Published online Nov 7, 2015. doi: 10.3748/wjg.v21.i41.11680
Peer-review started: May 4, 2015
First decision: July 14, 2015
Revised: August 12, 2015
Accepted: September 15, 2015
Article in press: September 15, 2015
Published online: November 7, 2015
Processing time: 189 Days and 17.6 Hours
Recent studies have showed that RNAs regulate each other with microRNA (miRNA) response elements (MREs) and this mechanism is known as “competing endogenous RNA (ceRNA)” hypothesis. Long non-coding RNAs (lncRNAs) are supposed to play important roles in cancer. Compelling evidence suggests that lncRNAs can interact with miRNAs and regulate the expression of miRNAs as ceRNAs. Several lncRNAs such as H19, HOTAIR and MEG3 have been found to be associated with miRNAs in gastric cancer (GC), generating regulatory crosstalk across the transcriptome. These MRE sharing elements implicated in the ceRNA networks (ceRNETs) are able to regulate mRNA expression. The ceRNA regulatory networks including mRNAs, miRNAs, lncRNAs and circular RNAs may play critical roles in tumorigenesis, and the perturbations of ceRNETs may contribute to the pathogenesis of GC.
Core tip: Competitive endogenous RNAs (ceRNAs) share microRNA (miRNA) response elements and compete common miRNAs, thereby regulating each other’s expression. The ceRNA regulatory networks including mRNAs, miRNAs, long non-coding RNAs and circular RNAs play critical roles in tumorigenesis, and the perturbations of ceRNA networks may contribute to the pathogenesis of gastric cancer.
- Citation: Guo LL, Song CH, Wang P, Dai LP, Zhang JY, Wang KJ. Competing endogenous RNA networks and gastric cancer. World J Gastroenterol 2015; 21(41): 11680-11687
- URL: https://www.wjgnet.com/1007-9327/full/v21/i41/11680.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i41.11680
Gastric cancer (GC) is the second leading cause of cancer-related death worldwide and is a major cause of cancer-related mortality in China[1]. Since the carcinogenesis in GC is a complex process with etiological factors, genetic and epigenetic alterations involved[2], the molecular basis of GC, especially efforts to identify clusters of predictive markers, has been widely studied. Previous studies have demonstrated that several genetic abnormalities such as aberrant genes, copy number variants (CNV), microRNAs (miRNAs) and long noncoding RNAs (lncRNAs) were involved in the initiation and progression of GC[3], but the pathogenic mechanisms contributing to biological feature of GC remain to be elucidated.
Non-coding RNAs (ncRNAs) refer to a class of RNAs with no protein-coding function that are widely expressed in organisms, including small ncRNAs such as miRNAs and lncRNAs, both of which play important roles in the post-transcriptional regulation[4]. In fact, miRNAs have been extensively studied in the field of oncology, and emerging evidence suggests that miRNA-mediated regulation plays crucial roles in tumor cell biological processes, such as cell proliferation, migration and invasion[5]. Furthermore, aberrantly expressed miRNAs have been discovered in diverse diseases including GC.
The competing endogenous RNA (ceRNA) hypothesis postulates that RNAs that share miRNA response elements (MREs) in their 3’-untranslated regions (UTRs) can influence the expression of miRNAs, inducing gene silencing[6]. While recent several studies have demonstrated that lncRNAs can harbor MREs and interact with other RNA transcripts such as ceRNAs[7,8]. The complex crosstalks of ceRNAs have been found in many different cancer types including GC. Above all, functional interactions and disequilibrium of ceRNA networks (ceRNETs) may contribute to disease pathogenesis[9]. This review discusses the features of ceRNETs and the functional roles and regulatory interactions of ceRNETs in the development of GC.
In view of ceRNA hypothesis, three studies were reported in 2011 from Columbia University, Harvard Medical School and the University of Roma La Sapienza, which verified the ceRNA hypothesis from many aspects and further confirmed the establishment of regulatory mechanism based on ceRNAs[10]. The discovery of ceRNA mechanism provocatively subverts traditional meaning of the mRNA function, which means that mRNAs not only have the function of encoding proteins, but also participate in the gene regulation processes[11,12]. Transcriptional regulatory networks based on ceRNAs, not only enrich the biological pathway in the existing networks, but also expand the function of the human genome. Regulation members of ceRNETs consist of mRNAs, miRNAs, lncRNAs and circular RNAs (circRNAs). Notably, miRNAs and MREs are considered two important elements in the ceRNA hypothesis. The former is core motivation, while the latter is structural foundation.
So far, the number of protein coding genes in human genome has been found to be approximately 20000[13]. And most of mRNAs are covered in MREs[14,15]. Recent studies have demonstrated that many mRNAs are validated as ceRNAs, so mRNAs play an essential role in ceRNETs.
The function of miRNAs can be influenced by their target mRNAs for limited MREs within each cell. For a given mRNA, its upregulation can lead to the increasing number of MREs, which exceeds their targeting miRNAs. So each mRNA can act as an inhibitor for shared miRNAs. To date, PTEN that competes with various ceRNAs has been widely validated in a variety of advanced and metastatic cancers[16]. This tumor suppressor gene is involved in the regulation of cell proliferation, migration and apoptosis. The occurrence of PTEN inactivation was closely related to GC stage[17]. Recently, a study has successfully validated that a protein-coding transcript ZEB2 plays a role as a PTEN ceRNA in melanoma, which suggests that ZEB2 is involved in regulation of PTEN expression in an miRNA-dependent manner[18]. In another study, Tay et al[19] have validated that endogenous protein-coding transcripts VAPA and CNOT6L possessing tumor-suppressive properties could regulate PTEN through the disturbance of the PI3K/AKT signaling pathway.
Studies utilizing high-throughput technologies such as microarray and NGS for gene expression profiling have increased the discovery of predictive and treatment biomarkers. So far, numerous driver genes have been involved in gastric tumorigenesis. P53 mutations, which were observed in a large proportion of tumors, had a crucial and early role in gastric carcinogenesis of intestinal type[20,21]. E-cadherin gene (CDH1) inactivating mutations were identified in diffuse GC and carriers with CDH1 mutation(s) were more likely to have an increased risk of developing GC[22,23]. Furthermore, previous studies found that frequent ARID1A alterations were detected by exome sequencing in two specific molecular subtypes of GC[24,25]. In addition to the previously known mutations, a recent study of whole-genome sequencing (WGS)[26] has identified new driver genes of gastric adenocarcinoma. MUC6, which encodes gastric mucin, was significantly mutated. And RHOA mutations were observed in diffuse-type GC. These emerging drivers, together with other genes including CTNNA2, GLI3, and RNF43, are potential players in the perturbed pathways of GC. Although dozens of genes have been found, their roles in tumorigenesis in GC remain to be elucidated. Concurrently, these driver genes could be ceRNAs, which act as mediators, involved in the regulation of ceRNETs.
MiRNAs are small noncoding RNAs that regulate the expression of various genes by inhibiting or degrading target mRNAs[27]. It is estimated that 30% of genes in the human genome were regulated by miRNAs[28,29]. MiRNAs containing MREs are shared by all ceRNAs. Accumulating evidence supports that a new layer of regulation of ceRNETs produces a tendency to be mediated by miRNAs. Multiple miRNAs can regulate different MREs in mRNA transcripts, and each miRNA can inhibit hundreds of transcripts, so miRNAs act as mediators in huge transcriptional and signaling networks[30]. This regulatory mechanism constitutes the basis of ceRNA interplay networks.
Emerging evidence suggests that aberrant miRNAs participate in the pathogenesis of GC, mainly by regulating the expression of oncogenes and tumor suppressors. Overexpression of miR-21, a known oncogenic miRNA, could enhance cell proliferation and inhibit apoptosis in cancer patients[31,32]. The target genes of miR-21 such as TMP1, PTEN and RECK were confirmed in several studies by different technological methods[33,34]. These findings support that miR-21 that function via the regulation of target genes mediates oncogenic processes in GC. Dysregulated miRNAs (miR-125a, miR- 199a, and miR-100) were considered to be important factors in the regulation of GC[35-37], suggesting that they may play different functions in different sites.
Gastric carcinogenesis is a multi-stage process, in which molecular expression and signaling pathway disturbances are involved[38]. Chronic inflammation is a driving factor promoting the malignant transformation. Specifically, Helicobacter pylori (HP)-induced gastritis is a risk factor for GC. The expression of certain miRNAs including miR-21, miR-155, miR-194, miR196, miR-218, and miR-223 has been found to be increased in GC with HP infection. Saito et al[39] noted that the overexpressed miR-155 acting as an important negative regulator modulates the inflammatory responses in GC induced by HP infection. Additionally, Wang et al[40] reported that a great dependence was confirmed between miR-106a and lymph node metastasis in GC. Another study also[41] discovered that HP infection could lead to decreased expression of Let-7, which increases the expression of oncogene Ras. As stated above, aberrant miRNAs play central roles in ceRNETs by regulating target genes.
LncRNAs play regulatory roles and are dysregulated in a variety of tumors. However, the potential mechanisms of how lncRNAs alter in GC remain largely undefined. An increasing number of lncRNA transcripts emerged recently as ceRNAs have been implicated in GC.
In the research of GC, some lncRNAs are upregulated and act as oncogenic genes, including H19 and HOTAIR, while others are downregulated and function as suppressor genes, such as growth arrest-specific transcript 5 (GAS5) and maternally expressed gene 3 (MEG3). H19, a typical onco-lncRNA, was dysregulated in many cancers[42-44]. Park et al[45] reported that upregulated H19 can promote the development of GC by regulating the activity of P53. Recently, several studies[46] have demonstrated that HOTAIR may participate in the progression and metastasis of GC, and can be used as a therapeutic target for GC. GAS5, another famous lncRNA, plays a tumor-suppressive role in tumor formation. Significant downregulation of GAS5 could promote tumor cell proliferation by regulating expression of p21 and E2F1 proteins[47]. In addition, MEG3 was frequently studied in GC. Decreased expression of MEG3 could regulate cell proliferation and differentiation by interacting with p53, Rb, and VEGF[48]. Additionally, MEG3 may be associated with poor prognosis of GC by increasing the spread of cancer cells[49].
The key step in cancer research is to discover lncRNAs associated with specific diseases. At present, the screening of lncRNAs via chip analysis is a quick and accurate method. Song et al[50] demonstrated that 135 lncRNAs were dysregulated in GC tissues by microarray analysis, and H19 and uc001lsz were markedly expressed. The use of qRT-PCR also confirmed that the overexpression of H19 was closely related to GC, and uc001lsz might be a potential diagnostic marker for early GC. By means of expression profile analysis, Cao et al[51] identified 88 abnormally expressed lncRNAs including LINC00152, SNHG3, GAS5 and LINC00261. Additionally, Park et al[52] detected 31 differentially expressed lncRNAs using transcriptomics data, which further suggested that down-regulated BM742401 was closely related to poor survival of GC, and could be used as a therapeutic target to improve the prognosis of GC.
CircRNAs are a special kind of endogenous RNAs featuring stable structure and high tissue-specific expression[53]. Instead of nonlinear RNAs, circRNAs are more common features[54]. So far, thousands of circRNAs have been found in human cells. The newly discovered circular RNAs can act as ceRNAs that affect the regulation of gene expression.
At present, studies on circRNAs are relatively few. CircRNAs functioning as miRNA sponges may play an important role at the level of miRNA fine tuning[55]. Hansen et al[56] suggested that CDR1 (cerebellar degeneration-related protein 1), known as ciRS-72011, was perceived as a ceRNA. Unlike other transcripts, CDR1 containing more than 70 MREs plays a role in regulation by interacting with miRNAs. By functional approaches, CDR1 was found to be overexpressed as a ceRNA that bound miRNAs, thus inhibiting the activity of miR-7[57]. Additionally, the study also discovered that 16 MREs were shared between miR-138 and a circRNA transcript derived from the testis determining gene (sex-determining region Y, Sry), which could have an miRNA sponge effect in regulating gene expression by inhibiting the activity of miR-138. In general, circRNAs are difficult to be degraded by enzyme for the feature of stable configuration and high abundance, which brings the regulatory function of cirRNA into full play.
Currently, circRNAs have been involved in several types of diseases[58,59] including GC. A study discovered that a typical circRNA, hsa_ circ_002059, is significantly downregulated and may be a potential diagnostic marker in GC[60]. Given the fact that the interactions between circRNA and miRNA may be very common, with the recognition of more molecules, circRNAs research is likely to have a giant leap, which will make contributions to tumor biology.
The availability of transcriptome data of diverse cancers, together with bioinformatic tools and computational approaches, enables the prediction of ceRNETs. At present, research on ceRNETs is certainly in its infancy, but still has made some progress.
By a novel multivariate analysis method, a huge miR-mediated ceRNET including 248000 crosstalks was first observed in glioblastoma[61]. Based on a special algorithm, a recent study has constructed a breast cancer-specific ceRNET using the expression profiles of miRNAs and mRNAs[62]. Similarly, a computational approach[63] was explored to predict miRNA- mediated sponge interactions (MMI-networks) based on both normal and breast cancer expression data, separately. This study also found that ceRNETs may be significantly altered between normal and pathological breast tissues and lncRNA PVT1 was a key factor in the tumorigenesis of breast cancer. Interestingly, based on lncRNA microarray data of GC, Xia et al[64] first constructed a ceRNA regulatory network including 8 lncRNAs and 9 miRNAs using bioinformatics methods and confirmed this network using the data from six types of other cancers. Additionally, Basia et al[65] proposed to analyze the equilibrium and non-equilibrium properties of ceRNETs based on a stochastic model, while emphasizing the robustness and response-time to external perturbations of the network.
At present, the most effective way to reveal ceRNAs’ function is constructing ceRNETs. As increasing attention has focused on ceRNA research, ceRNA databases are constantly established. Sarver et al[66] developed a putative human ceRNA database ceRDB, which aimed to predict specific miRNA target genes related to ceRNAs. In ceRDB, the competing mRNAs were sorted by an interaction score based on the number of shared MREs among ceRNAs. The higher the score was, the more likely to be affected by ceRNAs the target mRNAs were. However, unlike the ceRDB database, which excluded information about lncRNAs. lnCeDB[67] provided a database of human lncRNAs that could potentially act as ceRNAs by computing a ceRNA score, which was a novel algorithm. Noteworthily, lncRNA-mRNA pairs with common shared miRNAs were available in this database. Additionally, based on ceRNA hypothesis, a web resource Linc2GO database[68] was established to provide comprehensive function annotations for human lincRNAs. starBase v2.0[69] stored the information about regulatory networks based on broadest experimental support, and this database provided potential interactions between miRNAs, mRNAs and lncRNAs. A newly developed database miRcode[70] was described to collect putative microRNA target sites based on complete GENCODE gene annotations and was used to predict the targets of miRNAs, including mRNAs and lncRNAs. The latest version of this database contained 10419 lncRNA genes. DIANA-LncBase database[71] attempted to depict putative miRNA-lncRNA interactions, providing annotations of miRNA targets on lncRNAs. Furthermore, ChIPBase[72] database platform aimed to unravel transcriptional regulatory relationships between lncRNAs/lincRNAs and miRNAs through the integration of ChIP-Seq data. In short, the effective use of these databases will help us seek for biomarkers, avoiding the blindness in practice (Table 1).
| Database | Website | Ref. |
| ceRDB | http://www.oncomir.umn.edu/cefinder/ | [66] |
| lnCeDB | http://gyanxet-beta.com/lncedb/ | [67] |
| Linc2GO | http://www.bioinfo.tsinghua.edu.cn/~liuke/Linc2GO/index.html | [68] |
| starBase v2.0 | http://starbase.sysu.edu.cn/ | [69] |
| miRcode | http://www.mircode.org/ | [70] |
| DIANA-LncBase | http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=lncBase/index | [71] |
| ChIPBase | http://deepbase.sysu.edu.cn/chipbase/ | [72] |
It is well-known that ceRNETs play a role in cell culture. Recently, some conditions required for ceRNETs have been found. First, the concentration of the ceRNAs should be strongly emphasized. Expression changes of ceRNAs should be large enough to effectively eliminate or weaken the inhibition of miRNAs to ceRNAs. Second, the effectiveness of ceRNETs always depends on the number of shared miRNAs. It can be speculated that, in a network, the ceRNA having more binding preference to the shared miRNA will have more profound ceRNA effect on the components with less binding preference. In addition, taking tissue specificity into account, ceRNETs would also rest on density and subcellular distribution of RNAs in the cell[73]. The balance between shared miRNAs and targeted ceRNAs is critical for ceRNA activity and disruption of this balance can trigger internal crosstalks in ceRNETs. In general, alterations of one ceRNA may lead to joint consequences in huge ceRNETs and thus promote cancer.
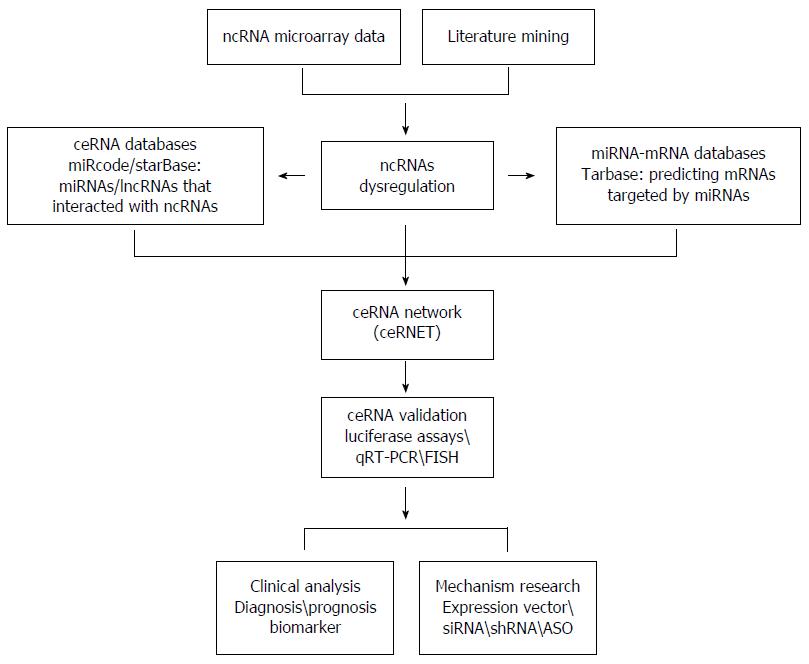
Although ceRNA research is in its infancy, the current progress has gained much attention. The availability of RNA-seq data, along with bioinformatics tools, enables the prediction of ceRNETs. As show in Figure 1, we display a way to research ceRNETs.
First, multiple strategies can be applied to obtain differentially expressed ncRNAs in cancers, including literature mining and microarray analysis. Then by means of computational algorithm and public databases, we can predict potential connections in ceRNETs. Some miRNA target prediction databases such as Tarbase, TargetScan and miRecords can provide experimentally verified miRNA-gene interactions, which are stable foundation for ceRNETs. As a supplement, the CLIP-Seq datasets come in handy. These ceRNA databases encompass information about miRNA, mRNA, lncRNA, circRNA and pseudogene associations. Taken together, ceRNETs including lncRNAs, miRNAs, and mRNAs are constructed by invoking bioinformatics analysis.
Second, the precondition to study ceRNETs should be expression correlations, regulatory relationships, and shared MREs of ceRNA pairs. The validation of ceRNETs is considered to be an experimental framework for the biochemical method of ceRNA interactions. Based on the ceRNETs, the differentially expressed ceRNAs could be confirmed by qRT-PCR or fluorescence in situ hybridization (FISH).
Finally, functional studies should be conducted to investigate the dysregulation of ceRNAs in carcinogenesis. In brief, the effect of overexpression/interference expression among ceRNAs was assessed by function gain/deficit experiments such as siRNAs, shRNAs, and antisense oligonucleotides (ASO). Furthermore, these experimentations for validating the perturbation of ceRNAs should be investigated in mouse models to get confirmed correlations.
In recent years, the mechanism of ncRNAs in tumors has become a hot research topic. At the same time, increasing evidence has indicated that ncRNAs can regulate each other and affect their function by binding to MREs of shared miRNAs[74]. Like the role of ceRNAs in GC, the disturbance of interactions between ceRNAs also plays a part.
Due to the ceRNA theory, the competition between lncRNAs and miRNAs makes indirect regulation possible. In light of the role in regulating target genes, miRNAs can exercise the similar function to negatively regulate the expression of lncRNAs, and thus exert a series of biological effects in GC. Yan et al[75] reported that MEG3 expression level was markedly reduced in both tissues and cell lines of GC, and further experiments found that transfection of MEG3 siRNA into cells could diminish the suppression of proliferation induced by overexpression of miR-148a, which suggested that miR-148a might decrease the expression of MEG3 by modulation of DNMT-1. Furthermore, another study[76] found that upregulated H19 could promote the proliferation of GC cells by binding miR-675, which inversely inhibited the tumor suppressor gene RUNX1. The interaction between H19/miR-675 and RUNX1 may serve as novel targets in the tumorigenesis of GC.
In addition to indirect regulation of ceRNAs, lncRNAs can have a direct interaction by invoking the “endogenous miRNA sponge” (miRNA sponge) to inhibit the activity of mRNAs, thus affecting the occurrence and development of tumors. Xu et al[77] discovered that upregulated lncRNA AC130710 played a crucial role during GC progression by targeting miR-129-5p. Liu et al[78] reported that the expression levels of upregulated HOTAIR and HER2 had a positive correlation in GC. And subsequent luciferase and RIP assays confirmed that HOTAIR served as an endogenous “sponge” to regulate the expression of HER2 by sinking miR-331-3p. These results indicate that possible crosstalks in ceRNETs may provide new clues for the mechanism of GC.
Recently, increasing evidence suggests that the dysregulation of ceRNA interactions including miRNAs and lncRNAs has been involved in disease etiology, including gastric cancer. In this review, we present and discuss the features of ceRNETs and crosstalks in GC, as well as the methods in the study of ceRNETs.
CeRNAs that function as key regulators have been implicated in many biological processes and the perturbation of ceRNETs may contribute to carcinogenesis. Given the complexity of ceRNETs, future works should focus on identifying the hubs that have significant influence on network balance or tumorigenesis. Despite some improvements in research field, the mechanisms of ceRNA crosstalks are still not fully elucidated. And there are still several considerations limiting the applications of ceRNETs. With the development of computational methods, research techniques and abundance of all components in ceRNETs, we anticipate that ceRNETs will provide a new avenue for the research of GC, and shed light on complex mechanisms underlying malignant processes.
| 1. | Catalano V, Labianca R, Beretta GD, Gatta G, de Braud F, Van Cutsem E. Gastric cancer. Crit Rev Oncol Hematol. 2009;71:127-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 282] [Cited by in RCA: 325] [Article Influence: 19.1] [Reference Citation Analysis (1)] |
| 2. | Crew KD, Neugut AI. Epidemiology of gastric cancer. World J Gastroenterol. 2006;12:354-362. [PubMed] |
| 3. | Chang WJ, Du Y, Zhao X, Ma LY, Cao GW. Inflammation-related factors predicting prognosis of gastric cancer. World J Gastroenterol. 2014;20:4586-4596. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 135] [Cited by in RCA: 157] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 4. | Li PF, Chen SC, Xia T, Jiang XM, Shao YF, Xiao BX, Guo JM. Non-coding RNAs and gastric cancer. World J Gastroenterol. 2014;20:5411-5419. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 98] [Cited by in RCA: 121] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 5. | Tan JY, Marques AC. The miRNA-mediated cross-talk between transcripts provides a novel layer of posttranscriptional regulation. Adv Genet. 2014;85:149-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 28] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 6. | Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4127] [Cited by in RCA: 5722] [Article Influence: 381.5] [Reference Citation Analysis (0)] |
| 7. | Arvey A, Larsson E, Sander C, Leslie CS, Marks DS. Target mRNA abundance dilutes microRNA and siRNA activity. Mol Syst Biol. 2010;6:363. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 250] [Cited by in RCA: 269] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 8. | Wang P, Ning S, Zhang Y, Li R, Ye J, Zhao Z, Zhi H, Wang T, Guo Z, Li X. Identification of lncRNA-associated competing triplets reveals global patterns and prognostic markers for cancer. Nucleic Acids Res. 2015;43:3478-3489. [PubMed] |
| 9. | Ergun S, Oztuzcu S. Oncocers: ceRNA-mediated cross-talk by sponging miRNAs in oncogenic pathways. Tumour Biol. 2015;36:3129-3136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 10. | Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013;3:1113-1121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 504] [Cited by in RCA: 693] [Article Influence: 53.3] [Reference Citation Analysis (0)] |
| 11. | de Giorgio A, Krell J, Harding V, Stebbing J, Castellano L. Emerging roles of competing endogenous RNAs in cancer: insights from the regulation of PTEN. Mol Cell Biol. 2013;33:3976-3982. [PubMed] |
| 12. | Su X, Xing J, Wang Z, Chen L, Cui M, Jiang B. microRNAs and ceRNAs: RNA networks in pathogenesis of cancer. Chin J Cancer Res. 2013;25:235-239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 13. | Baltimore D. Our genome unveiled. Nature. 2001;409:814-816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 158] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 14. | Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5833] [Cited by in RCA: 6651] [Article Influence: 369.5] [Reference Citation Analysis (0)] |
| 15. | Lee DY, Jeyapalan Z, Fang L, Yang J, Zhang Y, Yee AY, Li M, Du WW, Shatseva T, Yang BB. Expression of versican 3’-untranslated region modulates endogenous microRNA functions. PLoS One. 2010;5:e13599. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 121] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 16. | Poliseno L, Pandolfi PP. PTEN ceRNA networks in human cancer. Methods. 2015;77-78:41-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 97] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 17. | Zhang X, Park JS, Park KH, Kim KH, Jung M, Chung HC, Rha SY, Kim HS. PTEN deficiency as a predictive biomarker of resistance to HER2-targeted therapy in advanced gastric cancer. Oncology. 2015;88:76-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 32] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 18. | Karreth FA, Tay Y, Perna D, Ala U, Tan SM, Rust AG, DeNicola G, Webster KA, Weiss D, Perez-Mancera PA. In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147:382-395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 548] [Cited by in RCA: 565] [Article Influence: 37.7] [Reference Citation Analysis (0)] |
| 19. | Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344-352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2305] [Cited by in RCA: 3185] [Article Influence: 265.4] [Reference Citation Analysis (0)] |
| 20. | Busuttil RA, Zapparoli GV, Haupt S, Fennell C, Wong SQ, Pang JM, Takeno EA, Mitchell C, Di Costanzo N, Fox S. Role of p53 in the progression of gastric cancer. Oncotarget. 2014;5:12016-12026. [PubMed] |
| 21. | Fricke E, Keller G, Becker I, Rosivatz E, Schott C, Plaschke S, Rudelius M, Hermannstädter C, Busch R, Höfler H. Relationship between E-cadherin gene mutation and p53 gene mutation, p53 accumulation, Bcl-2 expression and Ki-67 staining in diffuse-type gastric carcinoma. Int J Cancer. 2003;104:60-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 34] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 22. | Chu CM, Chen CJ, Chan DC, Wu HS, Liu YC, Shen CY, Chang TM, Yu JC, Harn HJ, Yu CP. CDH1 polymorphisms and haplotypes in sporadic diffuse and intestinal gastric cancer: a case-control study based on direct sequencing analysis. World J Surg Oncol. 2014;12:80. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 23. | Sugimoto S, Yamada H, Takahashi M, Morohoshi Y, Yamaguchi N, Tsunoda Y, Hayashi H, Sugimura H, Komatsu H. Early-onset diffuse gastric cancer associated with a de novo large genomic deletion of CDH1 gene. Gastric Cancer. 2014;17:745-749. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 40] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 24. | Inada R, Sekine S, Taniguchi H, Tsuda H, Katai H, Fujiwara T, Kushima R. ARID1A expression in gastric adenocarcinoma: clinicopathological significance and correlation with DNA mismatch repair status. World J Gastroenterol. 2015;21:2159-2168. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 29] [Cited by in RCA: 46] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 25. | Yamamoto H, Watanabe Y, Maehata T, Morita R, Yoshida Y, Oikawa R, Ishigooka S, Ozawa S, Matsuo Y, Hosoya K. An updated review of gastric cancer in the next-generation sequencing era: insights from bench to bedside and vice versa. World J Gastroenterol. 2014;20:3927-3937. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 56] [Cited by in RCA: 66] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 26. | Wang K, Yuen ST, Xu J, Lee SP, Yan HH, Shi ST, Siu HC, Deng S, Chu KM, Law S. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46:573-582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 694] [Cited by in RCA: 861] [Article Influence: 71.8] [Reference Citation Analysis (0)] |
| 27. | Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390-7394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 787] [Cited by in RCA: 816] [Article Influence: 40.8] [Reference Citation Analysis (0)] |
| 28. | Ebert MS, Sharp PA. MicroRNA sponges: progress and possibilities. RNA. 2010;16:2043-2050. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 568] [Cited by in RCA: 596] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 29. | Ebert MS, Sharp PA. Emerging roles for natural microRNA sponges. Curr Biol. 2010;20:R858-R861. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 366] [Cited by in RCA: 386] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 30. | Hong L, Yang Z, Ma J, Fan D. Function of miRNA in controlling drug resistance of human cancers. Curr Drug Targets. 2013;14:1118-1127. [PubMed] |
| 31. | Wang Z, Cai Q, Jiang Z, Liu B, Zhu Z, Li C. Prognostic role of microRNA-21 in gastric cancer: a meta-analysis. Med Sci Monit. 2014;20:1668-1674. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 48] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 32. | Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799-2803. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1136] [Cited by in RCA: 1215] [Article Influence: 60.8] [Reference Citation Analysis (0)] |
| 33. | Xu LF, Wu ZP, Chen Y, Zhu QS, Hamidi S, Navab R. MicroRNA-21 (miR-21) regulates cellular proliferation, invasion, migration, and apoptosis by targeting PTEN, RECK and Bcl-2 in lung squamous carcinoma, Gejiu City, China. PLoS One. 2014;9:e103698. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 154] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 34. | Yang SM, Huang C, Li XF, Yu MZ, He Y, Li J. miR-21 confers cisplatin resistance in gastric cancer cells by regulating PTEN. Toxicology. 2013;306:162-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 177] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 35. | He XJ, Ma YY, Yu S, Jiang XT, Lu YD, Tao L, Wang HP, Hu ZM, Tao HQ. Up-regulated miR-199a-5p in gastric cancer functions as an oncogene and targets klotho. BMC Cancer. 2014;14:218. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 74] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 36. | Xu Y, Huang Z, Liu Y. Reduced miR-125a-5p expression is associated with gastric carcinogenesis through the targeting of E2F3. Mol Med Rep. 2014;10:2601-2608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 45] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 37. | Shi DB, Xing AY, Gao C, Gao P. [Expression of microRNA-100 in human gastric cancer]. Zhonghua Bing Li Xue Za Zhi. 2013;42:15-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 38. | Piazuelo MB, Correa P. Gastric cáncer: Overview. Colomb Med (Cali). 2013;44:192-201. [PubMed] |
| 39. | Saito Y, Suzuki H, Tsugawa H, Imaeda H, Matsuzaki J, Hirata K, Hosoe N, Nakamura M, Mukai M, Saito H. Overexpression of miR-142-5p and miR-155 in gastric mucosa-associated lymphoid tissue (MALT) lymphoma resistant to Helicobacter pylori eradication. PLoS One. 2012;7:e47396. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 92] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 40. | Wang Z, Liu M, Zhu H, Zhang W, He S, Hu C, Quan L, Bai J, Xu N. miR-106a is frequently upregulated in gastric cancer and inhibits the extrinsic apoptotic pathway by targeting FAS. Mol Carcinog. 2013;52:634-646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 59] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 41. | Hayashi Y, Tsujii M, Wang J, Kondo J, Akasaka T, Jin Y, Li W, Nakamura T, Nishida T, Iijima H. CagA mediates epigenetic regulation to attenuate let-7 expression in Helicobacter pylori-related carcinogenesis. Gut. 2013;62:1536-1546. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 103] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 42. | Zhu M, Chen Q, Liu X, Sun Q, Zhao X, Deng R, Wang Y, Huang J, Xu M, Yan J. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J. 2014;281:3766-3775. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 260] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 43. | Berteaux N, Lottin S, Monté D, Pinte S, Quatannens B, Coll J, Hondermarck H, Curgy JJ, Dugimont T, Adriaenssens E. H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1. J Biol Chem. 2005;280:29625-29636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 275] [Cited by in RCA: 296] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 44. | Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J, Fang G. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279:3159-3165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 325] [Cited by in RCA: 379] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 45. | Park IY, Sohn BH, Choo JH, Joe CO, Seong JK, Lee YI, Chung JH. Deregulation of DNA methyltransferases and loss of parental methylation at the insulin-like growth factor II (Igf2)/H19 loci in p53 knockout mice prior to tumor development. J Cell Biochem. 2005;94:585-596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 45] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 46. | Endo H, Shiroki T, Nakagawa T, Yokoyama M, Tamai K, Yamanami H, Fujiya T, Sato I, Yamaguchi K, Tanaka N. Enhanced expression of long non-coding RNA HOTAIR is associated with the development of gastric cancer. PLoS One. 2013;8:e77070. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 177] [Cited by in RCA: 217] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 47. | Sun M, Jin FY, Xia R, Kong R, Li JH, Xu TP, Liu YW, Zhang EB, Liu XH, De W. Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer. BMC Cancer. 2014;14:319. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 216] [Cited by in RCA: 268] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 48. | Benetatos L, Vartholomatos G, Hatzimichael E. MEG3 imprinted gene contribution in tumorigenesis. Int J Cancer. 2011;129:773-779. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 227] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 49. | Sun M, Xia R, Jin F, Xu T, Liu Z, De W, Liu X. Downregulated long noncoding RNA MEG3 is associated with poor prognosis and promotes cell proliferation in gastric cancer. Tumour Biol. 2014;35:1065-1073. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 238] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 50. | Song H, Sun W, Ye G, Ding X, Liu Z, Zhang S, Xia T, Xiao B, Xi Y, Guo J. Long non-coding RNA expression profile in human gastric cancer and its clinical significances. J Transl Med. 2013;11:225. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 166] [Cited by in RCA: 198] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 51. | Cao WJ, Wu HL, He BS, Zhang YS, Zhang ZY. Analysis of long non-coding RNA expression profiles in gastric cancer. World J Gastroenterol. 2013;19:3658-3664. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 144] [Cited by in RCA: 172] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 52. | Park SM, Park SJ, Kim HJ, Kwon OH, Kang TW, Sohn HA, Kim SK, Moo Noh S, Song KS, Jang SJ. A known expressed sequence tag, BM742401, is a potent lincRNA inhibiting cancer metastasis. Exp Mol Med. 2013;45:e31. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 49] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 53. | Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6253] [Cited by in RCA: 6221] [Article Influence: 478.5] [Reference Citation Analysis (0)] |
| 54. | Zlotorynski E. Non-coding RNA: Circular RNAs promote transcription. Nat Rev Mol Cell Biol. 2015;16:206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 44] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 55. | Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4631] [Cited by in RCA: 6219] [Article Influence: 478.4] [Reference Citation Analysis (0)] |
| 56. | Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414-4422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 650] [Cited by in RCA: 842] [Article Influence: 56.1] [Reference Citation Analysis (0)] |
| 57. | Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609-5612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 629] [Cited by in RCA: 763] [Article Influence: 58.7] [Reference Citation Analysis (0)] |
| 58. | Peng L, Yuan XQ, Li GC. The emerging landscape of circular RNA ciRS-7 in cancer (Review). Oncol Rep. 2015;33:2669-2674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 165] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 59. | Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW, Zeillinger R, Pils D. Correlation of circular RNA abundance with proliferation--exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 518] [Cited by in RCA: 622] [Article Influence: 56.5] [Reference Citation Analysis (0)] |
| 60. | Li P, Chen S, Chen H, Mo X, Li T, Shao Y, Xiao B, Guo J. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 543] [Cited by in RCA: 640] [Article Influence: 58.2] [Reference Citation Analysis (0)] |
| 61. | Sumazin P, Yang X, Chiu HS, Chung WJ, Iyer A, Llobet-Navas D, Rajbhandari P, Bansal M, Guarnieri P, Silva J. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147:370-381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 554] [Cited by in RCA: 591] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 62. | Zhou X, Liu J, Wang W. Construction and investigation of breast-cancer-specific ceRNA network based on the mRNA and miRNA expression data. IET Syst Biol. 2014;8:96-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 63] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 63. | Paci P, Colombo T, Farina L. Computational analysis identifies a sponge interaction network between long non-coding RNAs and messenger RNAs in human breast cancer. BMC Syst Biol. 2014;8:83. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 182] [Cited by in RCA: 210] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 64. | Xia T, Liao Q, Jiang X, Shao Y, Xiao B, Xi Y, Guo J. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci Rep. 2014;4:6088. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 246] [Cited by in RCA: 332] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 65. | Bosia C, Pagnani A, Zecchina R. Modelling Competing Endogenous RNA Networks. PLoS One. 2013;8:e66609. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 100] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 66. | Sarver AL, Subramanian S. Competing endogenous RNA database. Bioinformation. 2012;8:731-733. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 80] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 67. | Das S, Ghosal S, Sen R, Chakrabarti J. lnCeDB: database of human long noncoding RNA acting as competing endogenous RNA. PLoS One. 2014;9:e98965. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 129] [Cited by in RCA: 138] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 68. | Liu K, Yan Z, Li Y, Sun Z. Linc2GO: a human LincRNA function annotation resource based on ceRNA hypothesis. Bioinformatics. 2013;29:2221-2222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 90] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 69. | Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92-D97. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2607] [Cited by in RCA: 4299] [Article Influence: 330.7] [Reference Citation Analysis (0)] |
| 70. | Jeggari A, Marks DS, Larsson E. miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics. 2012;28:2062-2063. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 447] [Cited by in RCA: 594] [Article Influence: 42.4] [Reference Citation Analysis (0)] |
| 71. | Paraskevopoulou MD, Georgakilas G, Kostoulas N, Reczko M, Maragkakis M, Dalamagas TM, Hatzigeorgiou AG. DIANA-LncBase: experimentally verified and computationally predicted microRNA targets on long non-coding RNAs. Nucleic Acids Res. 2013;41:D239-D245. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 267] [Cited by in RCA: 298] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 72. | Yang JH, Li JH, Jiang S, Zhou H, Qu LH. ChIPBase: a database for decoding the transcriptional regulation of long non-coding RNA and microRNA genes from ChIP-Seq data. Nucleic Acids Res. 2013;41:D177-D187. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 250] [Cited by in RCA: 256] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 73. | Kartha RV, Subramanian S. Competing endogenous RNAs (ceRNAs): new entrants to the intricacies of gene regulation. Front Genet. 2014;5:8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 208] [Cited by in RCA: 316] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 74. | Jalali S, Bhartiya D, Lalwani MK, Sivasubbu S, Scaria V. Systematic transcriptome wide analysis of lncRNA-miRNA interactions. PLoS One. 2013;8:e53823. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 333] [Cited by in RCA: 366] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 75. | Yan J, Guo X, Xia J, Shan T, Gu C, Liang Z, Zhao W, Jin S. MiR-148a regulates MEG3 in gastric cancer by targeting DNA methyltransferase 1. Med Oncol. 2014;31:879. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 112] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 76. | Li H, Yu B, Li J, Su L, Yan M, Zhu Z, Liu B. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5:2318-2329. [PubMed] |
| 77. | Xu C, Shao Y, Xia T, Yang Y, Dai J, Luo L, Zhang X, Sun W, Song H, Xiao B. lncRNA-AC130710 targeting by miR-129-5p is upregulated in gastric cancer and associates with poor prognosis. Tumour Biol. 2014;35:9701-9706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 74] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 78. | Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD, Kong R, Xia R, Lu KH, Li JH. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 659] [Cited by in RCA: 799] [Article Influence: 66.6] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
P- Reviewer: Pandi NS S- Editor: Yu J L- Editor: Wang TQ E- Editor: Liu XM