymorphism in determining the severity of NAFLD: Owing to the significant association between CD14 C (-159) T polymorphism and NAFLD, we compared different parameters amongst NAFLD patients with (n = 94) and without this polymorphism (n = 106). There was a significant difference in serum ALT [95 (72-130) IU/L vs 83 (68-108) IU/L, P = 0.016] and TNFα levels [62 (40-112) pg/mL vs 56 (34-80) pg/mL, P = 0.04] amongst NAFLD patients with and without CD14 C (-159) T polymorphism (Table 7). However, no difference was observed between the two groups with regard to the degree of hepatic steatosis, hepatic fibrosis, NAS score, and presence of NASH, borderline NASH, and no-NASH amongst the biopsy proven patients with NAFLD in each group.
| CD14 C (-159) T polymorphism (NAFLD = 200) | |||
| Parameters | Without polymorphism (n = 106) | With polymorphism (n = 94) | P value |
| TNF-α (pg/mL) (n = 200) | 56 (34-80) | 62 (40-112) | 0.04 |
| Adiponectin (pg/mL) | 745 (649-893) | 745 (634-928) | 0.93 |
| (n = 200) | |||
| IL-1β (pg/mL) | 43 (32-47) | 43 (25-47) | 0.53 |
| (n = 200) | |||
| HOMA-IR (n = 200) | 1.9 (1.3-2.7) | 1.8 (1.4-3.8) | 0.34 |
| MS (n = 200) | |||
| ≥ 3 components | 43 (40.5%) | 35 (37.2%) | 0.63 |
| ALT (IU/L) | 83 (68-108) | 95 (-130) | 0.016 |
| NAS score (n = 60) | 8 | 6 | 0.94 |
| No NASH | 11 | 10 | |
| Borderline NASH | 13 | 12 | |
| NASH | |||
| Severity of steatosis (n = 60) | |||
| 1 | 9 | 6 | |
| 2 | 14 | 15 | 0.733 |
| 3 | 9 | 7 | |
| Severity of fibrosis (n = 60) | |||
| 0 | 13 | 11 | 0.90 |
| 1 | 16 | 13 | |
| 2 | 1 | 2 | |
| 3 | 2 | 2 | |
NAFLD is one of the most predominant causes of liver disease in the world, and is considered a hepatic manifestation of metabolic syndrome. Its histology spectrum ranges from steatosis to NASH, and can progress to cirrhosis and HCC[1-3]. NAFLD progression is governed by genetic susceptibility, environmental factors, SIBO, lifestyle, and features of metabolic syndrome. Gene expression and genome-wide association studies have identified novel disease pathways and polymorphisms in genes that may be potential biomarkers of NAFLD development and progression. Pathways that include SIBO and toll-like receptor signaling seem to be one of the contributors of NAFLD development. The primary focus of our study was to analyze the polymorphisms of TLR2, TLR4, and CD14 genes in NAFLD patients and to assess their contribution to the causation and severity of the disease.
The overgrowth of bacterial components is recognized by pathogen-associated molecular patterns, including TLRs. Toll-like receptor 2 (TLR2) are receptors for gram-positive bacterial components. In humans, due to a single nucleotide gene polymorphism at position 753, arginine is replaced by glutamine and the G allele replaced by A allele diminishes the ability of TLR2 to respond to bacterial cell wall components[24,25]. Although there are animal studies to show the protective role for TLR2-mediated signals in liver injury and occurrence of NASH with TLR2 deficiency[26,27], ours is the first human study to demonstrate the absence of an association of TLR2 Arg753Gln polymorphism with the risk of developing NAFLD.
In humans, TLR4 is the principal receptor for bacterial endotoxin recognition and functional variants in the gene confer endotoxin hyporesponsiveness[28]. The missense mutation (Asp299Gly) in the fourth exon of the TLR4 gene alters the extracellular domain of this receptor. An additional missense polymorphism (Thr399Ile) in the extracellular domain of the TLR4 receptor co-segregates with the Asp299Gly substitution in more than 95% of the Caucasian population[29]. There are conflicting reports on the effects of the Asp299Gly polymorphism on endotoxin responsiveness in vitro[30-34]; however, the authors of several clinical reports associated this polymorphism with the risk of gram-negative infections[35,36] or severe respiratory syncytial viral infection[37], as well as such chronic disorders as asthma[38], arteriosclerosis[39], and diabetic neuropathy[40]. We did not observe any association of TLR4 A299G and TLR4 C399T gene polymorphism with the risk of developing NAFLD or NASH. In addition, we did not find any association of haplotypes for TLR4 gene with NAFLD. Our results are in accordance with a study by Day et al[13], in which biopsy-proven patients with NAFLD were genotyped for Asp299Gly single nucleotide polymorphism (SNP) in the TLR4 gene and no association was found between the susceptibility of NASH and the Asp299Gly TLR4 SNP. A recent study by Kiziltas et al[41] demonstrated that subjects with a heterozygous mutation in genotype 299 (Asp299Gly) were significantly lower in the NAFLD group than in the control group. The authors concluded that this mutation may have had a protective role against the genesis of NAFLD.
We found a significant association of C (-159) T polymorphism with NAFLD on multiplicative and recessive models. Patients with NAFLD with C (-159) T polymorphism had a significantly higher prevalence of TT genotype with significantly high levels of TNF-α (P = 0.04) and ALT (P = 0.01) than those without this polymorphism. Patients with TT genotype had a 2.6 fold higher risk of developing NAFLD in comparison to the CC genotype of CD14 C (-159) T polymorphism. However, this polymorphism did not affect disease severity, as there was no difference in NAS score and the prevalence of NASH or borderline NASH amongst those with and without polymorphism. Brun et al[14] found that TT genotype distribution was significantly higher in NASH patients than in control subjects, while subjects carrying TT genotype had higher TNF-α levels than CT and CC genotypes. Another study demonstrated that “high” activity of TT genotype of C-159T SNP in the CD14 promoter gene was associated with NASH, and that patients with CD14 polymorphism had higher levels of serum TNF-α levels in comparison to those without C-159 T SNP[13].
In contrast to C (-159) T polymorphism, we did not find any association of CD14 C (-550) T polymorphism with NAFLD. Ours is the first study to demonstrate the lack of an association of CD14 C (-550) T polymorphism with the risk of developing NAFLD. However, haplotype analysis of genotypes for CD14 revealed that TC genotype had an increased risk of NAFLD. Haplotype study is now gaining attention because multiple linked SNPs have the potential to provide significantly more power for genetic analysis than individual SNPs[42]. There was no previously performed haplotype analysis for CD14 gene in patients with liver disease, with ours being the first study to demonstrate its utility in patients with NAFLD.
In conclusion, our study demonstrated that NAFLD patients with TT genotype of C (-159) T polymorphism in CD14 promoter gene have a higher risk of NAFLD development. However, this polymorphism did not affect the severity of liver disease. We did not find any association between TLR 2 ARG753, TLR 4 (Asp299Gly), TLR4 (Thr399Ile), or CD 14 C/T 550 polymorphisms and the risk of NAFLD development. Studies with a larger cohort of patients are required to confirm the results.
Environmental and genetic factors predispose individuals to the development of non-alcoholic fatty liver disease (NAFLD). In this study, the authors demonstrated that cluster of differentiation 14 (CD14) polymorphism could predict the development of NAFLD.
NAFLD is the one of the manifestation of the obesity-related complications and incidence of NAFLD-related hepatocellular carcinoma increasing worldwide. It is therefore very important to understand the molecular mechanism underlying the pathogenesis of NAFLD.
Individuals with TT genotype of C (-159) T polymorphism in CD14 promoter gene have a higher risk of NAFLD development.
Individuals with TT genotype of C (-159) T polymorphism in CD14 promoter gene have a higher risk of NAFLD development. It can be considered a marker for identifying a population at risk of NAFLD progression.
Individuals with CD14 C (-159) T polymorphism have a higher risk of NAFLD development.
| 1. | Alba LM, Lindor K. Review article: Non-alcoholic fatty liver disease. Aliment Pharmacol Ther. 2003;17:977-986. [PubMed] |
| 2. | Sevastianos VA, Hadziyannis SJ. Nonalcoholic fatty liver disease: from clinical recognition to treatment. Expert Rev Gastroenterol Hepatol. 2008;2:59-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 9] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 3. | Duseja A, Sharma B, Kumar A, Kapil S, Das A, Dhiman RK, Chawla YK. Nonalcoholic fatty liver in a developing country is responsible for significant liver disease. Hepatology. 2010;52:2248-2249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 38] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 4. | Day CP. Genes or environment to determine alcoholic liver disease and non-alcoholic fatty liver disease. Liver Int. 2006;26:1021-1028. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 120] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 5. | Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, Boerwinkle E, Cohen JC, Hobbs HH. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461-1465. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2701] [Cited by in RCA: 2696] [Article Influence: 149.8] [Reference Citation Analysis (2)] |
| 6. | Sookoian S, Pirola CJ. Meta-analysis of the influence of I148M variant of patatin-like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of nonalcoholic fatty liver disease. Hepatology. 2011;53:1883-1894. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 756] [Cited by in RCA: 756] [Article Influence: 50.4] [Reference Citation Analysis (1)] |
| 7. | Valenti L, Al-Serri A, Daly AK, Galmozzi E, Rametta R, Dongiovanni P, Nobili V, Mozzi E, Roviaro G, Vanni E. Homozygosity for the patatin-like phospholipase-3/adiponutrin I148M polymorphism influences liver fibrosis in patients with nonalcoholic fatty liver disease. Hepatology. 2010;51:1209-1217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 477] [Cited by in RCA: 538] [Article Influence: 33.6] [Reference Citation Analysis (1)] |
| 8. | Duseja A, Aggarwal R. APOC3 and PNPLA3 in non-alcoholic fatty liver disease: need to clear the air. J Gastroenterol Hepatol. 2012;27:848-851. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 9. | Petersen KF, Dufour S, Hariri A, Nelson-Williams C, Foo JN, Zhang XM, Dziura J, Lifton RP, Shulman GI. Apolipoprotein C3 gene variants in nonalcoholic fatty liver disease. N Engl J Med. 2010;362:1082-1089. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 340] [Cited by in RCA: 324] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 10. | Duseja A, Das R, Nanda M, Das A, Garewal G, Chawla Y. Nonalcoholic steatohepatitis in Asian Indians is neither associated with iron overload nor with HFE gene mutations. World J Gastroenterol. 2005;11:393-395. [PubMed] |
| 11. | Aderem A, Ulevitch RJ. Toll-like receptors in the induction of the innate immune response. Nature. 2000;406:782-787. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2272] [Cited by in RCA: 2261] [Article Influence: 87.0] [Reference Citation Analysis (1)] |
| 12. | Beutler BA. TLRs and innate immunity. Blood. 2009;113:1399-1407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 588] [Cited by in RCA: 648] [Article Influence: 36.0] [Reference Citation Analysis (2)] |
| 13. | Day CP, Julian B. Leathart. Genetic evidence for a role for gut flora in the pathogenesis of NASH in humans. Hepatology. 2006;44 Suppl 1:abstract 4. |
| 14. | Brun P, Castagliuolo I, Floreani AR, Buda A, Blasone L, Palù G, Martines D. Increased risk of NASH in patients carrying the C(-159)T polymorphism in the CD14 gene promoter region. Gut. 2006;55:1212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 15. | Duseja A, Das A, Das R, Dhiman RK, Chawla Y, Bhansali A, Kalra N. The clinicopathological profile of Indian patients with nonalcoholic fatty liver disease (NAFLD) is different from that in the West. Dig Dis Sci. 2007;52:2368-2374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 63] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 16. | Misra A, Wasir JS, Pandey RM. An evaluation of candidate definitions of the metabolic syndrome in adult Asian Indians. Diabetes Care. 2005;28:398-403. [PubMed] |
| 17. | Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults. Executive Summary of The Third Report of The National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, And Treatment of High Blood Cholesterol In Adults (Adult Treatment Panel III). JAMA. 2001;285:2486-2497. [PubMed] |
| 18. | Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, Ferrell LD, Liu YC, Torbenson MS, Unalp-Arida A. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313-1321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6807] [Cited by in RCA: 8559] [Article Influence: 407.6] [Reference Citation Analysis (9)] |
| 19. | Von Hahn T, Halangk J, Witt H, Neumann K, Müller T, Puhl G, Neuhaus P, Nickel R, Beuers U, Wiedenmann B. Relevance of endotoxin receptor CD14 and TLR4 gene variants in chronic liver disease. Scand J Gastroenterol. 2008;43:584-592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 20. | Marsik C, Jilma B, Joukhadar C, Mannhalter C, Wagner O, Endler G. The Toll-like receptor 4 Asp299Gly and Thr399Ile polymorphisms influence the late inflammatory response in human endotoxemia. Clin Chem. 2005;51:2178-2180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 21. | Chitturi S, Abeygunasekera S, Farrell GC, Holmes-Walker J, Hui JM, Fung C, Karim R, Lin R, Samarasinghe D, Liddle C. NASH and insulin resistance: Insulin hypersecretion and specific association with the insulin resistance syndrome. Hepatology. 2002;35:373-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 807] [Cited by in RCA: 821] [Article Influence: 34.2] [Reference Citation Analysis (0)] |
| 22. | Clarke GM, Morris AP. A comparison of sample size and power in case-only association studies of gene-environment interaction. Am J Epidemiol. 2010;171:498-505. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 23. | Lewis CM, Knight J. Introduction to genetic association studies. Cold Spring Harb Protoc. 2012;2012:297-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 120] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 24. | Lorenz E, Mira JP, Frees KL, Schwartz DA. Relevance of mutations in the TLR4 receptor in patients with gram-negative septic shock. Arch Intern Med. 2002;162:1028-1032. [PubMed] |
| 25. | Schröder NW, Hermann C, Hamann L, Göbel UB, Hartung T, Schumann RR. High frequency of polymorphism Arg753Gln of the Toll-like receptor-2 gene detected by a novel allele-specific PCR. J Mol Med (Berl). 2003;81:368-372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 112] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 26. | Szabo G, Velayudham A, Romics L, Mandrekar P. Modulation of non-alcoholic steatohepatitis by pattern recognition receptors in mice: the role of toll-like receptors 2 and 4. Alcohol Clin Exp Res. 2005;29:140S-145S. [PubMed] |
| 27. | Rivera CA, Gaskin L, Allman M, Pang J, Brady K, Adegboyega P, Pruitt K. Toll-like receptor-2 deficiency enhances non-alcoholic steatohepatitis. BMC Gastroenterol. 2010;10:52. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 68] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 28. | Guha M, Mackman N. LPS induction of gene expression in human monocytes. Cell Signal. 2001;13:85-94. [PubMed] |
| 29. | Arbour NC, Lorenz E, Schutte BC, Zabner J, Kline JN, Jones M, Frees K, Watt JL, Schwartz DA. TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nat Genet. 2000;25:187-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1504] [Cited by in RCA: 1481] [Article Influence: 57.0] [Reference Citation Analysis (0)] |
| 30. | Schwartz DA. Inhaled endotoxin, a risk for airway disease in some people. Respir Physiol. 2001;128:47-55. [PubMed] |
| 31. | Schwartz DA. TLR4 and LPS hyporesponsiveness in humans. Int J Hyg Environ Health. 2002;205:221-227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 37] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 32. | Heesen M, Bloemeke B, Kunz D. The cytokine synthesis by heterozygous carriers of the Toll-like receptor 4 Asp299Gly polymorphism does not differ from that of wild type homozygotes. Eur Cytokine Netw. 2003;14:234-237. [PubMed] |
| 33. | Imahara SD, Jelacic S, Junker CE, O’Keefe GE. The TLR4 +896 polymorphism is not associated with lipopolysaccharide hypo-responsiveness in leukocytes. Genes Immun. 2005;6:37-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 21] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 34. | Erridge C, Stewart J, Poxton IR. Monocytes heterozygous for the Asp299Gly and Thr399Ile mutations in the Toll-like receptor 4 gene show no deficit in lipopolysaccharide signalling. J Exp Med. 2003;197:1787-1791. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 137] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 35. | Agnese DM, Calvano JE, Hahm SJ, Coyle SM, Corbett SA, Calvano SE, Lowry SF. Human toll-like receptor 4 mutations but not CD14 polymorphisms are associated with an increased risk of gram-negative infections. J Infect Dis. 2002;186:1522-1525. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 424] [Cited by in RCA: 416] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 36. | Lorenz E, Hallman M, Marttila R, Haataja R, Schwartz DA. Association between the Asp299Gly polymorphisms in the Toll-like receptor 4 and premature births in the Finnish population. Pediatr Res. 2002;52:373-376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 149] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 37. | Tal G, Mandelberg A, Dalal I, Cesar K, Somekh E, Tal A, Oron A, Itskovich S, Ballin A, Houri S. Association between common Toll-like receptor 4 mutations and severe respiratory syncytial virus disease. J Infect Dis. 2004;189:2057-2063. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 238] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 38. | Werner M, Topp R, Wimmer K, Richter K, Bischof W, Wjst M, Heinrich J. TLR4 gene variants modify endotoxin effects on asthma. J Allergy Clin Immunol. 2003;112:323-330. [PubMed] |
| 39. | Kiechl S, Lorenz E, Reindl M, Wiedermann CJ, Oberhollenzer F, Bonora E, Willeit J, Schwartz DA. Toll-like receptor 4 polymorphisms and atherogenesis. N Engl J Med. 2002;347:185-192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 779] [Cited by in RCA: 771] [Article Influence: 32.1] [Reference Citation Analysis (0)] |
| 40. | Rudofsky G, Reismann P, Witte S, Humpert PM, Isermann B, Chavakis T, Tafel J, Nosikov VV, Hamann A, Nawroth P. Asp299Gly and Thr399Ile genotypes of the TLR4 gene are associated with a reduced prevalence of diabetic neuropathy in patients with type 2 diabetes. Diabetes Care. 2004;27:179-183. [PubMed] |
| 41. | Kiziltas S, Ata P, Colak Y, Mesçi B, Senates E, Enc F, Ulasoglu C, Tuncer I, Oguz A. TLR4 gene polymorphism in patients with nonalcoholic fatty liver disease in comparison to healthy controls. Metab Syndr Relat Disord. 2014;12:165-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 42. | Cambien F. Genes in population. In: Malcolm S, Goodship J, editors. Genotype to Phenotype. Guildford, UK: Academic Press, 2001: 31-53. . |
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: India
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
P- Reviewer: Zhou BJ S- Editor: Yu J L- Editor: Rutherford A E- Editor: Zhang FF