Published online Aug 28, 2016. doi: 10.3748/wjg.v22.i32.7175
Peer-review started: April 27, 2016
First decision: May 30, 2016
Revised: June 10, 2016
Accepted: July 6, 2016
Article in press: July 6, 2016
Published online: August 28, 2016
Processing time: 119 Days and 9 Hours
Pancreatic cancer is a disease that carries a poor prognosis. Accurate tissue diagnosis is required. Tumours contain a high content of stromal tissue and therefore biopsies may be inconclusive. Circulating tumour cells (CTCs) have been investigated as a potential “liquid biopsy” in several malignancies and have proven to be of prognostic value in breast, prostate and colorectal cancers. They have been detected in patients with localised and metastatic pancreatic cancer with sensitivities ranging from 38%-100% using a variety of platforms. Circulating tumour DNA (ctDNA) has also been detected in pancreas cancer with a sensitivity ranging from 26%-100% in studies across different platforms and using different genetic markers. However, there is no clear consensus on which platform is the most effective for detection, nor which genetic markers are the most useful to use. Potential roles of liquid biopsies include diagnosis, screening, guiding therapies and prognosis. The presence of CTCs or ctDNA has been shown to be of prognostic value both at diagnosis and after treatment in patients with pancreatic cancer. However, more prospective studies are required before this promising technology is ready for adoption into routine clinical practice.
Core tip: Pancreatic cancer is a difficult disease to diagnose and treat. Persistently poor outcomes mean that new biomarkers of disease and treatments are required. Circulating tumour cells and circulating tumour DNA have been investigated as liquid biopsies in pancreatic cancer. Sensitivity is variable but specificity promising. The most effective platform and most informative biomarkers are yet to be identified. There are many potential roles for this technology in the management of patients with pancreatic cancer, including screening, diagnosis, prognosis and monitoring of treatment efficacy; however based on current available evidence they are not yet ready for routine clinical practice.
- Citation: Lewis AR, Valle JW, McNamara MG. Pancreatic cancer: Are "liquid biopsies" ready for prime-time? World J Gastroenterol 2016; 22(32): 7175-7185
- URL: https://www.wjgnet.com/1007-9327/full/v22/i32/7175.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i32.7175
Pancreatic cancer possesses one of the worst prognoses of all malignancies. Overall 5 year survival rates are approximately 5% and mortality rates continue to rise in both sexes[1], in contrast to the survival trends seen in most other malignancies.
The only potentially curative treatment is surgery; however only 15%-20% of patients have resectable disease at presentation and even in those that undergo surgery and adjuvant chemotherapy, the 5-year survival is just 16.3%-28.9%[2]. In the majority of patients the disease presents as locally advanced or metastatic.
The median survival for patients with metastatic disease treated with FOLFIRINOX chemotherapy is 11.1 mo[3] and 5.6 mo in those treated with single-agent gemcitabine[4].
Known risk factors for developing pancreatic cancer include tobacco use, obesity, new-onset diabetes, chronic pancreatitis, hepatitis B and Helicobacter pylori (H. pylori) infection[5].
Other high-risk groups include: patients with ≥ 2 first-degree relatives with a history of pancreatic cancer; those with a known mutation of the BRCA2 gene or with other familial syndromes known to be associated with pancreatic cancer, e.g., hereditary pancreatitis, hereditary non-polyposis colorectal cancer, Li-Fraumeni, Peutz-Jeghers, familial melanoma; or those with a recognised pre-malignant lesion, e.g., Intraductal Papillary Mucinous Neoplasms (IPMN)[6,7].
Approximately 95% of pancreatic cancers are ductal adenocarcinomas[8] and tumours contain high percentages of stromal tissue[9]. Over 90% are Kirsten rat sarcoma viral oncogene homolog (KRAS)-mutated[10]. Other commonly mutated oncogenes include p16, TP53 and SMAD4[10] and high levels of epidermal growth factor receptor (EGFR) mutations have also been noted[11]. The serum biomarker CA19-9 is used in the monitoring of treatment response of pancreatic cancer but it has low specificity and is not recommended for use in primary diagnosis[8]. It is subject to false negatives; where the patient does not produce the Lewis enzyme[12] and false positives; with any cause of a raised bilirubin[8] or other malignancies.
Diagnosis of pancreatic cancer can prove challenging. Histological diagnosis often requires invasive tests because of the anatomical position of the pancreas. Due to the high content of stromal cells within the tumour tissue, biopsies do not always provide sufficient material to confirm a diagnosis.
Endoscopic ultrasound with fine-needle aspiration (EUS-FNA) is recommended in the work-up of patients with pancreatic cancer and is the only recommended method of obtaining a biopsy in patients with potentially resectable disease; although there are concerns about tumour seeding along the biopsy tract[8], the traversed duodenum is resected at the time of surgery, abrogating this risk. It has been shown that EUS has better sensitivity than computed tomography (CT) for detection of pancreatic masses; the sensitivity of EUS-FNA is approximately 85%[2]. However, EUS-FNA is an invasive test that requires the use of sedation and can be difficult to tolerate in a patient group who often present with a poor performance status due to the aggressive nature of the disease.
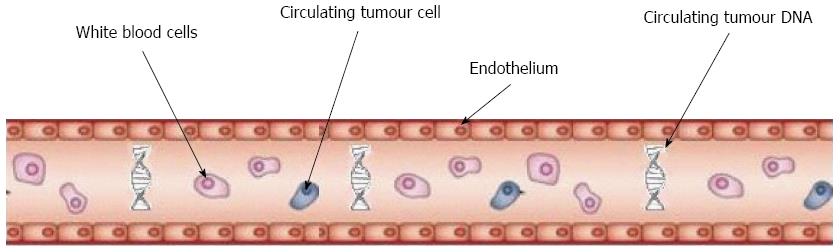
The use of circulating tumour cells (CTCs) isolated in the peripheral blood of patients with cancer as a potential “liquid biopsy” has been under investigation for some time. Their utility was first demonstrated in breast, prostate and more recently lung cancer where they have been shown to be a prognostic marker both in limited stage and metastatic disease[13].
Given the challenges in the investigation and treatment of pancreatic cancer and the need for better biomarkers, many articles have looked at the potential role of CTCs in the disease management pathway (Figure 1).
Circulating tumour cells are present at a rate of approximately 1 CTC per billion blood cells[14] in patients with a malignancy, therefore enumeration requires a process of enrichment prior to detection. There are several ways of detecting CTCs in peripheral blood.
CellSearch is the only US Food and Drug Administration (FDA) approved method for CTC analysis and therefore the most widely studied. It is dependent on the expression of epithelial markers by the CTC, specifically the Epithelial Cell Adhesion Molecule (EpCAM)[12].
It uses 7.5 mL of peripheral blood drawn into an ethylene-diamine-tetra-acetic acid (EDTA) blood sampling tube. A cellular preservative is immediately added. Specific EpCAM, CD45, and cytokeratin fluorescent antibody labels are applied. Samples are then analysed to give a count per 7.5 mL blood[15]. High levels of CTC heterogeneity with variable expression or down-regulation of EpCAM is a technical challenge within the process [14].
An alternative method is isolation by size of epithelial tumour cells (ISET) where blood collected in an EDTA tube is divided into aliquots and diluted with a red cell lysis buffer and then passed through a filtration module. Following washing and staining, CD45-negative cells with features of high nuclear-to-cytoplasmic ratio and hyperchromatic nuclei are designated as CTCs and enumerated[12].
There have been a number of studies describing CTC detection in patients with pancreatic cancer. Zhang et al[16] reported CTCs in 16/22 patients diagnosed with pancreatic cancer at all disease stages, using a combined immunostaining/fluorescence in situ hybridisation (FISH) method. They also included 9 patients with pre-malignant or benign lesions and 30 healthy controls. Overall, they reported a sensitivity of 68.18% and specificity of 94.87% using a cut-off of 4 CTCs/7.5 mL of blood.
Rhim et al[17] detected CTCs in 73% of patients with pancreatic cancer at all stages at the time of diagnosis and 33% of patients with pre-malignant lesions, using a cut-off of 3 cells/7.5 mL of blood. No CTCs were detected in controls.
Nagrath et al[18] were able to detect CTCs in 15/15 patients with a diagnosis of pancreatic cancer across all stages of disease using microchip technology, whilst Khoja et al[12] reported detection of CTCs in 21/53 patients with all stages of pancreatic cancer using CellSearch and 24/27 different patients with pancreatic cancer using ISET. Patients were tested at the time of diagnosis or at the time of disease progression following previous treatment. The mean number of cells detected using ISET was 26 compared to a mean of 6 using CellSearch[12].
Allard et al[15] found CTCs in 6 of 16 patients with metastatic pancreatic cancer using the CellSearch method. Gall et al[19] detected CTCs in 4/75 (5%) patients with locally advanced pancreatic cancer before starting chemotherapy and 5/59 (9%) from the original 75 patients two months after commencing treatment. Ren et al[20] detected CTCs in 80.5% of 41 patients with locally advanced or metastatic pancreatic cancer before commencing 5-fluorouracil chemotherapy and in 29.3% after 1 cycle of chemotherapy. These combined results are summarised in Table 1.
| Study | CTC platform | Markers | Stage of disease | Time of sampling | Number of patients, n | Cut-off for positivity per 7. 5 mL blood | CTCs detected, n |
| Zhang et al[16] 2014 | Immunocy-togenetics | CK, CD45, DAPI, CEP8 | All stages | Pre-operative | 22 | 4 | 16/22 (72.7%) |
| Rhim et al[17] 2014 | Micro-fluidic "GEM" Chip | DAPI, CD45, CK, PDX-1 | All stages | Diagnosis | 11 | ≥ 1 | 8/11 (73%) |
| Khoja et al[12] 2011 | CellSearch and ISET | EpCAM, CK, vimentin, E-cadherin | All stages | Diagnosis or at time of diagnosis of progressive disease > 6 wk from therapy | 53 | ≥ 1 | 21/53 (40%) using CellSearch |
| 24/27 (93%) using ISET | |||||||
| Nagrath et al[18] 2013 | CTC-Chip | Cks | Metastatic | Not stated | 15 | 37.5 (5/mL) | 15/15 (100%) |
| Allard et al[15] 2004 | CellSearch | CK8, CK18, CK19 | Metastatic | Not stated | 16 | ≥ 1 | 6/16 ((38%) |
| Gall et al[19] 2013 | CellSearch | EpCam, EGFR | Locally advanced | Pre-treatment | 75 | ≥ 1 | 4/75 (5%) |
| Ren et al[20] 2011 | Immuno-cyto-chemistry | CA 19-9, CK8, CK18 | Locally advanced and metastatic | Pre- and post- treatment | 41 | ≥ 1 | 33/41 (80.5%) pre-treatment and 12/41 (29.3%) post-treatment |
Although the sensitivity of CTCs in the trials discussed is variable, the specificity seems to be significantly better. Trials that have compared samples from patients with pancreatic cancer to healthy controls have shown that healthy controls do not have detectable CTCs[15,18,21,22].
Patients with pancreatic cancer seem to have relatively low numbers of CTCs compared to patients with other tumours including breast, colorectal and prostate cancer[11,14]. This is thought to be due to lower numbers of cells, rather than reduced detection, and it has been hypothesised that this may be related to CTC sequestration as cells pass through the portal circulation in patients with pancreatic cancer[15]. There seems to be no clear consensus on the optimal cut-off for number of CTCs detected in the peripheral circulation of patients with cancer to count as “positive” and similarly there are no significant differences in rate of detection between patients with localised and metastatic disease[2,14,23-25].
Clear data on the absolute sensitivity and specificity of utilising CTCs as a liquid biopsy in patients with pancreatic cancer is still lacking. Studies looking at CTCs in a range of malignancies including breast, colorectal, prostate, and hepatocellular carcinoma have reported very high specificity of around 99%[14,17,22]. In addition, few of the studies including patients with pancreatic cancer have analysed samples from healthy controls, and where they have done, these have always proved negative[14,17,19,21].
An alternative method for obtaining a liquid biopsy is to isolate circulating tumour DNA (ctDNA) detected by polymerase chain reaction (PCR) or next generation sequencing (NGS) as a proxy measure.
A multi-disease study (including 155 patients with pancreatic cancer) using PCR assays to search for ctDNA reported that ctDNA was often present where CTCs were not. The study reported detection of ctDNA in > 80% of patients with advanced pancreatic cancer and 48% of patients with localised pancreatic cancer. In a further sub-analysis of the study population, KRAS mutations were detected, using this method, with a sensitivity of 87% and specificity of 99.2% in patients with colorectal cancer[26]. In a pilot study of patients with pancreatic cancer, Earl et al[27] were only able to detect KRAS mutations in ctDNA in 26% of patients but they did find a strong correlation between the presence of a KRAS mutation and worse overall survival. Sausen et al[28] used digital PCR and detected KRAS mutations in 43% of 51 patients at time of diagnosis with a specificity of > 99.9%. The presence of ctDNA was also analysed following resection and it was reported that disease relapse was detectable at 3.1 mo using ctDNA compared to 9.6 months using CT. Kinugasa et al[29] detected KRAS mutations in ctDNA of 47/75 patients (62%) and reported a concordance in KRAS mutation detections with tissue biopsy of 77.3%. Presence of a KRAS mutation in ctDNA in this study was associated with poorer prognosis.
Zill et al[30] compared ctDNA from peripheral blood samples with tumour biopsy samples in 17 patients with both localised and advanced pancreatic cancer and reported a concordance of 90% in genetic mutations; they also demonstrated a correlation between CA19-9 levels and ctDNA percentage across time in a group of 8 patients. Sergeant et al[31] used RT-PCR to isolate EpCAM and were able to detect this in 25% of patients with stage I and II pancreatic cancer prior to undergoing surgery. They detected EpCAM in 65% of patients immediately post-operatively and then in 28.6% at day 1, 23.1% at day 7 and 23.5% at 6 wk post-surgery, but they did not demonstrate an association with survival.
Zhou et al[32] were able to detect a range of tumour markers including CK20, CEA and C-MET in between 80 and 100% of 25 patients with pancreatic cancer. de Albuquerque et al[22] used a 5-marker panel and were able to detect these in 47.1% of patients with locally advanced or metastatic pancreatic cancer using immunomagnetic RT-PCR and Chausovsky et al[33] detected CK20 in 22 of 28 patients with pancreatic cancer using RT-PCR. These results are summarised in Table 2.
| Study | CtDNA platform | Markers | Stage of disease | Time of sampling | Number of patients, n | Markers detected, n |
| Zhou et al[32] 2011 | Nested RT-PCR | H-tert, C-MET, CK20, CEA mRNAs | All stages | Pre-treatment | 25 | H-TERT 25/25 (100%) |
| CMET 20/25(80%), CK20 21/25 (84%), CEA 20/25 (80%) | ||||||
| de Albuquerque et al[22] 2012 | Immunomagnetic RT-PCR | KRT19, MUC1, EpCAM, CEACAM5, BIRC5 | Stage III and IV | Pre-treatment | 34 | 16/34 (47%) |
| Chausovsky et al[33] 1999 | RT-PCR | CK20 | Metastatic | Not stated | 28 | 22/28 (79%) |
| Sausen et al[28] 2015 | Digital PCR | KRAS | Localised | Pre-treatment | 51 | 22/51 (43%) |
| Earl et al[27] 2015 | Digital PCR | KRAS | All stages | Pre-treatment (NB 7 patients had received chemotherapy) | 31 | 8/31 (26%) |
| Sergeant et al[31] 2011 | RT-PCR | EpCAM | Stage I,II and 4 | Pre and post treatment | 40 | 10/40 (25%) preoperatively, |
| 10/35(28.6%) D1, 9/39 (23.1%) at D7, 8/34 (23.5%) at 6 wk | ||||||
| Bettegowda et al[26] 2014 | Digital PCR | KRAS, NRAS, PIK3CA, BRAF | Localised and metastatic | Not stated | 155 | 48% of those with localised and > 80% metastatic disease |
| Zill et al[30] 2015 | NGS | KRAS, TP53, APC, SMAD4, FBXW7 | Localised and metastatic | Post-treatment | 17 | 17/17 (100%) |
| Kinugasa et al[29] 2015 | Digital PCR | KRAS | All stages | Pre-treatment | 75 | 47/75 (62%) |
Overall, ctDNA appears to be a promising method for use as a liquid biopsy. However, the sensitivity of tests used is variable and there is no consensus as yet on which is the best marker or group of markers to use for most accurate detection.
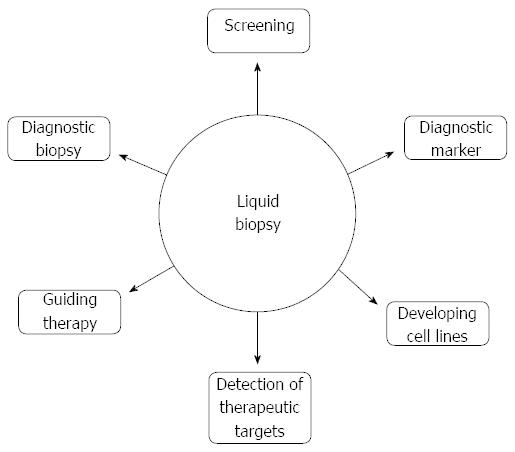
Trial evidence to date suggests that identification of CTCs and ctDNA could have roles at many different stages of pancreatic cancer management (Figure 2).
Given the challenges in obtaining suitable biopsy samples from patients with suspected pancreatic cancer, a “liquid biopsy” using a peripheral blood sample would be a highly attractive alternative.
This is a minimally invasive test and the costs are also significantly lower than those of an EUS-FNA: US$371.99 for a CellSearch analysis[34] vs US$1405 for EUS-FNA[35].
To be truly useful as a diagnostic tool, a liquid biopsy in patients with pancreas cancer would need to have proven utility in both localised as well as metastatic disease. Much of the evidence in favour of CTCs comes from studies including patients with metastatic tumours[13], and indeed the CellSearch system gained FDA approval following use in a trial which included patients with metastatic breast cancer[36]. There is evidence for their use in early breast[24,25,34] and prostate[37] cancer detection, though the evidence in other disease sites is less clear[13]. Widespread clinical application is not prevalent. However, multiple studies have used the system of other methods of ctDNA detection in patients with localised, curative-intent pancreatic cancer[12,19,38] and two meta-analyses discussed below incorporated data from studies including patients with resectable and non-resectable disease.
Allard et al[15] commented that patients with pancreatic cancer had relatively low levels of CTCs detected compared to other malignancies, even in the presence of widespread metastatic disease. Whether patients with pancreatic cancer produce fewer peripheral CTCs than patients with other malignancies remains unclear but this could account for the relatively low detection rates seen in the studies discussed above and may represent a significant challenge in the use of liquid biopsies.
Furthermore, high levels of heterogeneity have been demonstrated in CTCs[39-43] from patients with a range of malignancies. This appears to be both spatial and temporal as detectable CTCs may change in appearance over time and with treatment[11]. For example, expression of EpCAM or other tumour markers appears to be variable from cell to cell both in different patients and within the same patient, but also the expression may vary over time within the same patient. This provides a further challenge in the use of CTCs and ctDNA as a biopsy or screening tool as ctDNA might be detected in a serum sample that may not then be found in the tumour sample. This could lead to confusion around the diagnosis and perhaps necessitate more tests and delays to treatment.
Heitzer et al[40] demonstrated that mutations were found in CTCs from the peripheral circulation that weren’t found in the primary tumour of patients with colorectal cancer. On further NGS of the primary tumour, it was demonstrated that these mutations were present at subclonal level, potentially demonstrating that issues around heterogeneity can be overcome.
A successful liquid biopsy (for diagnosis) would therefore need to rely on a combination of CTC enumeration and genetic analysis to provide the most accurate result and better sensitivity and, from currently-available evidence, the best combination of genetic markers is not well defined.
Some patient groups are at higher risk of developing pancreatic cancer[6]. A recent review of trials into screening high-risk individuals found that screening these patients resulted in a higher curative resection rate and longer median survival time[44]. For example, a recent study by Vasen et al[45] demonstrates a screening benefit for patients with the CDKN2A mutation. However other studies have not found screening using a combination of clinical examination, blood tests, EUS and magnetic resonance cholangio-pancreatography (MRCP) in high risk individuals useful[46] and in current guidance it is not recommended[8]. The only existing biomarker for pancreatic cancer: CA19-9, has not been shown to be a reliable screening marker[8].
Evidence showing high levels of specificity combined with a relatively low cost would favour the use of CTCs as a screening tool. The variable sensitivity of this test means that based on current evidence, CTCs would not be suitable for screening and there is no current clinical trial evidence demonstrating the benefit of CTCs as a screening tool in any malignancy.
Another potential use would be as a biomarker of prognosis either before or at the time of curative-intent surgery. Sausen et al[28] detected CTCs in 22 patients (43% of cohort) with localised pancreatic cancer at the time of diagnosis and found that presence of CTCs did predict relapse after resection and worse outcome. They also observed that ctDNA could be detected 6 mo before a radiological confirmation of recurrence, suggesting that CTCs could play an important role in detection of residual disease post-operatively or in the detection of early recurrence. Bissolati et al[47] performed intra-operative collection of blood samples from both the systemic circulation and portal vein. They found that CTC positivity in the portal circulation predicted liver metastases but not any significant difference in Disease Free Survival (DFS) or Overall Survival (OS). Sergeant et al[31] detected EpCAM from peripheral blood pre- and post-operatively in 40 patients undergoing pancreatectomy and also assessed peritoneal lavage fluid for EpCAM. Although detectable in 65% of patients post-operatively, EpCAM positivity was not associated with a worse prognosis. These findings indicate that a significant majority of CTCs will not survive, and as few as 0.01% may go on to form metastases[48,49].
A liquid biopsy could theoretically be a useful marker of disease response to chemotherapy.
Many studies have shown that CTCs can be used to predict treatment outcomes in a range of malignancies including breast, bladder, prostate and bowel[50,51] but not necessarily as a form of monitoring. Obermayr et al[52] revealed that the ongoing presence of CTCs during follow-up in patients with ovarian cancer was more common in those with platinum-resistant disease. The South-West Oncology Group[53] investigated the use of CTCs to guide treatment change in patients with metastatic breast cancer. Patients were treated with one cycle of standard chemotherapy and those who continued to test positive for CTCs after one cycle of chemotherapy were switched to an alternative regimen. However, a benefit in overall survival was not demonstrated. In a recent study by Tie et al[54] Patients with colorectal cancer had serum samples tested for ctDNA and compared with mutations within the primary tumour sample in the post-operative period. Six patients out of a cohort of 52 who went on to have adjuvant chemotherapy had detectable ctDNA. Samples were tested again every 3 mo; all patients who were ctDNA positive became ctDNA negative during chemotherapy. However two patients later became ctDNA positive again and both these patients relapsed.
There is little evidence for the use of liquid biopsies in monitoring of chemotherapy response in patients with pancreatic cancer. Ren et al[20] measured CTCs in patients with pancreatic cancer before and after their first cycle of 5-fluorouracil chemotherapy. The CTCs were detected in 80.5% of patients prior to chemotherapy and in 29.3%, one week after the first cycle of chemotherapy. Gall et al[19] detected CTCs in patients with locally advanced pancreatic cancer before, and then after 2 mo of chemotherapy. They reported that 4 of 75 (5%) patients had detectable CTCs before commencing chemotherapy and 5 of 59 (9%) had detectable CTCs after two months of treatment, however there was no crossover between the two groups. The trial was too small to demonstrate a significant difference in survival in these patients.
Two meta-analyses have reviewed the potential roles of CTCs as a prognostic biomarker: Ma et al[55] analysed 9 papers with a total of 603 patients included, with a range of different stages of pancreatic cancer. Four of the included papers examined prognosis after commencement of systemic treatment. The hazard ratio (HR) for DFS and OS in patients before treatment were 1.82 and 1.93 respectively (P < 0.003) and post treatment were 8.36 and 2.20, suggesting that the presence of CTCs after completing treatment has better predictive value for disease relapse and worse OS compared to pre-treatment.
The estimated pooled HR for OS across all 9 papers was 1.64 (95%CI: 1.39-1.94, P < 0.00001), showing that CTC positivity was associated with worse OS. This meta-analysis did find evidence of publication bias, however[55].
Han et al[56] explored 9 papers, 7 of which were included in the Ma et al[55] meta-analysis, but did not report any publication bias. Overall, the study included 623 patients with a range of stages of pancreatic cancer, having either surgery or chemotherapy. The HR for progression free survival (PFS) was 1.89 (95%CI: 1.25-4.00, P < 0.001) and the HR for OS was 1.23 (95%CI: 0.88-2.08, P < 0.001), suggesting that CTC positivity did predict poorer outcomes. This seems to be consistent whether sampling was performed before or after treatment. On sub-group analysis, no variations between ethnicity or between different methods (CellSearch vs RT-PCR) of CTC detection were reported.
Cell lines cultured from tumour biopsy samples have been used in cancer research for many years. A number of small studies have developed cell cultures from CTCs of patients with metastatic breast, lung and prostate cancer[57-59] and a pilot study by Cayrefourcq et al[60] demonstrated the development of permanent cell lines from CTC samples of patients with colorectal cancer. This had not previously been reported, perhaps in part because patients with colon cancer, similar to those with pancreatic cancer, seem to have relatively low levels of CTCs in the peripheral circulation.
This technology is at an early stage of development and has not yet been reported for patients with pancreatic cancer. However, these studies indicate that liquid biopsy could form the basis of cell lines upon which investigation of genetic mutations and targets for therapies can take place for patients with pancreatic cancer. A potential issue with this technology would be temporal heterogeneity of the cancer and thus it may be possible that repeated cultures would be required.
Studies in patients with a range of malignancies have begun to use CTCs and ctDNA to identify mechanisms of resistance and potential therapeutic targets. Murtaza et al[61] analysed ctDNA from patients with advanced cancer and using PCR were able to identify mutations known to be associated with acquired drug resistance. Heitzer et al[40] used CTC analysis and next-generation sequencing of the primary tumour in patients with colorectal cancer to identify mutations that could be of therapeutic interest. However, the issue of heterogeneity remains problematic; mutations were identified in CTCs that were present in the primary tumour, only on sub-clone analysis of the primary tumour, or were unique to that CTC. This makes the relevance of individual mutations difficult to quantify in the context of a full tumour genome.
Genetic analysis of liquid biopsy samples could, in the future, form part of a “personalised” mutation profile for a patient with pancreatic cancer and identify which targeted agents would be suitable for that patient. However, studies evaluating this approach are small and have not yet included patients with pancreatic cancer. Therefore, more studies and on a larger scale are required.
Although not addressed thus far in this article it is worth noting the increasing evidence supporting the potential roles for detection of micro-RNA (miRNA) in pancreatic cancer.
miRNAs are small molecules consisting of chains of RNA, typically around 20 nucleotides in length[62]. There is evidence that miRNAs play a role in modulating gene expression and thus biological processes[62].
This role seems to be highly variable as miRNAs can act as both an oncogene and a tumour suppressor gene[63]. miRNAs have been implicated in all tumour types including pancreatic cancer[64] and have been isolated in the bloodstream of patients with pancreatic cancer[65].
Small studies have shown that miRNA isolated from serum or biopsy samples can differentiate between pancreatic cancer and chronic pancreatitis[66] and IPMN[67] and small studies have shown they may be prognostic markers[62,65].
Technology for the detection of miRNAs is early in development and there is currently not sufficient evidence to make recommendations but they may in the future work concurrently with ctDNA and CTCs in the management of pancreatic cancer.
Pancreatic cancer is a disease with a prognosis that remains poor in contrast to the improvements in survival noted in other cancers over recent years[1]. The challenges faced in obtaining a diagnosis and the poor survival following treatment mean that new biomarkers and treatment strategies are necessary.
Current clinical and retrospective trial evidence indicates that CTCs are detectable in patients with pancreatic cancer, at both limited and advanced stages. Many of the studies available are hampered by their small sample size, however meta-analyses have performed statistical analysis on over 600 patients and demonstrated a clear correlation between CTC or ctDNA positivity and poorer outcome[55,56].
There are many potential applications of liquid biopsies in the care pathway of patients with pancreatic cancer. Their strength lies in being a relatively non-invasive test that can be repeated at any time. Therefore, one role could include diagnosis, particularly for patients too unwell to undergo invasive tests, or where these have proven inconclusive. A significant limitation to the use of CTCs and ctDNA as a liquid biopsy is their relatively low sensitivity and a lack of clarity on which is the most effective method of detection in this disease group.
Very few studies have directly compared CTCs to ctDNA. Dawson et al[68] analysed ctDNA and CTCs in patients with metastatic breast cancer and reported that ctDNA levels were more closely related to disease burden. In contrast, Maheswaran et al[69] detected CTCs and ctDNA in patients with non-small cell lung cancer and reported that detection of CTCs was more sensitive. Apart from Khoja et al[12], few trials have directly compared methods of CTC or ctDNA analysis and therefore the most sensitive method is not clear. It seems likely that the most effective test would use both CTCs and ctDNA, but data is lacking in this area both around efficacy and feasibility on a larger scale.
Another challenge is the high levels of heterogeneity found in CTCs, that has been shown to be both temporal and spatial[11]. This could impact on the sensitivity and reliability of liquid biopsies. Heterogeneity in CTCs has been found in many malignancies. Indeed it has been reported that between 63% and 69% of mutations are not present at every disease site in patients with metastatic renal cell cancer[70]. This could potentially be overcome using NGS of tumour samples in addition to CTC samples to identify more mutations. However, this has only been demonstrated in a small study[40] and may prove too costly and time-consuming to be feasible in routine practice. More work is required to evaluate this approach in pancreatic cancer.
Although only a small trial, the fact that ctDNA became detectable months before a radiologically-detectable relapse in patients who had undergone a resection[28] suggests that in future, liquid biopsies could potentially form an important role in the monitoring of patients after surgery. A larger trial would be needed to validate these results and then a future area of research might look at whether instituting chemotherapy at the point of “liquid biopsy relapse” could alter the outlook for patients with recurrent disease after surgery. Similarly, liquid biopsy could play a potential role as a non-invasive marker of treatment response to chemotherapy or as a marker of disease progression following chemotherapy in advanced disease. However, none of the trials that have detected CTCs following chemotherapy in patients with pancreatic cancer have proven that patients with detectable CTCs after chemotherapy have a worse survival[31], although trials studying this question only included small numbers of patients. Furthermore, as yet there is no evidence to show that moving directly onto second-line chemotherapy in patients with advanced pancreatic cancer with CTC-positivity following first-line chemotherapy would result in a survival benefit. A trial looking at switching chemotherapy in response to presumed resistance (based on on-going detection of CTCs following one cycle of chemotherapy) has not proven to have an impact on survival in patients with metastatic breast cancer[53]. It may well be that sufficient time for a cytotoxic effect to eliminate all CTCs from peripheral circulation requires more than one cycle of chemotherapy and there is no comparative data exploring this. A useful trial might measure CTCs following each cycle of chemotherapy and correlate on-going positivity to PFS and OS in an effort to identify a cut-off point for switching regimen.
Ultimately, liquid biopsies can only be of limited utility whilst there is still a lack of more effective treatments for pancreatic cancer. There is little purpose in demonstrating that a patient with pancreatic cancer is not responding to chemotherapy if there are limited alternatives available, therefore, on-going prospective studies developing novel therapeutic strategies are imperative.
Disappointingly, the addition of targeted therapies to conventional chemotherapies to date has failed to result in survival benefits in patients with pancreatic cancer. Despite the fact that the majority of pancreatic adenocarcinomas are KRAS positive, and a significant proportion have EGFR mutations, agents targeting this mutation have yet to demonstrate a clinically meaningful benefit.
Although Moore et al[71] reported a benefit to the use of the tyrosine kinase inhibitor, erlotinib, in addition to gemcitabine in patients with advanced pancreatic cancer for both OS and PFS, this has not been replicated in other trials[72-74]. Nor has efficacy been demonstrated with other agents; for example, mitogen-activated protein kinase inhibitors[75] or VEGF inhibitors either alone or in combination with other targeted therapies or standard chemotherapy[76,77]. In the study by Infante et al[75], outcomes were independent of KRAS mutations determined by circulating free DNA and archival tumour tissue. Therefore, the potential of CTC utilisation in the identification of resistance mechanisms to these and similar agents may have clinical utility.
In summary, despite offering great promise to alter the outlook of this challenging disease, a significant amount of further data, in many different areas of the management pathway, is needed before liquid biopsies are ready for prime-time.
| 1. | Malvezzi M, Bertuccio P, Rosso T, Rota M, Levi F, La Vecchia C, Negri E. European cancer mortality predictions for the year 2015: does lung cancer have the highest death rate in EU women? Ann Oncol. 2015;26:779-786. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 212] [Cited by in RCA: 203] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 2. | Neoptolemos J, Palmer D, Ghaneh P, Valle JW, Buchler MW. ESPAC-4: A multicentre, international, open-label randomized controlled phase III trial of adjuvant combination chemotherapy of gemcitabine (GEM) and capecitabine (CAP) versus monotherapy gemcitabine in patients with resected pancreatic ductal adenocarcinoma. J Clin Oncol. 2016;34 Suppl:abstr LBA4006. |
| 3. | Conroy T, Desseigne F, Ychou M, Bouché O, Guimbaud R, Bécouarn Y, Adenis A, Raoul JL, Gourgou-Bourgade S, de la Fouchardière C. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;364:1817-1825. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4838] [Cited by in RCA: 5913] [Article Influence: 394.2] [Reference Citation Analysis (24)] |
| 4. | Burris HA, Moore MJ, Andersen J, Green MR, Rothenberg ML, Modiano MR, Cripps MC, Portenoy RK, Storniolo AM, Tarassoff P. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J Clin Oncol. 1997;15:2403-2413. [PubMed] |
| 5. | Yeo TP. Demographics, epidemiology, and inheritance of pancreatic ductal adenocarcinoma. Semin Oncol. 2015;42:8-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 101] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 6. | Lavu H, Yeo CJ. Pancreatic ductal adenocarcinoma treatment--the past, present, and future. Semin Oncol. 2015;42:4-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 7. | Ghiorzo P. Genetic predisposition to pancreatic cancer. World J Gastroenterol. 2014;20:10778-10789. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 80] [Cited by in RCA: 80] [Article Influence: 6.7] [Reference Citation Analysis (1)] |
| 8. | Ducreux M, Cuhna AS, Caramella C, Hollebecque A, Burtin P, Goéré D, Seufferlein T, Haustermans K, Van Laethem JL, Conroy T. Cancer of the pancreas: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2015;26 Suppl 5:v56-v68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 905] [Cited by in RCA: 956] [Article Influence: 86.9] [Reference Citation Analysis (1)] |
| 9. | Drumm B, Neumann AW, Policova Z, Sherman PM. Bacterial cell surface hydrophobicity properties in the mediation of in vitro adhesion by the rabbit enteric pathogen Escherichia coli strain RDEC-1. J Clin Invest. 1989;84:1588-1594. [PubMed] |
| 10. | Rishi A, Goggins M, Wood LD, Hruban RH. Pathological and molecular evaluation of pancreatic neoplasms. Semin Oncol. 2015;42:28-39. [PubMed] |
| 11. | Ignatiadis M, Lee M, Jeffrey SS. Circulating Tumor Cells and Circulating Tumor DNA: Challenges and Opportunities on the Path to Clinical Utility. Clin Cancer Res. 2015;21:4786-4800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 281] [Article Influence: 28.1] [Reference Citation Analysis (0)] |
| 12. | Khoja L, Backen A, Sloane R, Menasce L, Ryder D, Krebs M, Board R, Clack G, Hughes A, Blackhall F. A pilot study to explore circulating tumour cells in pancreatic cancer as a novel biomarker. Br J Cancer. 2012;106:508-516. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 179] [Cited by in RCA: 212] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 13. | Lianidou ES, Strati A, Markou A. Circulating tumor cells as promising novel biomarkers in solid cancers. Crit Rev Clin Lab Sci. 2014;51:160-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 95] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 14. | Bell K, McKenzie HA, Shaw DC. Haemoglobin, serum albumin and transferrin variants of Bali (Banteng) cattle, Bos (Bibos) javanicus. Comp Biochem Physiol B. 1990;95:825-832. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 102] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 15. | Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, Tibbe AG, Uhr JW, Terstappen LW. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10:6897-6904. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1810] [Cited by in RCA: 1980] [Article Influence: 94.3] [Reference Citation Analysis (0)] |
| 16. | Zhang Y, Wang F, Ning N, Chen Q, Yang Z, Guo Y, Xu D, Zhang D, Zhan T, Cui W. Patterns of circulating tumor cells identified by CEP8, CK and CD45 in pancreatic cancer. Int J Cancer. 2015;136:1228-1233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 80] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 17. | Rhim AD, Thege FI, Santana SM, Lannin TB, Saha TN, Tsai S, Maggs LR, Kochman ML, Ginsberg GG, Lieb JG. Detection of circulating pancreas epithelial cells in patients with pancreatic cystic lesions. Gastroenterology. 2014;146:647-651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 183] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 18. | Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235-1239. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3036] [Cited by in RCA: 2612] [Article Influence: 137.5] [Reference Citation Analysis (0)] |
| 19. | Gall TM, Frampton AE, Krell J, Jacob J, Stebbing J, Jiao LR. Is the detection of circulating tumor cells in locally advanced pancreatic cancer a useful prognostic marker? Expert Rev Mol Diagn. 2013;13:793-796. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 20. | Ren C, Han C, Zhang J, He P, Wang D, Wang B, Zhao P, Zhao X. Detection of apoptotic circulating tumor cells in advanced pancreatic cancer following 5-fluorouracil chemotherapy. Cancer Biol Ther. 2011;12:700-706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 48] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 21. | Cauley CE, Pitman MB, Zhou J, Perkins J, Kuleman B, Liss AS, Fernandez-Del Castillo C, Warshaw AL, Lillemoe KD, Thayer SP. Circulating Epithelial Cells in Patients with Pancreatic Lesions: Clinical and Pathologic Findings. J Am Coll Surg. 2015;221:699-707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 22. | de Albuquerque A, Kubisch I, Breier G, Stamminger G, Fersis N, Eichler A, Kaul S, Stölzel U. Multimarker gene analysis of circulating tumor cells in pancreatic cancer patients: a feasibility study. Oncology. 2012;82:3-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 89] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 23. | Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781-791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3360] [Cited by in RCA: 3450] [Article Influence: 156.8] [Reference Citation Analysis (0)] |
| 24. | Xenidis N, Markos V, Apostolaki S, Perraki M, Pallis A, Sfakiotaki G, Papadatos-Pastos D, Kalmanti L, Kafousi M, Stathopoulos E. Clinical relevance of circulating CK-19 mRNA-positive cells detected during the adjuvant tamoxifen treatment in patients with early breast cancer. Ann Oncol. 2007;18:1623-1631. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 54] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 25. | Xenidis N, Ignatiadis M, Apostolaki S, Perraki M, Kalbakis K, Agelaki S, Stathopoulos EN, Chlouverakis G, Lianidou E, Kakolyris S. Cytokeratin-19 mRNA-positive circulating tumor cells after adjuvant chemotherapy in patients with early breast cancer. J Clin Oncol. 2009;27:2177-2184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 195] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 26. | Berchtold D, Akovbiantz A. [The nonresectable pancreatic tumor, an interdisciplinary problem. The surgeon’s viewpoint]. Schweiz Med Wochenschr. 1988;118:773-774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2770] [Cited by in RCA: 3728] [Article Influence: 310.7] [Reference Citation Analysis (1)] |
| 27. | Earl J, Garcia-Nieto S, Martinez-Avila JC, Montans J, Sanjuanbenito A, Rodríguez-Garrote M, Lisa E, Mendía E, Lobo E, Malats N. Circulating tumor cells (Ctc) and kras mutant circulating free Dna (cfdna) detection in peripheral blood as biomarkers in patients diagnosed with exocrine pancreatic cancer. BMC Cancer. 2015;15:797. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 151] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 28. | Sausen M, Phallen J, Adleff V, Jones S, Leary RJ, Barrett MT, Anagnostou V, Parpart-Li S, Murphy D, Kay Li Q. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat Commun. 2015;6:7686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 295] [Cited by in RCA: 382] [Article Influence: 34.7] [Reference Citation Analysis (0)] |
| 29. | Kinugasa H, Nouso K, Miyahara K, Morimoto Y, Dohi C, Tsutsumi K, Kato H, Matsubara T, Okada H, Yamamoto K. Detection of K-ras gene mutation by liquid biopsy in patients with pancreatic cancer. Cancer. 2015;121:2271-2280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 186] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 30. | Zill OA, Greene C, Sebisanovic D, Siew LM, Leng J, Vu M, Hendifar AE, Wang Z, Atreya CE, Kelley RK. Cell-Free DNA Next-Generation Sequencing in Pancreatobiliary Carcinomas. Cancer Discov. 2015;5:1040-1048. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 221] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 31. | Sergeant G, Roskams T, van Pelt J, Houtmeyers F, Aerts R, Topal B. Perioperative cancer cell dissemination detected with a real-time RT-PCR assay for EpCAM is not associated with worse prognosis in pancreatic ductal adenocarcinoma. BMC Cancer. 2011;11:47. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 60] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 32. | Zhou J, Hu L, Yu Z, Zheng J, Yang D, Bouvet M, Hoffman RM. Marker expression in circulating cancer cells of pancreatic cancer patients. J Surg Res. 2011;171:631-636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 48] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 33. | Chausovsky G, Luchansky M, Figer A, Shapira J, Gottfried M, Novis B, Bogelman G, Zemer R, Zimlichman S, Klein A. Expression of cytokeratin 20 in the blood of patients with disseminated carcinoma of the pancreas, colon, stomach, and lung. Cancer. 1999;86:2398-2405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 34. | Medicaid CfMa. 2015 Medicare Clinical Laboratory Fee Schedule. : Centre for Medicare and Medicaid 2015; . |
| 35. | Chen VK, Arguedas MR, Kilgore ML, Eloubeidi MA. A cost-minimization analysis of alternative strategies in diagnosing pancreatic cancer. Am J Gastroenterol. 2004;99:2223-2234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 50] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 36. | Xenidis N, Perraki M, Kafousi M, Apostolaki S, Bolonaki I, Stathopoulou A, Kalbakis K, Androulakis N, Kouroussis C, Pallis T. Predictive and prognostic value of peripheral blood cytokeratin-19 mRNA-positive cells detected by real-time polymerase chain reaction in node-negative breast cancer patients. J Clin Oncol. 2006;24:3756-3762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 207] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 37. | Doyen J, Alix-Panabières C, Hofman P, Parks SK, Chamorey E, Naman H, Hannoun-Lévi JM. Circulating tumor cells in prostate cancer: a potential surrogate marker of survival. Crit Rev Oncol Hematol. 2012;81:241-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 55] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 38. | Kuligowski ME, Chang A, Rath R. Multiple fixed drug eruption due to ibuprofen. Contact Dermatitis. 1991;25:259-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 121] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 39. | Frenel JS, Carreira S, Goodall J, Roda D, Perez-Lopez R, Tunariu N, Riisnaes R, Miranda S, Figueiredo I, Nava-Rodrigues D. Serial Next-Generation Sequencing of Circulating Cell-Free DNA Evaluating Tumor Clone Response To Molecularly Targeted Drug Administration. Clin Cancer Res. 2015;21:4586-4596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 161] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 40. | Heitzer E, Auer M, Gasch C, Pichler M, Ulz P, Hoffmann EM, Lax S, Waldispuehl-Geigl J, Mauermann O, Lackner C. Complex tumor genomes inferred from single circulating tumor cells by array-CGH and next-generation sequencing. Cancer Res. 2013;73:2965-2975. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 406] [Cited by in RCA: 401] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 41. | Stoecklein NH, Klein CA. Genetic disparity between primary tumours, disseminated tumour cells, and manifest metastasis. Int J Cancer. 2010;126:589-598. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 139] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 42. | Klein CA, Blankenstein TJ, Schmidt-Kittler O, Petronio M, Polzer B, Stoecklein NH, Riethmüller G. Genetic heterogeneity of single disseminated tumour cells in minimal residual cancer. Lancet. 2002;360:683-689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 368] [Cited by in RCA: 350] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 43. | Powell AA, Talasaz AH, Zhang H, Coram MA, Reddy A, Deng G, Telli ML, Advani RH, Carlson RW, Mollick JA. Single cell profiling of circulating tumor cells: transcriptional heterogeneity and diversity from breast cancer cell lines. PLoS One. 2012;7:e33788. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 407] [Cited by in RCA: 422] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 44. | Lu C, Xu CF, Wan XY, Zhu HT, Yu CH, Li YM. Screening for pancreatic cancer in familial high-risk individuals: A systematic review. World J Gastroenterol. 2015;21:8678-8686. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 67] [Cited by in RCA: 64] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 45. | Vasen H, Ibrahim I, Ponce CG, Slater EP, Matthäi E, Carrato A, Earl J, Robbers K, van Mil AM, Potjer T. Benefit of Surveillance for Pancreatic Cancer in High-Risk Individuals: Outcome of Long-Term Prospective Follow-Up Studies From Three European Expert Centers. J Clin Oncol. 2016;34:2010-2019. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 212] [Cited by in RCA: 277] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 46. | Langer P, Kann PH, Fendrich V, Habbe N, Schneider M, Sina M, Slater EP, Heverhagen JT, Gress TM, Rothmund M. Five years of prospective screening of high-risk individuals from families with familial pancreatic cancer. Gut. 2009;58:1410-1418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 174] [Article Influence: 10.2] [Reference Citation Analysis (2)] |
| 47. | Bissolati M, Sandri MT, Burtulo G, Zorzino L, Balzano G, Braga M. Portal vein-circulating tumor cells predict liver metastases in patients with resectable pancreatic cancer. Tumour Biol. 2015;36:991-996. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 92] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 48. | Fidler IJ. Metastasis: quantitative analysis of distribution and fate of tumor emboli labeled with 125 I-5-iodo-2’-deoxyuridine. J Natl Cancer Inst. 1970;45:773-782. [PubMed] |
| 49. | Gupta PB, Mani S, Yang J, Hartwell K, Weinberg RA. The evolving portrait of cancer metastasis. Cold Spring Harb Symp Quant Biol. 2005;70:291-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 70] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 50. | Goodman OB, Symanowski JT, Loudyi A, Fink LM, Ward DC, Vogelzang NJ. Circulating tumor cells as a predictive biomarker in patients with hormone-sensitive prostate cancer. Clin Genitourin Cancer. 2011;9:31-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 88] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 51. | de Albuquerque A, Kubisch I, Stölzel U, Ernst D, Boese-Landgraf J, Breier G, Stamminger G, Fersis N, Kaul S. Prognostic and predictive value of circulating tumor cell analysis in colorectal cancer patients. J Transl Med. 2012;10:222. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 49] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 52. | Obermayr E, Sanchez-Cabo F, Tea MK, Singer CF, Krainer M, Fischer MB, Sehouli J, Reinthaller A, Horvat R, Heinze G. Assessment of a six gene panel for the molecular detection of circulating tumor cells in the blood of female cancer patients. BMC Cancer. 2010;10:666. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 91] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 53. | Smerage JB, Barlow WE, Hortobagyi GN, Winer EP, Leyland-Jones B, Srkalovic G, Tejwani S, Schott AF, O’Rourke MA, Lew DL. Circulating tumor cells and response to chemotherapy in metastatic breast cancer: SWOG S0500. J Clin Oncol. 2014;32:3483-3489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 418] [Cited by in RCA: 505] [Article Influence: 42.1] [Reference Citation Analysis (0)] |
| 54. | Tie J, Tomasetti C, Li L, Springer S, Kinde I, Wong R et al. (2016) The potential of circulating tumor DNA (ctDNA) to reshape the design of clinical trials testing adjuvant therapy in patients with early stage cancer. J Clin Oncol. 2016;34 Suppl:abstr 3511. |
| 55. | Ma XL, Li YY, Zhang J, Huang JW, Jia HY, Liu L, Li P. Prognostic role of circulating tumor cells in patients with pancreatic cancer: a meta-analysis. Asian Pac J Cancer Prev. 2014;15:6015-6020. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 56. | Han L, Chen W, Zhao Q. Prognostic value of circulating tumor cells in patients with pancreatic cancer: a meta-analysis. Tumour Biol. 2014;35:2473-2480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 93] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 57. | Zhang L, Ridgway LD, Wetzel MD, Ngo J, Yin W, Kumar D, Goodman JC, Groves MD, Marchetti D. The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci Transl Med. 2013;5:180ra48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 358] [Cited by in RCA: 419] [Article Influence: 32.2] [Reference Citation Analysis (0)] |
| 58. | Yu M, Bardia A, Aceto N, Bersani F, Madden MW, Donaldson MC, Desai R, Zhu H, Comaills V, Zheng Z. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014;345:216-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 740] [Cited by in RCA: 753] [Article Influence: 62.8] [Reference Citation Analysis (0)] |
| 59. | Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, Dowling C, Wanjala JN, Undvall EA, Arora VK. Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159:176-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 882] [Cited by in RCA: 1203] [Article Influence: 100.3] [Reference Citation Analysis (0)] |
| 60. | Cayrefourcq L, Mazard T, Joosse S, Solassol J, Ramos J, Assenat E, Schumacher U, Costes V, Maudelonde T, Pantel K. Establishment and characterization of a cell line from human circulating colon cancer cells. Cancer Res. 2015;75:892-901. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 307] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 61. | Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, Parkinson C, Chin SF, Kingsbury Z, Wong AS. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1288] [Cited by in RCA: 1334] [Article Influence: 102.6] [Reference Citation Analysis (0)] |
| 62. | Passadouro M, Faneca H. Managing Pancreatic Adenocarcinoma: A Special Focus in MicroRNA Gene Therapy. Int J Mol Sci. 2016;17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 63. | Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1765] [Cited by in RCA: 1996] [Article Influence: 99.8] [Reference Citation Analysis (0)] |
| 64. | Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. J Cell Mol Med. 2009;13:39-53. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 705] [Cited by in RCA: 806] [Article Influence: 47.4] [Reference Citation Analysis (0)] |
| 65. | Kishikawa T, Otsuka M, Ohno M, Yoshikawa T, Takata A, Koike K. Circulating RNAs as new biomarkers for detecting pancreatic cancer. World J Gastroenterol. 2015;21:8527-8540. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 122] [Cited by in RCA: 120] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 66. | Cao Z, Liu C, Xu J, You L, Wang C, Lou W, Sun B, Miao Y, Liu X, Wang X. Plasma microRNA panels to diagnose pancreatic cancer: Results from a multicenter study. Oncotarget. 2016; Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 67. | Vila-Navarro E, Vila-Casadesús M, Moreira L, Duran-Sanchon S, Sinha R, Ginés À, Fernández-Esparrach G, Miquel R, Cuatrecasas M, Castells A, Lozano JJ, Gironella M. MicroRNAs for Detection of Pancreatic Neoplasia: Biomarker Discovery by Next-generation Sequencing and Validation in 2 Independent Cohorts. Ann Surg. 2016; Epub ahead of print. [PubMed] |
| 68. | Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, Dunning MJ, Gale D, Forshew T, Mahler-Araujo B. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368:1199-1209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1713] [Cited by in RCA: 1759] [Article Influence: 135.3] [Reference Citation Analysis (0)] |
| 69. | Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, Inserra E, Diederichs S, Iafrate AJ, Bell DW. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366-377. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1363] [Cited by in RCA: 1329] [Article Influence: 73.8] [Reference Citation Analysis (0)] |
| 70. | Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883-892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6102] [Cited by in RCA: 6090] [Article Influence: 435.0] [Reference Citation Analysis (0)] |
| 71. | Moore MJ, Goldstein D, Hamm J, Figer A, Hecht JR, Gallinger S, Au HJ, Murawa P, Walde D, Wolff RA. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol. 2007;25:1960-1966. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2835] [Cited by in RCA: 2793] [Article Influence: 147.0] [Reference Citation Analysis (0)] |
| 72. | Van Cutsem E, Vervenne WL, Bennouna J, Humblet Y, Gill S, Van Laethem JL, Verslype C, Scheithauer W, Shang A, Cosaert J. Phase III trial of bevacizumab in combination with gemcitabine and erlotinib in patients with metastatic pancreatic cancer. J Clin Oncol. 2009;27:2231-2237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 469] [Cited by in RCA: 506] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 73. | Philip PA, Benedetti J, Corless CL, Wong R, O’Reilly EM, Flynn PJ, Rowland KM, Atkins JN, Mirtsching BC, Rivkin SE. Phase III study comparing gemcitabine plus cetuximab versus gemcitabine in patients with advanced pancreatic adenocarcinoma: Southwest Oncology Group-directed intergroup trial S0205. J Clin Oncol. 2010;28:3605-3610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 446] [Cited by in RCA: 504] [Article Influence: 31.5] [Reference Citation Analysis (0)] |
| 74. | Heinemann V, Vehling-Kaiser U, Waldschmidt D, Kettner E, Märten A, Winkelmann C, Klein S, Kojouharoff G, Gauler TC, von Weikersthal LF. Gemcitabine plus erlotinib followed by capecitabine versus capecitabine plus erlotinib followed by gemcitabine in advanced pancreatic cancer: final results of a randomised phase 3 trial of the ‘Arbeitsgemeinschaft Internistische Onkologie’ (AIO-PK0104). Gut. 2013;62:751-759. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 90] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 75. | Infante JR, Somer BG, Park JO, Li CP, Scheulen ME, Kasubhai SM, Oh DY, Liu Y, Redhu S, Steplewski K. A randomised, double-blind, placebo-controlled trial of trametinib, an oral MEK inhibitor, in combination with gemcitabine for patients with untreated metastatic adenocarcinoma of the pancreas. Eur J Cancer. 2014;50:2072-2081. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 286] [Article Influence: 23.8] [Reference Citation Analysis (1)] |
| 76. | Ko AH, Youssoufian H, Gurtler J, Dicke K, Kayaleh O, Lenz HJ, Keaton M, Katz T, Ballal S, Rowinsky EK. A phase II randomized study of cetuximab and bevacizumab alone or in combination with gemcitabine as first-line therapy for metastatic pancreatic adenocarcinoma. Invest New Drugs. 2012;30:1597-1606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 45] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 77. | Ko AH, Venook AP, Bergsland EK, Kelley RK, Korn WM, Dito E, Schillinger B, Scott J, Hwang J, Tempero MA. A phase II study of bevacizumab plus erlotinib for gemcitabine-refractory metastatic pancreatic cancer. Cancer Chemother Pharmacol. 2010;66:1051-1057. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 57] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: United Kingdom
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
P- Reviewer: Furukawa T, Garg PK, Ghiorzo P S- Editor: Yu J L- Editor: A E- Editor: Wang CH