Published online Apr 28, 2016. doi: 10.3748/wjg.v22.i16.4168
Peer-review started: December 2, 2015
First decision: January 13, 2016
Revised: February 16, 2016
Accepted: March 1, 2016
Article in press: March 2, 2016
Published online: April 28, 2016
Processing time: 140 Days and 13.6 Hours
AIM: To develop a mathematical model for the early detection of hepatocellular carcinoma (HCC) with a panel of serum proteins in combination with α-fetoprotein (AFP).
METHODS: Serum levels of interleukin (IL)-8, soluble intercellular adhesion molecule-1 (sICAM-1), soluble tumor necrosis factor receptor II (sTNF-RII), proteasome, and β-catenin were measured in 479 subjects categorized into four groups: (1) HCC concurrent with hepatitis C virus (HCV) infection (n = 192); (2) HCV related liver cirrhosis (LC) (n = 96); (3) Chronic hepatitis C (CHC) (n = 96); and (4) Healthy controls (n = 95). The R package and different modules for binary and multi-class classifiers based on generalized linear models were used to model the data. Predictive power was used to evaluate the performance of the model. Receiver operating characteristic curve analysis over pairs of groups was used to identify the best cutoffs differentiating the different groups.
RESULTS: We revealed mathematical models, based on a binary classifier, made up of a unique panel of serum proteins that improved the individual performance of AFP in discriminating HCC patients from patients with chronic liver disease either with or without cirrhosis. We discriminated the HCC group from the cirrhotic liver group using a mathematical model (-11.3 + 7.38 × Prot + 0.00108 × sICAM + 0.2574 ×β-catenin + 0.01597 × AFP) with a cutoff of 0.6552, which achieved 98.8% specificity and 89.1% sensitivity. For the discrimination of the HCC group from the CHC group, we used a mathematical model [-10.40 + 1.416 × proteasome + 0.002024 × IL + 0.004096 × sICAM-1 + (4.251 × 10-4) × sTNF + 0.02567 ×β-catenin + 0.02442 × AFP] with a cutoff 0.744 and achieved 96.8% specificity and 89.7% sensitivity. Additionally, we derived an algorithm, based on a binary classifier, for resolving the multi-class classification problem by using three successive mathematical model predictions of liver disease status.
CONCLUSION: Our proposed mathematical model may be a useful method for the early detection of different statuses of liver disease co-occurring with HCV infection.
Core tip: Hepatocellular carcinoma is one of the most common liver malignancies. We sought to create a mathematical model from a panel of serum proteins (intercellular adhesion molecules, beta catenin, interleukin-8, the proteasome, and soluble tumor necrosis factor receptor II) in combination with α-fetoprotein to aid in early detection of hepatocellular carcinoma. This panel was measured in 384 subjects infected with hepatitis C virus (HCV) as well as 95 healthy control subjects negative for HCV. Finally, we created mathematical models that may be valuable tools for the early detection of different statuses of liver disease co-occurring with HCV infection.
- Citation: Zekri ARN, Youssef ASED, Bakr YM, Gabr RM, Ahmed OS, Elberry MH, Mayla AM, Abouelhoda M, Bahnassy AA. Early detection of hepatocellular carcinoma co-occurring with hepatitis C virus infection: A mathematical model. World J Gastroenterol 2016; 22(16): 4168-4182
- URL: https://www.wjgnet.com/1007-9327/full/v22/i16/4168.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i16.4168
Hepatocellular carcinoma (HCC) is one of the most common primary liver malignancies and the third leading cause of cancer related mortality, and it is predominant in males[1,2]. Among the risk factors for developing HCC is the hepatitis C virus (HCV), which has been considered to be the second most common cause of HCC and the most common cause in Egypt, Japan, and the United States[3]. Egypt has the highest prevalence of HCV (genotype 4, predominately) worldwide, estimated to be 14.7% nationally[4].
Using the serum α-fetoprotein (AFP) level, the most widely used tumor biomarker, for the early diagnosis of HCC has several limitations[5]. Because of its low sensitivity, only 44% of patients are diagnosed at a localized disease stage, and only 30% of HCC patients are candidates for potentially curative treatments[6]. Moreover, the heterogeneity of HCC due to the coexistence of inflammation and cirrhosis makes the early detection of HCC difficult[7]. This complication highlights the need to identify valuable biomarkers for the early detection of HCC. Although single markers lack sensitivity and specificity for accurate cancer detection[8], specific panels of markers may offer an improvement in diagnostic performance. The ability of novel biomarkers to accurately detect HCC depends on their capacity to discriminate HCC from benign diseases of the liver, such as chronic hepatitis and liver cirrhosis.
Novel biomarkers for early diagnosis should meet the following criteria: first, they should achieve high accuracy; second, sample collection for detecting the markers should be easily operable and non-invasive; and third, the detection method should be cost effective[9].
Emerging key candidate biomarkers include mediators of the tumor microenvironment and the host response, notably cytokines involved in the immune system, inflammation, tumor development, and metastasis, e.g., interleukin (IL)-8 and tumor necrosis factor (TNF)-α[10-12]; enzymes and isozymes involved in basic cellular processes, such as cell-cycle regulation, apoptosis, transcriptional regulation, and antigen processing, e.g., proteasomes[13,14]; proteantigen mediated adhesion-dependent cell-cell interactions facilitating cell metastasis and evading the immune system, e.g., soluble intercellular adhesion molecule-1 (sICAM-1)-1[15,16]; and proteantigen mediated Wnt/wingless (Wg) signaling pathway, which is involved in a large variety of developmental processes, including cell fate regulation, proliferation and self-renewal of stem and progenitor cells, e.g., β-catenin[17,18].
Our current study was conducted as a confirmatory study of our four previously published papers: Zekri et al[19-22], to address the potential roles of sICAM-1, β-catenin, the proteasome, IL-8 and sTNFR-II in the early detection of HCC. Additionally, it was conducted as a complementary study to our previously published paper by Zekri et al[23], to address the potential role of combinations of sICAM-1, β-catenin, IL-8, the proteasome, and soluble soluble tumor necrosis factor receptor II (sTNF-RII) or subsets of these biomarkers with AFP, using logistic disease predictor models, in the early detection of HCC in a normal population and in high risk patients (patients with chronic hepatitis C and liver cirrhosis). We revealed that mathematical models identified a unique panel of biomarkers that improved the individual performance of AFP for the discrimination of HCC patients from patients with benign liver disease. Moreover, in the presence of inflammation and cirrhosis, whereas AFP offered relatively poor discrimination of HCC patients from benign disease patients, our mathematical model afforded better discrimination.
This is a retrospective case-control study conducted on 384 adult patients with HCV related chronic diseases classified into: 192 patients with HCC recruited from the multidisciplinary HCC clinic, Tropical Medicine Department, Faculty of Medicine and National Cancer Institute (NCI) Outpatients Clinic, Cairo University; 96 patients with liver cirrhosis (LC) recruited from the Endemic Medicine Department, Faculty of Medicine, Cairo University and 96 chronic hepatitis C (CHC) patients recruited from the Kasr El Aini Viral Hepatitis Center, Faculty of Medicine, Cairo University, in addition to 95 healthy subjects enrolled as the control group from May 2012 to April 2013. The study was approved by the Investigation and Ethics Committee of NCI and written informed consent was obtained from all persons involved.
Patients with HCC were diagnosed by abdominal ultrasonography and triphasic computed tomography (CT) of the abdomen, and serum AFP and were confirmed histopathologically. The patients also showed no evidence of local invasion or distant metastasis. Notably, diabetes mellitus is a risk factor for HCC development. All of the 192 HCC patients, diabetic or not, were included in the mathematical modeling process. However, patients with HCV related liver cirrhosis were diagnosed by abdominal ultrasonography, and the diagnoses were confirmed histopathologically. Patients with chronic hepatitis C were characterized by persistent increases of the alanine aminotransferase (ALT) values to more than three times normal level for at least 6 mo. The control group showed no clinical or biochemical evidence of liver disease with normal abdominal ultra-sonography.
The exclusion criteria included being free of HBV and HCV infection, as confirmed by ELISA and PCR, as well as being free of diabetes. Regarding the groups with HCV liver related diseases, patients having HBV infection or those who received previous treatment or antiviral therapy for HCV were excluded from the study. In addition, all alcoholic subjects were excluded.
Five milliliters of venous blood was collected and allowed to coagulate for 30 min before centrifugation at 5000 rpm for 10 min. The serum fraction was aliquoted into cryotubes and stored at -80 °C until use.
Serum levels of sICAM-1, IL-8, and sTNF-RII were measured with an enzyme linked immunosorbent assay (ELISA) kit from R&D Systems Inc, Minneapolis, MN, United States, Proteasome levels were measured by an ELISA kit from Enzo Life Sciences, Inc., Switzerland and β-catenin was measured by an ELISA kit from Glory Science Co., Ltd, United States according to the manufacturers’ instructions. These assays used the quantitative sandwich enzyme immunoassay technique. A monoclonal antibody specific for sICAM-1, IL-8, sTNF-RII, β-catenin, or proteasomes was used. The average of the duplicate readings for each standard, control, and sample was taken, and the average zero standard optical density was subtracted. A standard curve was constructed by plotting the mean absorbance for each standard on the Y-axis against the concentration on the X-axis, and a best fit curve was drawn through the points on the graph. By interpolation, the unknown concentrations of sICAM-1, IL-8, sTNF-RII, β-catenin, and proteasomes could be determined.
The SPSS software package (version 15) and the R programming environment were used to analyze the data. Continuous variables were expressed as the mean ± SD, median and interquartile range. Comparisons between groups were analyzed by non-parametric one-way ANOVA for continuous variables and by the χ2 test for categorical variables. P values ≤ 0.05 were considered significant. Correlations between the variables were analyzed using Spearman’s correlation coefficient.
In our model construction, we followed two strategies: The first was using the glmnet function as a multi-class classifier based on multinomial logit regression to differentiate between the four classes in our study including the control, cirrhotic, non-cirrhotic, and HCC groups. The second was using the glm function to study different pairwise combinations among the classes. The details are as follows:
Use of the multi-class classifier: We used the glmnet function to analyze the input data based on the multinomial logit mode. To find the best fit, we used the cross validation version of the glmnet, which tested different values of the lambda parameter. The four classes were labeled with discrete values between 1 and 4. To evaluate the performance of the model, we used the predict function to compute the response after applying the model. The predicted values were float values ranging between 1 and 5. To find the best cutoffs differentiating among different classes, we used receiver operating characteristic (ROC) curve analysis over pairs of classes.
The use of the binary-class classifier over pairs of classes: We tested different pairwise class combinations in nine combinations: (1) disease vs control; (2) HCV vs control; (3) HCC vs HCV; (4) HCC vs LC; (5) LC vs CHC; (6) CHC vs control; (7) HCC vs non-HCC; (8) LC vs control; and (9) HCC vs LC. For each of these combinations, we used the glm function to analyze the input data based on the binomial logit mode. We used the predict function in combination with ROC curve analysis to find the best cutoffs differentiating the two classes in each combination. Visualization of the results was performed using different R packages and functions.
The clinical data of the studied groups are shown in (Table 1). The mean age of the HCC group was significantly higher than that in other groups (P < 0.001). Thus, there was a trend of increasing age with the progression of the disease from chronic hepatitis through liver cirrhosis to hepatocellular carcinoma.
| HCC (n = 192) | Cirrhotic (n = 96) | Non-cirrhotic (n = 96) | Control (n = 95) | P value | |
| Age, mean ± SD (range) | 56.7 ± 7.7a (29-80) | 54.01 ± 8.3b (27-66) | 40.54 ± 8.82c (22-61) | 33.37 ± 11d (19-62) | < 0.001 |
| Gender | |||||
| Male | 152a (79) | 67a (70) | 78a (81) | 21b (22) | < 0.001 |
| Female | 40 a (21) | 29a (30) | 18a (19) | 74b (78) | |
| Smoker | |||||
| Yes | 75a (39) | 26ab (27) | 20b (21) | 3c (3) | < 0.001 |
| No | 117a (61) | 70ab (73) | 76b (79) | 92c (97) | |
| DM | |||||
| Yes | 43a (22) | 22a (23) | 8b (8) | 0c (0) | < 0.001 |
| No | 149a (78) | 74a (77) | 88b (92) | 95c (100) | |
| HCV Ab | |||||
| Present | 168a (88) | 90a (94) | 96b (100) | 0c | < 0.001 |
| Absent | 24a (12) | 6a (6) | 0b (0) | 95c (100) | |
| HBs Ag | |||||
| Present | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0.57 |
| Absent | 191 (100) | 96 (100) | 96 (100) | 95 (100) | |
| Ascites | |||||
| Yes | 86a (45) | 75b (78) | 0c (0) | 0c (0) | < 0.001 |
| No | 106a (55) | 21b (22) | 96c (100) | 95c (100) | |
| Child score | |||||
| A | 69a (36) | 12b (12) | < 0.001 | ||
| B | 69a (36) | 27a (28) | |||
| C | 49a (28) | 57b (60) | |||
| ALT, | |||||
| mean | 64.79 ± 52.737 | 63.6 ± 42.54 | 63.41 ± 44.22 | 21.84 ± 5.04 | < 0.001 |
| (range) | (5-395) | (6-290) | (10-223) | (11-33) | |
| median | 51a | 60a | 54a | 22b | |
| AST, | |||||
| mean | 94.39 ± 105.61 | 53.91 ± 31.72 | 47.53 ± 29.04 | 26.68 ± 5.07 | < 0.001 |
| (range) | (16-1155) | (16-176) | (8-167) | ( 15-37) | |
| median | 77a | 46.5b | 39.5b | 27c | |
| T-Bil, | |||||
| mean | 2.4 ± 2.63 | 3.25 ± 2.51 | 0.88 ± 0.49 | 0.79 ± 0.16 | < 0.001 |
| (range) | (0.3-25.8) | (0.2-19.7) | (0.3-5) | (0.5-1.1) | |
| median | 1.7a | 2.86b | 0.84c | 0.8c | |
| Albumin, | |||||
| mean | 3.02 ± 0.63 | 2.55 ± 0.51 | 4.28 ± 0.4 | 4.45 ± 0.34 | < 0.001 |
| (range) | (1.8-4.8) | (1.7-4.3) | (3.2-5.4) | (3.8-5.2) | |
| median | 3.1a | 2.5b | 4.2c | 4.5d | |
| AFP, | |||||
| mean | 3933.25 ± 16142 | 35.21 ± 40.1 | 18.19 ± 29.62 | < 0.001 | |
| (range) | (1.5-114170) | (1.7-190) | (0.65-112) | 0 | |
| median | 152a | 17.95b | 4.6c | 0d |
Regarding gender differences, in the majority of the HCV related liver disease patients in the three groups were male. Risk factors for HCC such as diabetes mellitus (DM) were reported in 22.5% of HCC patients, 23% of LC patients, 8% of CHC patients, and 0% of controls. The percentage of diabetic patients was significantly higher in patients with HCC and LC compared to the CHC group and the control group (P < 0.001).
Cigarette smokers comprised 39% of the HCC group, 27% of the LC group, 21% of the CHC group, and 3% of the control group, with a significant difference between patients with (HCC and CHC) and the control group (P < 0.001). There was no significance between the LC group and either the HCC group or the CHC group.
Regarding liver function, the median value of ALT was significantly higher in the HCV related liver disease patients in the three patient groups than in the control group (P < 0.001). The median value of AST was significantly higher in the HCC group than in the other groups (P < 0.001), whereas there was no significant difference in the median AST value between the LC group and the CHC group. The median value of T-bilirubin was significantly higher in the LC group than in the other groups (P < 0.001). The median value of albumin was significantly higher in the normal group than in the HCV related liver disease patients in the three patient groups (P < 0.001). The median value of albumin in patients with LC was significantly lower than in the patients (HCC and CHC) and healthy controls (P < 0.001).
Regarding ascites, 42% of the HCC group and 78% of the LC group showed ascites, and there was a significant difference between the two groups (P < 0.001). The severity of liver disease in the HCC and cirrhotic groups was assessed using the child score; 36% of HCC group scored child A, 36% scored child B, and 28% scored child C compared to 12.5%, 28%, and 59.5% in the LC group, respectively (P < 0.001), consistently with the conclusion that the LC group had a more decompensated pattern than the HCC group.
Spearman’s rank analysis of the studied serological markers for the control group showed a maximum rho of -0.218, indicating little or no correlation between age and analyte level in the control group, because the rho value was in the range of 0-(-0.25), as shown in Supplementary Table 1.
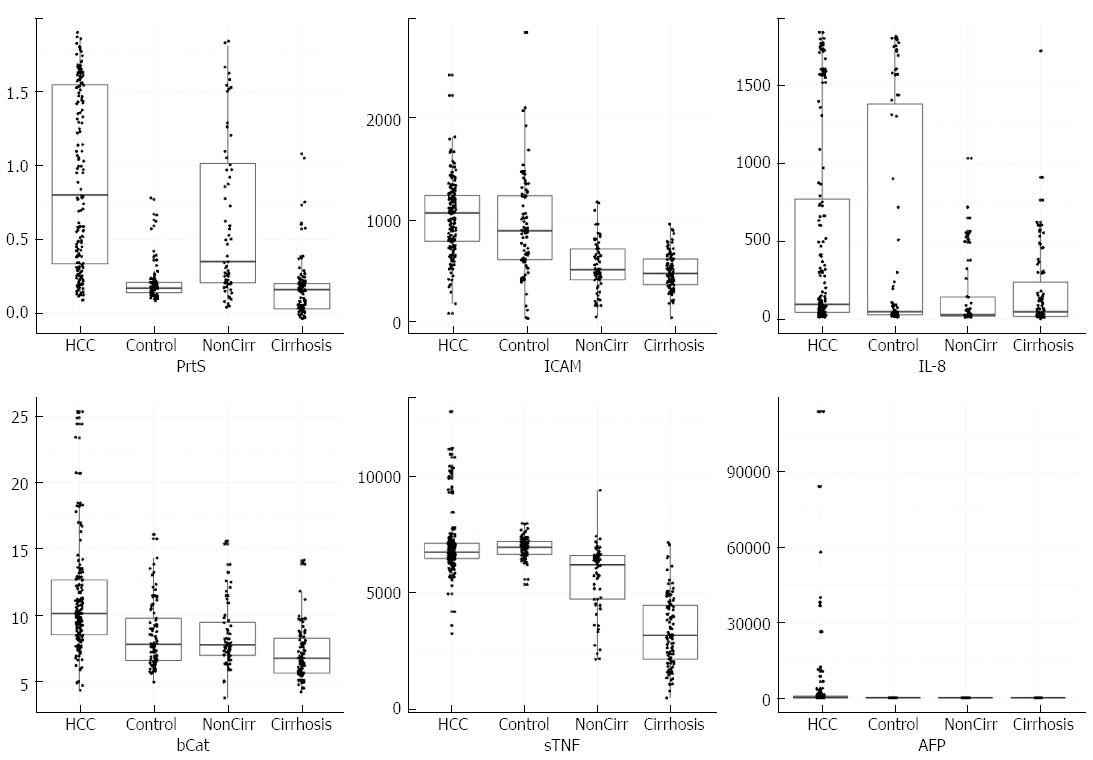
The levels of the studied biomarkers in the different studied groups were expressed as box plots with scattered measurement points (Figure 1); more details such as the mean ± SD, median and range are shown in (Table 2).
| HCC (n = 192) | Cirrhotic (n = 96) | Non-Cirrhotic (n = 96) | Control (n = 95) | P value | |
| Proteosome, | |||||
| median | 0.8a | 0.17b | 0.4c | 0.16d | < 0.001 |
| mean ± SD | 0.91 ± 0.6 | 0.25 ± 0.25 | 0.68 ± 0.58 | 0.18 ± 0.3 | |
| range | 0.13-1.87 | 0.12-1.67 | 0.09-1.83 | 0.01-2.22 | |
| IL-8, | |||||
| median | 107a | 55a | 36.5b | 47b | < 0.001 |
| mean ± SD | 518.19 ± 656.23 | 552.9 ± 732.2 | 283.76 ± 442.7 | 238.8 ± 431.6 | |
| range | 14-1837 | 14-1811 | 8-1734 | 3-1719 | |
| sICAM-1, | |||||
| median | 1072.5a | 892b | 510.5c | 473c | < 0.001 |
| mean ± SD | 1034.73 ± 372.7 | 978.17 ± 540.17 | 570 ± 287 | 493 ± 188 | |
| range | 79-2419 | 31-2838 | 46-1654 | 37-1078 | |
| sTNF-RII, | |||||
| median | 6785a | 7011.5b | 5948.5c | 3190.5d | < 0.001 |
| mean ± SD | 7058.31 ± 1338.78 | 6984.5 ± 626.8 | 5456 ± 1531.7 | 3400 ± 1519 | |
| range | 3250-12776 | 4351-9919 | 1772-9403 | 495-7157 | |
| β-catenin, | |||||
| median | 10.1a | 7.95b | 7.85b | 6.8c | < 0.001 |
| mean ± SD | 11.26 ± 5.49 | 9.34 ± 5.13 | 8.81 ± 4.07 | 7.5 ± 3.2 | |
| range | 4.3-55.4 | 5.0-49.2 | 3.8-41.6 | 4.3-28.4 |
The median proteasome serum concentration was significantly elevated in patients with HCC compared with patients with LC and CHC and healthy controls (P < 0.001) with mean values 0.91 ± 0.6, 0.25 ± 0.25, 0.68 ± 0.58, and 0.19 ± 0.3 μg/mL, respectively. However, the median proteasome concentration was significantly elevated in patients with CHC compared with patients with LC and the healthy controls (P < 0.001).
Moreover, the median concentration of serum IL-8 was significantly higher in patients with HCC and LC than in patients with CHC and the healthy controls, with mean values of 518.19 ± 656.23 pg/mL, 552.9 ± 732.2 pg/mL, 283.76 ± 442.7 pg/mL, and 238.8 ± 431.6 pg/mL, respectively (P < 0.001). However, there was no significant difference in the median proteasome concentrations in patients with HCC and LC (P = 0.09). Additionally, no significant difference was observed in median proteasome concentrations in patients with CHC and healthy controls.
Additionally, the median serum concentration of sICAM-1 was significantly elevated in patients in the HCC group compared to patients with LC and CHC and the healthy controls with mean values of 1034.73 ± 373.7 ng/mL, 978.17 ± 540.17 ng/mL, 570 ± 287 ng/mL, and 493 ± 188 ng/mL, respectively (P < 0.001). However, there was no significant difference in median sICAM-1 concentrations in patients with CHC and the healthy controls (P = 0.07). Moreover, the median serum concentration of sTNF-RII was significantly higher in patients with LC compared to patients with HCC, CHC, and the control group with mean values of 6984.5 ± 626.8 pg/mL, 7058.31 ± 1338.78 pg/mL, 5456 ± 1531.7 pg/mL, and 3400 ± 1519 pg/mL, respectively (P < 0.001). Also, the median sTNR-RII serum concentration was significantly elevated in patients with HCC compared with patients with CHC and the healthy controls (P < 0.001).
Additionally, the median concentration of β-catenin was significantly higher in patients with HCC than in patients with (LC and CHC) and healthy controls (P < 0.001) with mean values 11.26 ± 5.49 ng/mL, 9.34 ± 5.13 ng/mL, 8.8 ± 4.07 ng/mL, and 7.5 ± 3.2 ng/mL, respectively, whereas there were no significant differences in its median β-catenin concentrations in patients with LC and CHC (P = 0.76).
Correlation analysis of the studied serological markers performed with Spearman’s correlation coefficient (Table 3) revealed a moderately positive correlation between sTNF-RII and sICAM-1 or β-catenin (r = 0.482, 0.264; P < 0.01), respectively, and the rho values were in the range of (0.25-0.5). However, there was little positive correlation or no correlation between the levels of IL-8 and the proteasome, sICAM-I, sTNF-RII, or β-catenin levels (r = 0.240, 0.233, 0.184, 0.141; P < 0.01), respectively. Additionally, there was little positive correlation or no correlation between the proteasome levels and sICAM-I, β-catenin, and sTNF-RII (r = 0.240, 230, 0.18; P < 0.01), respectively. Moreover, there was little positive correlation or no correlation between the levels of β-catenin and sICAM-1 (r = 0.222; P < 0.01), because the rho values were in the range of (0-0.25).
| Proteosome | IL-8 | sICAM-1 | sTNF-RII | β-catenin | |
| Proteosome | 1 | 0.2401 | 0.2401 | 0.1851 | 0.2301 |
| 0.000 | 0.000 | 0.000 | 0.000 | ||
| IL-8 | 0.2401 | 1 | 0.2331 | 0.1841 | 0.1411 |
| 0.000 | 0.000 | 0.000 | 0.003 | ||
| sICAM-1 | 0.2401 | 0.2331 | 1 | 0.5471 | 0.2221 |
| 0.000 | 0.000 | 0.000 | 0.000 | ||
| sTNF-RII | 0.1851 | 0.1841 | 0.5471 | 1 | 0.2641 |
| 0.000 | 0.000 | 0.000 | 0.000 | ||
| β-catenin | 0.2301 | 0.1411 | 0.2221 | 0.2641 | 1 |
| 0.000 | 0.003 | 0.000 | 0.000 |
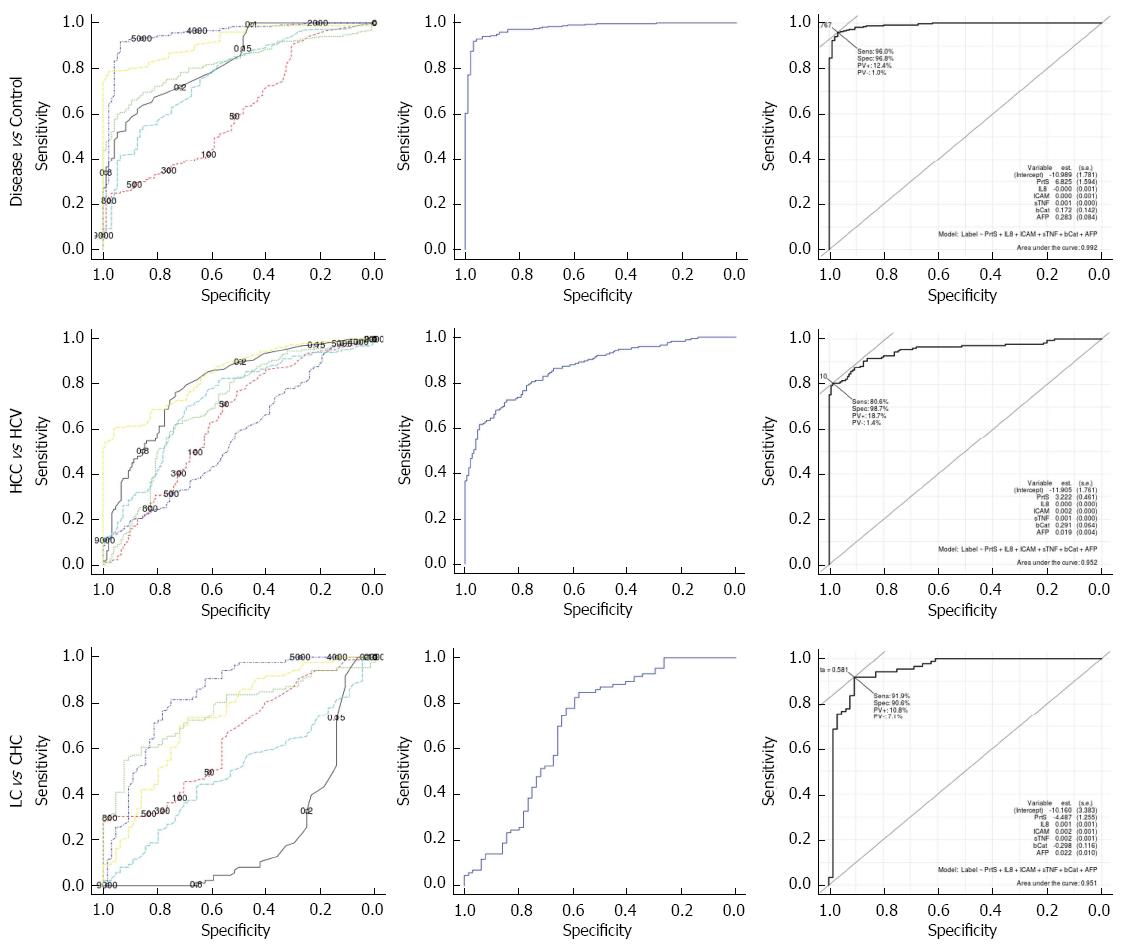
Further analyses of the data using ROC analysis curves and the corresponding area under the curve were performed to investigate the diagnostic accuracy of the individual studied markers and combined markers with AFP.
Table 4 summarizes the area under the curve (AUC) for individual serum markers and combined markers, using two different methods, in different comparisons of the studied groups. Method 1 was the implementation of a multi-class classifier based on logistic regression, whereas method 2 was the implementation of a binary-class classifier based on logistic regression. Individual serum marker results revealed that sTNF-RII had the highest diagnostic performance in discriminating LC cases from healthy controls (AUC = 0.981); CHC cases from healthy controls (AUC = 0.857); LC cases from CHC cases (AUC = 0.829); diseased cases (patients with HCC, LC, and CHC) from healthy controls (AUC = 0.951); and HCV cases (patients with LC and CHC) from healthy controls (AUC = 0.928). However, the proteasome levels had the highest diagnostic performance in discriminating HCC cases from LC cases (AUC = 0.981). AFP had the highest diagnostic performance in discriminating HCC cases from HCV cases (AUC = 0.849); HCC cases from non-HCC cases (healthy controls, LC group, CHC group) (AUC = 0.902); HCC cases from CHC cases (AUC = 0.908), and HCC cases from healthy controls (AUC = 0.986).
| Method 1 | Method 2 | Proteasome | IL-8 | sICAM-1 | sTNF-RII | β-catenin | AFP | |
| Disease vs Control | 0.979 | 0.992 | 0.834 | 0.612 | 0.825 | 0.951 | 0.768 | 0.923 |
| HCC vs LC | 0.841 | 0.962 | 0.911 | 0.576 | 0.585 | 0.424 | 0.718 | 0.804 |
| HCC vs HCV | 0.866 | 0.952 | 0.799 | 0.639 | 0.705 | 0.568 | 0.723 | 0.849 |
| HCC vs Non-HCC | 0.917 | 0.970 | 0.848 | 0.652 | 0.787 | 0.724 | 0.771 | 0.902 |
| HCV vs Control | 0.957 | 0.983 | 0.726 | 0.543 | 0.716 | 0.928 | 0.677 | 0.850 |
| LC vs CHC | 0.695 | 0.951 | 0.220 | 0.648 | 0.769 | 0.829 | 0.494 | 0.736 |
| CHC vs Control | 0.917 | 0.968 | 0.846 | 0.471 | 0.571 | 0.857 | 0.678 | 0.749 |
| LC vs Control | 0.990 | 1.000 | 0.637 | 0.597 | 0.825 | 0.981 | 0.676 | 0.924 |
| HCC vs Control | 0.998 | 1.000 | 0.927 | 0.672 | 0.917 | 0.970 | 0.846 | 0.986 |
| HCC vs CHC | 0.967 | 0.971 | 0.648 | 0.724 | 0.867 | 0.762 | 0.730 | 0.908 |
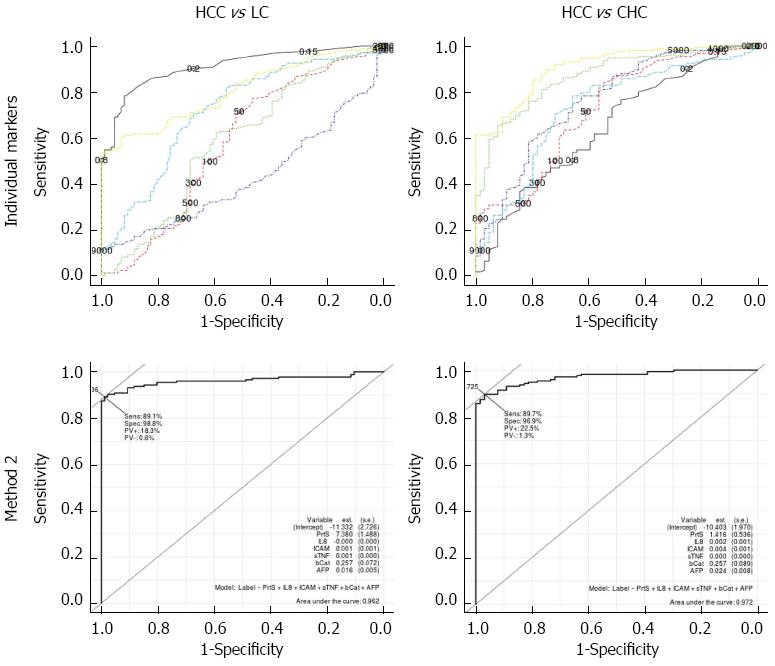
Additionally, the results in the table showed that Method 2 (the binary-class classifier) over the different pairwise comparison of the studied groups was superior to both Method 1 (the multi-class one) and individual markers, as shown in (Figures 2 and 3). For the discrimination of HCC cases from LC cases; Method 2 offered improved diagnostic performance (AUC = 0.962) over Method 1 (AUC = 0.840) and the top performing marker, proteasome levels (AUC = 0.911). For the discrimination of HCC cases from CHC cases, Method 2 offered improved diagnostic performance (AUC = 0.972) over Method 1 (AUC = 0.967) and the top performing marker, AFP (AUC = 0.908). For the discrimination of HCC cases from healthy controls, Method 2 offered improved diagnostic performance (AUC = 1) over Method 1(AUC = 0.998) and the top performing marker, AFP (AUC = 0.986).
Table 5 presents the best cutoff based on Youden statistics for each model and the related statistical measures of the performance for the binary-class classifier. All models over different pairwise comparisons of the studied groups achieved high diagnostic performance using all measures: the sensitivity ranged from 80.5% to 100%; the specificity ranged from 90.5% to 100%; the accuracy ranged from 88.9% to 100%; the PPV (positive predictive values) ranged from 87.9% to 100%; and the NPV (negative predictive values) ranged from 81.3% to 100%.
| AUC | BT_Y | Spec (%) | Sen (%) | Acc (%) | TN | TP | FN | FP | NPV (%) | PPV (%) | |
| Disease vs Control | 0.992 | 0.764 | 96.8 | 96.0 | 96.2 | 92 | 312 | 13 | 3 | 87.6 | 99.0 |
| HCC vs LC | 0.962 | 0.655 | 98.8 | 89.1 | 92.3 | 85 | 156 | 19 | 1 | 81.7 | 99.3 |
| HCC vs HCV | 0.952 | 0.712 | 98.7 | 80.5 | 88.9 | 148 | 141 | 34 | 2 | 81.3 | 98.6 |
| HCC vs Others | 0.970 | 0.378 | 91.0 | 91.4 | 91.2 | 223 | 160 | 15 | 22 | 93.7 | 87.9 |
| HCV vs Control | 0.983 | 0.764 | 96.8 | 91.3 | 93.5 | 92 | 137 | 13 | 3 | 87.6 | 97.8 |
| LC vs CHC | 0.950 | 0.581 | 90.6 | 91.9 | 91.3 | 58 | 79 | 7 | 6 | 89.2 | 92.9 |
| CHC vs Control | 0.968 | 0.341 | 90.5 | 93.7 | 91.8 | 86 | 60 | 4 | 9 | 95.5 | 86.9 |
| LC vs Control | 1.000 | 0.500 | 100.0 | 100.0 | 100.0 | 95 | 86 | 0 | 0 | 100.0 | 100.0 |
| HCC vs Control | 1.000 | 0.500 | 100.0 | 100.0 | 100.0 | 95 | 175 | 0 | 0 | 100.0 | 100.0 |
| HCC vs CHC | 0.971 | 0.745 | 96.8 | 89.7 | 91.6 | 62 | 157 | 18 | 2 | 77.5 | 98.7 |
Table 6 presents the different binary-class classifier based mathematical models that best fit the data for each pairwise comparison. Table 7 presents the reduced models with the most significant markers (significance level at P < 0.05) and with the best cut-off. From these models, only three markers (sTNF-RII, proteasome, and AFP) were able to discriminate healthy controls from all other groups. For further discrimination between the other studied groups; sICAM-1, IL-8, and β-catenin were also needed in addition to sTNF-RII, the proteasome, and AFP.
| Combination | Model |
| Disease vs Control | [-11 + 6.83 × Prot + (-5.99 × 10-5) × IL-8 + 3.12 × 10-4× sICAM + 0.0013 × sTNF-RII + 0.172aβ-catenin + 0.283 × AFP][P values = 6.83E-10, 1.86E-05, 0.938962, 0.764414, 6.79E-08, 0.225761, 0.000818] |
| HCC vs LC | [11.33 + 7.380 × Prot + (-2.047 × 10-4) × IL-8 + 0.0011 × sICAM + 6.299 × 104× sTNF + 0.26 ×β-catenin + 0.016 × AFP][P values = 3.22E-05, 7.11E-07, 0.591834, 0.041838, 0.056595, 0.000357, 0.000638] |
| HCC vs HCV | [-11.9 + 3.222 × Prot + (3.813 × 10-4) × IL + 0.00152 × sICAM + (6.481 × 10-4) × sTNF + 0.0291 ×β-catenin + 0.019 × AFP][P values = 1.39E-11, 2.63E-12, 0.231248, 0.000928, 0.000952, 5.41E-06, 5.09E-06] |
| HCC vs Non-HCC | [-12.27 + 3.3 × Prot + 0.0004 × IL + 0.00155 × sICAM + (6.803 × 10-4) × sTNF + 0.294 ×β-catenin + 0.02 × AFP][P values = 5.26E-13, 4.14E-13, 0.211689, 0.000767, 0.000388, 3.89E-06, 3.42E-06] |
| HCV vs Control | [-10.84 + 6.80 × Prot + (2.68 × 10-5) × IL-8 + 0.00017 × ICAM + 0.00126 × sTNF + 0.1742 ×β-catenin + 0.278 × AFP][P values = 1.12E-09, 1.87E-05, 0.972804, 0.875503, 9.02E-08, 0.227858, 0.000935] |
| LC vs CHC | [-10.16 + (-4.487) × Prot + 0.00134 × IL-8 + 0.0021 × sICAM + 0.0018 × sTNF + (-0.2984) ×β-catenin + 0.0217 × AFP][P values = 0.002674, 0.000351, 0.052275., 0.021958, 0.000211, 0.009838, 0.030621] |
| CHC vs Control | [-9.354 + 6.634 × Proteasome + (-0.00166) × IL-8 + (-0.00126) × sICAM + 0.001111 × sTNF + 0.208624 ×β-catenin + 0.24604 × AFP][P values = 1.40E-07, 1.64E-05, 0.2627, 0.3799, 8.55E-06, 0.1477, 0.0016] |
| LC vs Control | [-515.7 + 108.7 × Proteasome + 0.01295 × IL-8 + 0.0194 × sICAM + 0.05745 × sTNF + 4.485 ×β-catenin + 12.59 × AFP][P values = 0.997, 0.998, 0.999, 0.999, 0.998, 0.998, 0.995] |
| HCC vs Control | [-375.640 + 122.598 × Prot + (-0.0577) × IL + 0.0368 × sICAM + 0.0427 × sTNF + 1.1533 ×β-catenin + 13.584 × AFP][P values = 0.737, 0.802, 0.823, 0.917, 0.812, 0.923, 0.66] |
| HCC vs CHC | [-10.40 + 1.416 × Prot + 0.002024 × IL-8 + 0.0041 × sICAM + (4.251 × 10-4) × sTNF + 0.2.67 ×β-catenin + 0.0244 × AFP][P values = 1.28E-07, 0.00826, 0.01827, 5.52E-05, 0.04942, 0.00391, 0.00354] |
| Final reduced model (with relevant terms) | Best threshold | |
| Disease vs Control | (-11 + 6.83 × Prot + 0.00129 × sTNF + 0.283 × AFP) | 0.764 |
| HCC vs LC | (-11.3 + 7.38 × Prot + 0.00108 × sICAM + 0.2574 ×β-catenin + 0.01597 × AFP) | 0.655 |
| HCC vs HCV | [-11.91 + 3.222 × Prot + 0.001518 × sICAM + (6.481 × 10-4) × sTNF + 0.291 ×β-catenin + 0.0193 × AFP] | 0.712 |
| HCC vs Non-HCC | [-12.27 + 3.299 × Prot + 0.001548 × sICAM + (6.803 × 10-4) × sTNF + 0.2936 ×β-catenin + 0.0198 × AFP] | 0.378 |
| HCV vs Control | (-10.84 + 6.803 × Prot + 0.00126 × sTNF + 0.2783 × AFP) | 0.764 |
| LC vs CHC | [-10.16 + (-4.487) × Prot + 0.002086 × sICAM + 0.001858 × sTNF + (-0.2984) ×β-catenin + 0.02169 × AFP] | 0.582 |
| CHC vs Control | (-9.353476 + 6.63414 × Prot + 0.001111 × sTNF + 0.24604 × AFP) | 0.341 |
| HCC vs CHC | [-10.40 + 1.416 × Prot + 0.002024 × IL-8 + 0.004096 × sICAM + (4.251 × 10-4) × sTNF + 0.2567 ×β-catenin + 0.02442 × AFP] | 0.745 |
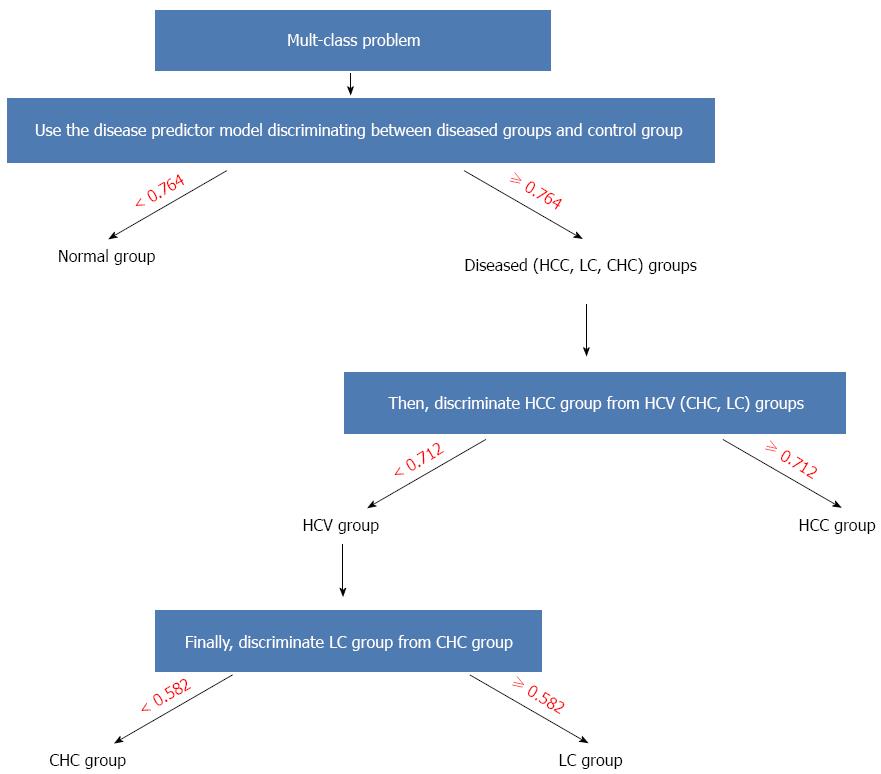
The algorithmic use of different models for making a final prediction among different groups (accordingly solving the multi-class classification problem) could be performed as follows: First, use the mathematical model involving sTNF-RII, the proteasome, and AFP with a cut-off of 0.764 to discriminate disease cases from healthy controls. Second, for disease cases, discriminate HCC cases from HCV cases by using the corresponding mathematical model in the table involving the levels of sTNF-RII, the proteasome, sICAM-1, β-catenin, and AFP with a cut-off of 0.712. Third, for HCV cases, proceed to discriminate LC cases from CHC cases by using the respective mathematical model with a cut-off of 0.582. The decision tree representing this algorithm is shown in (Figure 4).
Hepatocellular carcinoma (HCC) is a common disorder worldwide that ranks as the fifth and eighth most common cancer among men and women, respectively[24]. In Egypt, HCC is the first and second most common cancer among men and women, respectively[24]. The rising incidence of HCC in Egypt may be explained by the high prevalence of HCV[25], estimated nationally to be approximately 14%[4].
Early diagnosis is crucial for improving the survival rate of patients. The serum level of AFP is often not significantly elevated in patients with early-stage, potentially curable, HCC[5].
Therefore, finding a new appropriate panel of serum markers rather than a single marker is necessary for the early detection of HCC in normal populations (healthy controls) and in high risk patients (CHC and LC patients).
Our study was conducted as a complementary study to our previously published papers by Zekri et al[23], 2015 to investigate whether combinations of serum sICAM-1, β-catenin, IL-8, proteasome, and sTNF-RII levels or subsets of these biomarkers with AFP, using logistic disease predictor models, could facilitate the early detection of HCC.
Our study revealed that the serum level of AFP was significantly elevated in the HCC group than the other groups, a finding in concordance with those from previous studies by many authors[26,27].
The serum level of sICAM-1 was significantly elevated in the HCC group compared with other groups (P < 0.001). However, there was no significant difference between the CHC group and the control group. These data are consistent with those from previous studies by Shimizu et al[28], 1995 and Moriyama et al[29], 2006, which have reported that increasing levels of sICAM-1 over time represent a significant HCC risk factor in patients with HCV-associated CH or LC. ROC curve analysis of the individual markers revealed that serum levels of sICAM-1 had excellent diagnostic accuracy for the discrimination of the HCC group from the control group, because the AUC was in the range of (0.9-1), and had good diagnostic accuracy in discriminating the HCC group from the CHC group as AUC was in the range of (0.8-0.89), whereas it failed to have diagnostic accuracy in discriminating the HCC group from the LC group, because the AUC was in the range of (0.5-0.59). Thus, regular measurements of sICAM-1 concentrations may be used as a diagnostic marker for the early detection of HCC in healthy controls and CHC patients.
Serum levels of β-catenin were significantly higher in the HCC group than in the other groups (P < 0.001), whereas there was no significant difference in the levels in the LC group and the CHC group. These results are is consistent with those from our previous study, Zekri et al[22], 2011, which has found that serum levels of β-catenin are higher in patients with HCC compared with CH, ASC (chronic HCV with persistent normal alanine aminotransferase levels) and healthy control groups. In agreement with our results, previous studies by Li et al[30] and Guan et al[27] have reported higher nuclear accumulation of β-catenin in HCC tissue than in the corresponding para-carcinoma, cirrhotic, and normal tissues. ROC curve analysis of the individual markers revealed that serum levels of β-catenin had good diagnostic accuracy for the discrimination of the HCC group from the control group, because the AUC was in the range of (0.8-0.89), and had fair diagnostic accuracy in discriminating between the HCC group and the LC group, and between the HCC group and the CHC group, because the AUC was in the range of (0.7-0.79). These data suggested that serum levels of β-catenin may be used as a diagnostic marker for the early detection of HCC in the normal population, CHC, and LC patients.
Serum levels of proteasome were significantly higher in the HCC group than in the other groups (P < 0.001). This result is consistent with results from a previous study by Henry et al[13], which has found that plasma proteasome levels are significantly higher in patients with HCC compared with the LC group and the control group. However, serum levels of proteasome were also significantly elevated in the CHC group than in the LC group and the control group (P < 0.001). ROC curve analysis of the individual markers revealed that serum proteasome levels showed excellent diagnostic accuracy in discriminating between the HCC group and the LC group, and also between the HCC group and the control group, because the AUC was in the range of (0.9-1). However, it showed poor diagnostic accuracy between in discriminating the HCC group from the CHC group, with an AUC in the range of (0.6-0.69). These data suggested that serum levels of proteasome are a reliable diagnostic marker for the early detection of HCC in normal populations and LC patients.
Serum levels of sTNF-RII were significantly higher in patients with HCC than in the CHC group and the control group (P < 0.001). This result is concordant with results from our previous study, Zekri et al[21], which has reported significantly elevated serum levels of sTNF-RII in patients with HCC than in patients with CLD (chronic liver disease associated with elevated liver enzyme levels), ASC (a symptomatic liver disease associated with normal liver enzyme levels), and the control group. However, the serum level of sTNF-RII was significantly higher in patients with LC than in HCC patients in our current study (P < 0.001). The ROC curve analysis of the individual markers revealed that the serum level of sTNF-RII had excellent diagnostic accuracy in discriminating the HCC group from the control group, because the AUC was in the range of (0.9-1), and fair diagnostic accuracy in discriminating the HCC group from the CHC group, because the AUC was in the range of (0.7-0.79), whereas its diagnostic accuracy in discriminating between the HCC group and LC group was insignificant, because the AUC was below 0.5. This result suggests that sTNF-RII may be used as a diagnostic marker for the early detection of HCC in the normal population and CHC patients.
The serum level of IL-8 was significantly elevated in patients with HCC and LC compared with the CHC group and healthy controls (P < 0.001). This result is concordant with results from a previous study by Elewa et al[31], which has reported that the serum level of IL-8 is significantly higher in patients with HCV-associated HCC than in patients with chronic hepatitis C without HCC and healthy controls, as well as a previous study by Ren et al[10], which has reported that the IL-8 serum level is significantly higher in patients with HCC than in the control group. Moreover, this result is in agreement with those from a previous study by Zimmermann et al[32], which has reported that the level of serum IL-8 is significantly higher in patients with LC than in those with CH and healthy controls. However, in the present study, there was no significant difference between the HCC group and the LC group, or between the CHC group and control group. ROC curve analysis of the individual markers revealed that the serum level of IL-8 had fair diagnostic accuracy in discriminating the HCC group from the CHC group, because the AUC was in the range of (0.7-0.79), and had poor diagnostic accuracy in discriminating the HCC group from the control group, because the AUC was in the range of (0.6-0.69), whereas it failed to have diagnostic accuracy in discriminating between the HCC group and the LC group, because the AUC was in the range of (0.5-0.59). This result suggested that the serum level of IL-8 may be used as diagnostic marker for the early detection of HCC in CHC patients.
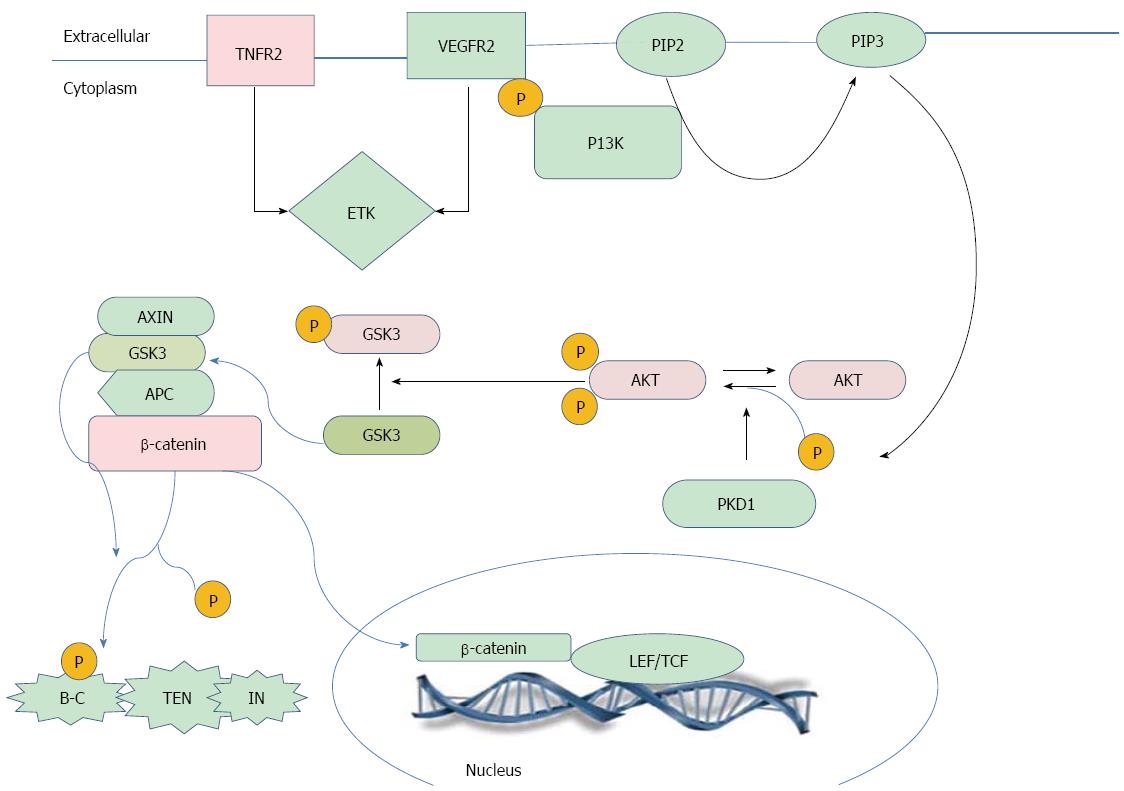
Correlation studies (Figure 5) showing the interplay between the studied markers revealed a moderately positive correlation between sTNF-RII and sICAM-1. A possible explanation of this result is that ICAM-1 expression is regulated through four primary pathways: NF-κB, JAK/STAT and IFN-g, AP-1 and MAP Kinase, and PKC pathways. Ultimately, ICAM-1 is regulated at the level of transcription by one of these signaling cascades[33]. It has previously been reported that binding of TNF-α causes trimerization of TNFR-II, enabling its direct interaction with TRAF-II, resulting in the activation of transcription factors NF-κB or AP-1[34], and in turn leading to subsequent expression of ICAM-1. Additionally, there was a moderately positive correlation between sTNF-RII and β-catenin. A possible explanation for this result is that TNF-RII can also activate endothelial/epithelial tyrosine kinase (Etk) independently of TRAF2. Active Etk mediates crosstalk with vascular endothelial growth factor receptor 2 (VEGFR2) through reciprocal phosphorylation, resulting in the activation of the phosphatidylinositol 3-kinase (PI3K)-Akt angiogenic pathway[34]. PI3K (phosphatidylinositol 3-Kinase)-mediated activation of Akt/PKB (protein kinase-B) negatively regulates GSK3[35]. When GSK3 is active, it phosphorylates APC and β-catenin and stimulates an interaction between β-catenin and Beta-TRCP (Beta-transducin repeat-containing protein), a regulator of E3 ubiquitin ligase, which degrades β-catenin in proteasomes[36]. Thus, the inactivation of GSK3 inhibits β-catenin phosphorylation and subsequent degradation, leading to the nuclear accumulation of β-catenin. Moreover, our previous study, Zekri et al[20] has used correlation analysis of the expressed genes in HCC cases associated with HCV infection to reveal a significant negative correlation between GSK3B and AKT (P = 0.04), characterized by GSK3B downregulation with overexpression of AKT at the m-RNA level.
Further analysis of the data, using the R package and different modules for binary and multi-class classifiers based on generalized linear models, was carried out to model the top performing markers with AFP, in order to improve the diagnostic performance and the predictive power for the early detection of HCC. ROC curve analysis of the different modules revealed that using a binary classifier over different pairwise comparisons was superior to using the multi-class classification. Additionally, using a binary classifier based on a linear model offered an improvement in the diagnostic performance over using the top performing individual marker in each pairwise comparison. The performance of the binary classifier based linear model was evaluated by measuring the predictive power (PPV). The performance of the model was high, because the PPV was in the range of (87%-100%) over different pairwise comparisons.
From these models (Table 7), only three markers (sTNF-RII, proteasome, and AFP) were able to discriminate the control group from all other groups. To discriminate among different other groups, sICAM-1, β-catenin, and IL-8 were required. The use of different models for resolving the multi-class classification problem may be performed as follows: First, use the mathematical model based on the binary classifier (-11 + 6.83 × proteasome + 0.00129 × sTNF + 0.283 × AFP) with the cutoff of 0.764 to discriminate controls from disease cases. For disease cases, first discriminate HCC cases from HCV cases (cases with LC and CHC) using the mathematical model based on the binary classifier [-11.91 + 3.222 × proteasome + (3.813 × 10-4) × IL-8 + 0.001518 × sICAM + (6.481 × 10-4) × sTNF + 0.2906 ×β-catenin + 0.01931 × AFP] with the cutoff of 0.712. If the cases have HCV, proceed to discriminate LC cases from CHC cases using the mathematical model based on the binary classifier [-10.16+ (-4.487) × proteasome + 0.002086 × sICAM + 0.001858 × sTNF + (-0.2984) ×β-catenin + 0.02169 × AFP] with a cutoff of 0.582.
In general, for the early detection of hepatocellular carcinoma in high risk patients; using the mathematical model based on the binary classifier (-11.33 + 7.38 × proteasome + 0.001081 × sICAM + 0.2574 ×β-catenin + 0.01597 × AFP) with a cutoff of 0.655 improved the diagnostic accuracy of the discrimination of the HCC group from the LC group, with an AUC value of 0.961, 98.8% specificity, and 89.1% sensitivity. Additionally, using the mathematical model based on the binary classifier (-11.33 + 7.38 × Proteasome + 0.00108 × sICAM + 0.2574 ×β-catenin + 0.01597 × AFP) with a cutoff of 0.655 improved the diagnostic performance in discriminating the HCC group from the CHC group, with an AUC value of 0.971, 96.8% specificity, and 89.7% sensitivity.
In conclusion, this disease predictor model may be a valuable tool for the early detection of the statuses of different liver diseases co-occurring HCV infection in an accurate, non-invasive, inexpensive, and rapid manner. Our recommendation is to perform further studies to evaluate the mathematical model on a larger scale (validation dataset) and to generalize the early detection of HCC for other etiologic agents rather than HCV.
We acknowledge Mai Lotfy for helping format the manuscript and the virology and immunology staff members for their help in sample collection, as well as the NCI for financing the study.
Hepatocellular carcinoma (HCC) is considered to be a heterogeneous tumor due to the presence of inflammation and most probably cirrhosis, making the early detection of HCC as well as its treatment complicated. Early detection of HCC is usually associated with a proper clinical response; however, the existing markers differentiate only a very limited number of cases and lack sensitivity and specificity. Therefore, this study used a panel of serum proteins in association with AFP to build a mathematical model for the early detection of HCC and examined the cascade of complicated liver disease.
This is the first time that a proper mathematical model has been developed from a serum protein panel for early disease detection of different states of liver disease co-occurring with hepatitis C virus (HCV) infection.
Although these studied markers have been previously addressed in relation to liver disease, a mathematical model had yet to be built from this panel in combination with standard α-fetoprotein (AFP) for the differentiation between different states of liver disease co-occurring with HCV infection.
The proposed mathematical model using a panel of serum proteins in combination with AFP may be a useful method for the early detection of different liver disease states co-occurring with HCV infection.
This study was designed to explore a mathematical model for the early detection of hepatocellular carcinoma co-occurring with HCV infection, the authors provide a potential novel biomarker combination useful for the early diagnosis for patients with HCV. This is an interesting study.
| 1. | Hiotis SP, Rahbari NN, Villanueva GA, Klegar E, Luan W, Wang Q, Yee HT. Hepatitis B vs. hepatitis C infection on viral hepatitis-associated hepatocellular carcinoma. BMC Gastroenterol. 2012;12:64. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 2. | Iakova P, Timchenko L, Timchenko NA. Intracellular signaling and hepatocellular carcinoma. Semin Cancer Biol. 2011;21:28-34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 61] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 3. | Yamashita T, Honda M, Kaneko S. Molecular mechanisms of hepatocarcinogenesis in chronic hepatitis C virus infection. J Gastroenterol Hepatol. 2011;26:960-964. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 4. | El-Zanaty F, Way AA. Knowledge and prevalence of hepatitis C. In: Egypt Demographic and Health Survey 2008. Egyptian Ministry of Health, El-Zanaty and Associates and Macro International 2009; (accessed April 10, 2015) Available from: http://www.dhsprogram.com/2008edhs/Knowledge and prevalence of hepatitis C. |
| 5. | Zhao YJ, Ju Q, Li GC. Tumor markers for hepatocellular carcinoma. Mol Clin Oncol. 2013;1:593-598. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 157] [Cited by in RCA: 176] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 6. | Bruix J, Llovet JM. Prognostic prediction and treatment strategy in hepatocellular carcinoma. Hepatology. 2002;35:519-524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 837] [Cited by in RCA: 857] [Article Influence: 35.7] [Reference Citation Analysis (0)] |
| 7. | Zhu K, Dai Z, Zhou J. Biomarkers for hepatocellular carcinoma: progression in early diagnosis, prognosis, and personalized therapy. Biomark Res. 2013;1:10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 59] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 8. | Claudino WM, Quattrone A, Biganzoli L, Pestrin M, Bertini I, Di Leo A. Metabolomics: available results, current research projects in breast cancer, and future applications. J Clin Oncol. 2007;25:2840-2846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 166] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 9. | McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies (REMARK). J Natl Cancer Inst. 2005;97:1180-1184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1072] [Cited by in RCA: 1169] [Article Influence: 55.7] [Reference Citation Analysis (1)] |
| 10. | Ren Y, Poon RT, Tsui HT, Chen WH, Li Z, Lau C, Yu WC, Fan ST. Interleukin-8 serum levels in patients with hepatocellular carcinoma: correlations with clinicopathological features and prognosis. Clin Cancer Res. 2003;9:5996-6001. [PubMed] |
| 11. | Bradley JR. TNF-mediated inflammatory disease. J Pathol. 2008;214:149-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1178] [Cited by in RCA: 1455] [Article Influence: 80.8] [Reference Citation Analysis (1)] |
| 12. | McConnell BB, Yang VW. The Role of Inflammation in the Pathogenesis of Colorectal Cancer. Curr Colorectal Cancer Rep. 2009;5:69-74. [PubMed] |
| 13. | Henry L, Lavabre-Bertrand T, Vercambre L, Ramos J, Carillo S, Guiraud I, Pouderoux P, Bismuth M, Valats JC, Demattei C, Duny Y, Chaze I, Funakoshi N, Bureau JP, Daurès JP, Blanc P. Plasma proteasome level is a reliable early marker of malignant transformation of liver cirrhosis. Gut. 2009;58:833-838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 35] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 14. | Sorokin AV, Kim ER, Ovchinnikov LP. Proteasome system of protein degradation and processing. Biochemistry (Mosc). 2009;74:1411-1442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 94] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 15. | Lawson C, Wolf S. ICAM-1 signaling in endothelial cells. Pharmacol Rep. 2009;61:22-32. [PubMed] |
| 16. | Bernier AJ, Zhang J, Lillehoj E, Shaw AR, Gunasekara N, Hugh JC. Non-cysteine linked MUC1 cytoplasmic dimers are required for Src recruitment and ICAM-1 binding induced cell invasion. Mol Cancer. 2011;10:93. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 32] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 17. | Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469-480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4088] [Cited by in RCA: 4528] [Article Influence: 226.4] [Reference Citation Analysis (1)] |
| 18. | Laurent-Puig P, Zucman-Rossi J. Genetics of hepatocellular tumors. Oncogene. 2006;25:3778-3786. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 254] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 19. | Zekri AR, Ashour MS, Hassan A, Alam El-Din HM, El-Shehaby AM, Abu-Shady MA. Cytokine profile in Egyptian hepatitis C virus genotype-4 in relation to liver disease progression. World J Gastroenterol. 2005;11:6624-6630. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 39] [Cited by in RCA: 46] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 20. | Zekri AR, Bahnassy AA, Abdel-Wahab SA, Khafagy MM, Loutfy SA, Radwan H, Shaarawy SM. Expression of pro- and anti-inflammatory cytokines in relation to apoptotic genes in Egyptian liver disease patients associated with HCV-genotype-4. J Gastroenterol Hepatol. 2009;24:416-428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 21. | Zekri AR, Alam El-Din HM, Bahnassy AA, Zayed NA, Mohamed WS, El-Masry SH, Gouda SK, Esmat G. Serum levels of soluble Fas, soluble tumor necrosis factor-receptor II, interleukin-2 receptor and interleukin-8 as early predictors of hepatocellular carcinoma in Egyptian patients with hepatitis C virus genotype-4. Comp Hepatol. 2010;9:1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 23] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 22. | Zekri AR, Bahnassy AA, Alam El-Din HM, Morsy HM, Shaarawy S, Moharram NZ, Daoud SS. Serum levels of β-catenin as a potential marker for genotype 4/hepatitis C-associated hepatocellular carcinoma. Oncol Rep. 2011;26:825-831. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 23. | Zekri AR, Youssef AS, Bakr YM, Gabr RM, El-Rouby MN, Hammad I, Ahmed EA, Marzouk HA, Nabil MM, Hamed HA. Serum biomarkers for early detection of hepatocellular carcinoma associated with HCV infection in egyptian patients. Asian Pac J Cancer Prev. 2015;16:1281-1287. [PubMed] |
| 24. | GLOBOCAN database. IARC, France. 2012 (accessed April 21, 2015). Available from: http://globocan.iarc.fr. |
| 25. | Shaker MK, Abdella HM, Khalifa MO, El Dorry AK. Epidemiological characteristics of hepatocellular carcinoma in Egypt: a retrospective analysis of 1313 cases. Liver Int. 2013;33:1601-1606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 52] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 26. | Mittal A, Sathian B, Chandrashekharan N, Farooqui SM, Hussain A. Diagnostic significance of alpha fetoprotein in carcinomas of liver and biliary tract - a comparative study from the western region of Nepal. Asian Pac J Cancer Prev. 2011;12:3475-3478. [PubMed] |
| 27. | Guan CN, Chen XM, Lou HQ, Liao XH, Chen BY, Zhang PW. Clinical significance of axin and β-catenin protein expression in primary hepatocellular carcinomas. Asian Pac J Cancer Prev. 2012;13:677-681. [PubMed] |
| 28. | Shimizu Y, Minemura M, Tsukishiro T, Kashii Y, Miyamoto M, Nishimori H, Higuchi K, Watanabe A. Serum concentration of intercellular adhesion molecule-1 in patients with hepatocellular carcinoma is a marker of the disease progression and prognosis. Hepatology. 1995;22:525-531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 29. | Moriyama M, Matsumura H, Shioda J, Aoki H, Nakamura H, Arakawa Y, Nirei K, Yamagami H, Kaneko M, Tanaka N. Measurement of human intercellular adhesion molecule 1 in the blood is useful for predicting the occurrence of hepatocellular carcinomas from chronic hepatitis C and liver cirrhosis. Intervirology. 2006;49:327-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 30. | Li P, Cao Y, Li Y, Zhou L, Liu X, Geng M. Expression of Wnt-5a and β-catenin in primary hepatocellular carcinoma. Int J Clin Exp Pathol. 2014;7:3190-3195. [PubMed] |
| 31. | Elewa H, Abd-Elmeneem M, Hashem AM, Alshehaby A. Study of interleukin 8 (IL8) serum level in patients with chronic liver disease due to hepatitis C virus (HCV) with and without hepatocellular carcinoma (HCC). Int J Hepatol. 2010;1:9-17. |
| 32. | Zimmermann HW, Seidler S, Gassler N, Nattermann J, Luedde T, Trautwein C, Tacke F. Interleukin-8 is activated in patients with chronic liver diseases and associated with hepatic macrophage accumulation in human liver fibrosis. PLoS One. 2011;6:e21381. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 219] [Cited by in RCA: 223] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 33. | Roebuck KA, Finnegan A. Regulation of intercellular adhesion molecule-1 (CD54) gene expression. J Leukoc Biol. 1999;66:876-888. [PubMed] |
| 34. | Moelants EA, Mortier A, Van Damme J, Proost P. Regulation of TNF-α with a focus on rheumatoid arthritis. Immunol Cell Biol. 2013;91:393-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 158] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 35. | Clodfelder-Miller B, De Sarno P, Zmijewska AA, Song L, Jope RS. Physiological and pathological changes in glucose regulate brain Akt and glycogen synthase kinase-3. J Biol Chem. 2005;280:39723-39731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 100] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 36. | Jia J, Zhang L, Zhang Q, Tong C, Wang B, Hou F, Amanai K, Jiang J. Phosphorylation by double-time/CKIepsilon and CKIalpha targets cubitus interruptus for Slimb/beta-TRCP-mediated proteolytic processing. Dev Cell. 2005;9:819-830. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 113] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
P- Reviewer: Gao C, Sunami Y S- Editor: Yu J L- Editor: A E- Editor: Zhang DN