Published online Aug 21, 2015. doi: 10.3748/wjg.v21.i31.9348
Peer-review started: January 29, 2015
First decision: March 10, 2015
Revised: April 7, 2015
Accepted: June 16, 2015
Article in press: June 16, 2015
Published online: August 21, 2015
Processing time: 204 Days and 1 Hours
AIM: To detect linc00675 expression in pancreatic ductal adenocarcinoma (PDAC), to analyze the relationship between the expression level of linc00675 and the clinical pathological characteristics, to explore the biological functions of linc00675, and to determine whether linc00675 has independent prognostic value in PDAC.
METHODS: We studied linc00675 expression among eight histologically confirmed PDAC tissue samples and four chronic pancreatitis tissue samples through microarray screening. RT-qPCR was conducted to further investigate linc00675 expression in PDAC cell lines as well as archived tissues from a large cohort of PDAC patients. The correlations between the level of lnc00675 and clinicopathological characteristics and survival in patients with pancreatic cancer were evaluated using Correlation analysis. Univariate and multivariate analyses were conducted to predict whether lnc00675 expression is an independent prognostic and recurrence factor in patients with pancreatic cancer. After downregulating the expression of linc00675 through siRNA, MTT assay, flow cytometry, transwell assay and Western blot were used to explore the biological function of linc00675 in proliferation, invasion, and cell cycle progression of pancreatic cancer cells. The relative molecular expression levels of epithelial-mesenchymal transition were determined by reverse transcription-polymerase chain reaction (RT-PCR) and Western blot.
RESULTS: The expression of Linc00675 in PDAC tissue samples was shown to be 672 times that in chronic pancreatitis tissue samples by microarray screening (P = 3.69 × 10-5). This finding was confirmed in tumor tissues from 90 patients with PDAC compared with adjacent normal tissue samples by quantitative RT-PCR. We found that linc00675 overexpression positively correlated with lymph node metastasis (P = 0.005), perineural invasion (P = 0.006), and poor survival (P < 0.001). Univariate and multivariate analyses showed that linc00675 expression served as an independent predictor of overall survival (P = 0.009). Additionally, receiver operating characteristic curve analysis showed that high linc00675 might serve as a predictor of tumor progression within 6 mo to a year after surgery. In vitro functional analysis demonstrated that knockdown of linc00675 attenuated pancreatic cancer cell proliferation and invasion as well as induced S phase arrest. Suppression of linc00675 in pancreatic cancer cells resulted can reverse the progress of epithelial-mesenchymal transition.
CONCLUSION: Linc00675 may function as an oncogene during PDAC development, and its expression is an independent predictor of unfavorable prognosis in patients with PDAC.
Core tip: This is the first study to report that linc00675 is more highly expressed in pancreatic ductal adenocarcinoma (PDAC) tissues than in adjacent normal tissues. Overexpression of linc00675 in PDAC tissues positively correlated with short survival and tumor progression. The prominent finding in this study is that linc00675 is an independent prognostic marker for predicting the survival of PDAC patients after surgery.
- Citation: Li DD, Fu ZQ, Lin Q, Zhou Y, Zhou QB, Li ZH, Tan LP, Chen RF, Liu YM. Linc00675 is a novel marker of short survival and recurrence in patients with pancreatic ductal adenocarcinoma. World J Gastroenterol 2015; 21(31): 9348-9357
- URL: https://www.wjgnet.com/1007-9327/full/v21/i31/9348.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i31.9348
Pancreatic ductal adenocarcinoma (PDAC) that originates in the glandular epithelium accounts for approximately 90% of all pancreatic tumors and exhibits a high grade of malignancy[1]. PDAC patients have an extremely poor prognosis, with a 5-year survival rate of approximately 6%. Even in patients who undergo surgical resection, the disease commonly recurs and the 5-year survival rate remains low at 15%-25%[2]. This dismal prognosis is due to the aggressive nature of this disease, and its resistance to traditional therapeutic strategies. Therefore, the development of an effective treatment for PDAC requires further research to reveal the molecular mechanisms underlying its aggressive pathogenesis.
Long noncoding RNAs (lncRNAs) are RNA molecules over 200 nucleotides in length with little protein-coding potential. Long intergenic noncoding RNAs (lincRNAs) have transcription loci that fall between two protein-coding genes and function to regulate gene expression at various levels, including transcription, epigenetic regulation and post-transcriptional processing[3-7]. Accumulating evidence suggests that deregulation of lincRNAs may contribute to many types of human diseases, including cancer[8,9]. Moreover, they play critical roles in cancer initiation, progression and metastasis[10-12]. LincRNA expression signatures have been associated with patient survival and may be useful in the patient management and the design of anticancer treatments[13]. Several lincRNAs have been implicated in tumorigenesis. However, the biological functions and prognostic value of lincRNAs in pancreatic cancer remain largely unexplored. Thus, there is an urgent need to identify the etiology and biological function of lincRNAs that may serve as markers of diagnosis and prognosis in PDAC to improve survival in this disease.
In the present study, based on microarray analysis, we focused on a long intergenic noncoding RNA named linc00675 that showed 672-fold upregulation in PDAC compared with normal pancreatic tissues. Building on this finding, we determined the significance of linc00675 in PDAC by investigating the relationship between aberrantly expressed linc00675 and patients’ clinicopathological features, as well as performing further in vitro study of PDAC cell lines. We found that upregulation of linc00675 was associated with short survival. In addition to affecting the cell cycle, overexpression of linc00675 could therefore promote cancer cell proliferation, migration and invasion. Thus, our study revealed that linc00675 is a promising prognostic biomarker in pancreatic cancer, and could be useful in pancreatic cancer risk assessment and future therapeutic targeting.
Samples of fresh frozen cancer tissues, together with normal adjacent tissues, were obtained during surgical resection from Sun Yat-sen Memorial Hospital of Sun Yat-sen University. Informed consent was obtained from the patients before sample collection, and approved by the hospital’s Ethics Review Committee. All samples were confirmed by pathological examination.
The human pancreatic cancer cell lines PANC1, Capan2, BXPC-3, Mia PaCa2, SW1990, and immortalized human pancreatic ductal epithelial cells (HPDE6) were purchased from the American Type Culture Collection and grown in complete growth medium with 10% FBS and 1% penicillin/streptomycin as recommended by the manufacturer. All the cells were cultured in a humidified 5% CO2 incubator at 37 °C.
Total mRNA was extracted, purified using the mRNA-ONLYTM Eukaryotic mRNA Isolation Kit (Epicentre, Madison, CA). Total RNA was fragmented and then labeled (One-Color, Cy3, Agilent). After purification, the labeled RNA was hybridized to probes on the Hybridization Chamber gasket slides (Agilent). After being washed, the slides were scanned using an Agilent Microarray Scanner. The raw data were extracted with the Feature Extraction software (Agilent Technology). This software utilizes the robust multiarray average algorithm to adjust the background signals. Normalized data were obtained using the quantile method of intra-microarray normalization and median method of baseline transformation between the microarrays. Differentially expressed genes with a raw expression level of over 400 in more than 4 of the 12 samples used for profiling were extracted. Then they were ordered by P value. The 10 most significantly de-regulated genes (those with the smallest P values) were selected for validation. We also computed the maximum false discovery rate based on a single gene-probe P value threshold of 0.05. We considered as significant signatures with a false discovery rate ≤ 0.1. The microarray platform and data were submitted to the Gene Expression Omnibus public database at the National Center for Biotechnology Information (accession number: GSE61166, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE61166).
Real-time quantitative PCR (RT-qPCR) was performed for linc00675 and EMT marker (E-cadherin, N-cadherin, and Vimentin) mRNAs, with β-actin as an internal control. The total RNA was then converted to cDNA by reverse-transcription using oligodT primers and SuperScript II reverse transcriptase (Invitrogen). Quantitative PCR was performed with SYBR green master mix (Roche). Relative expression values were calculated (ΔΔCT method) using β-actin as a normalizer. The primer sequences used in the study are listed in Supplementary Table 1.
| Factor | Linc00675 expression | P value1 | |
| High (n = 45) | Low (n = 45) | ||
| Age (yr) | |||
| < 60 | 22 | 23 | 0.833 |
| ≥ 60 | 23 | 22 | |
| Sex | |||
| Male | 30 | 27 | 0.512 |
| Female | 15 | 18 | |
| Differentiation | |||
| Well | 16 | 17 | 0.304 |
| Moderate | 15 | 20 | |
| Poor | 14 | 8 | |
| UICC stage | |||
| pI | 9 | 15 | 0.153 |
| pII | 36 | 30 | |
| T stage | |||
| T1 | 6 | 9 | 0.697 |
| T2 | 16 | 15 | |
| T3 | 23 | 21 | |
| N stage | |||
| N0 | 12 | 25 | 0.005 |
| N1 | 33 | 20 | |
| Perineural invasion | |||
| Negative | 17 | 30 | 0.006 |
| Positive | 28 | 15 | |
siRNA oligos targeting linc00675 (CTGATGGAGGAGAATCAATT, GTCCGAGAATGGCT GTGATT, and GTTCCAGACTCCATCACAATT), and nontargeting siRNAs (UUCUCCGAACGUG UCACGUTT) were purchased from Sigma Aldrich. siRNA transfections were done with 80 nmol/L siRNA and Lipofectamine 2000 (Life Technologies) following the manufacturer’s instructions.
After transfection, 2 × 103 cells (SW1990 or Mia PaCa-2) were plated in 96-well plates. A cell proliferation reagent kit (Roche) was used to assess cell proliferation. Transfected cells were assessed every 24 h according to the manufacturer’s instructions. For cell cycle analysis, transfected cells were collected, washed in PBS, stained with propidium oxide using the Cell Cycle Analysis Kit (Beyotime, Haimen, China), and then subjected to FACS analysis. In vitro cell invasion assay was performed using the BD BioCoat™ Matrigel™ Invasion Chamber (Becton Dickinson) according to manufacturer’s instructions, with 3 × 104 cells seeded in the upper chamber. At least three biological replicates of the experiments were performed.
Cells were washed in PBS and lysed with RIPA buffer (Invitrogen, Carlsbad, CA, United States), and a bicinchoninic acid protein assay kit (Pierce) was used to calculate the protein concentration of each sample. Equivalent amounts of proteins were separated by SDS-PAGE and transferred to polyvinylidene fluoride membranes for immunoblotting. The membranes were blocked in 5% fat-free milk for 1 h at room temperature, then incubated with the following primary antibodies: anti-CyclinA, anti-CyclinE, anti-Cyclin D1, anti-CDK2 and anti-β-actin (Abcam, Cambridge, MA); anti-Vimentin, Anti-E-cadherin, anti-N-cadherin, and anti-GAPDH (Pro-teinTech Group, Chicago, IL, United States). GAPDH was used as a loading control. Horseradish peroxidase-conjugated secondary antibodies (Cell Signaling Technology) and an ECL chemiluminescence kit (Pierce) were used to detect bound antibody.
Statistical analyses were performed using SPSS Statistics 17.0 (SPSS Inc). All in vitro experiment quantitative data are presented as the mean ± SD from at least three independent experiments, unless otherwise noted. The differences between two groups were analyzed using a Student’s t-test. The correlation between linc00675 and clinical and pathological characteristics was assessed using Pearson’s χ2 test. Survival was evaluated using the Kaplan-Meier method. Cox proportional hazard analysis was performed to calculate the hazard ratio and 95% confidence interval (CI) to evaluate the association between linc00675 and other clinicopathologic factors and survival. All tests performed were two-sided. Differences were considered statistically significant if P < 0.05.
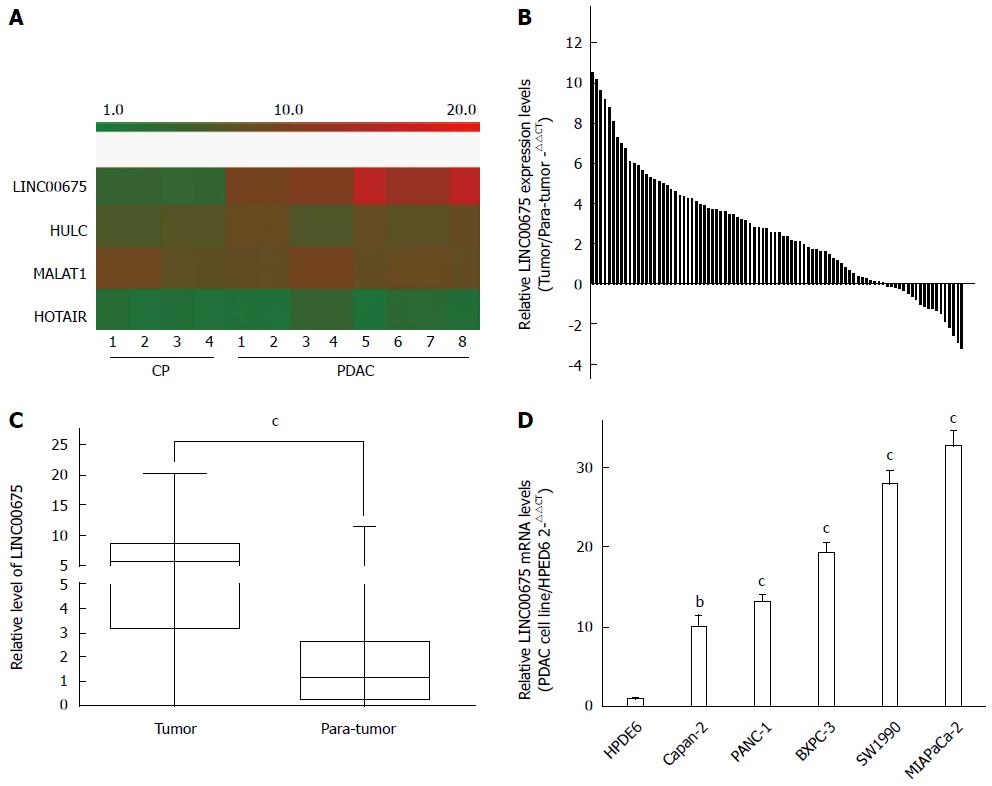
We conducted an analysis of tissues from eight PDAC cases and four cases of chronic pancreatitis (CP) using a microarray targeting 7419 lncRNAs (Agilent). We discovered that the expression of the long intergenic noncoding RNA linc00675 (LOC100289255) in PDAC tissues was 672 times that in CP tissues (P = 3.69 × 10-5, Figure 1A). The hybridization signals of another three long noncoding RNAs, HULC, MALAT1, and HOTAIR, which have previously been reported to be upregulated in pancreatic cancer, are also shown in Figure 1A. The expression of linc00675 had the most obvious difference. Next, we investigated whether linc00675 was upregulated in PDAC cell lines and a large cohort of PDAC tissues. As shown in Figure 1B and 1C, RT-qPCR revealed that expression of linc00675 was significantly higher in tumor tissues compared with matched adjacent non-tumor tissues (P < 0.001). We also found that the expression of linc00675 in each PDAC cell line was significantly higher than in the HPDE6 cell line (Figure 1D).
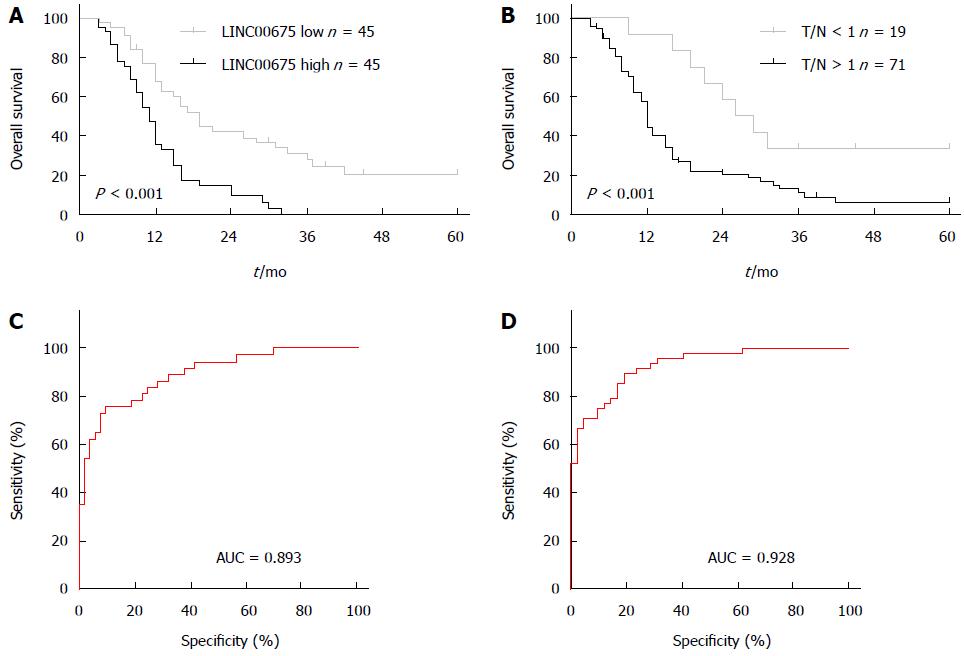
We assessed the correlation between linc00675 expression and clinical characteristics using expression levels obtained from qRT-PCR data of a cohort of 90 patients (Figure 1B). We found that linc00675 expression level was significantly associated with both lymph node metastasis (P = 0.005) and perineural invasion (P = 0.006) (Table 1). Log-rank analysis demonstrated that overall survival was significantly worse in patients with higher linc00675 expression (P < 0.001) (Figure 2A and B). Further multivariate analysis confirmed that linc00675 expression level was an independent prognostic indicator for overall survival of patients with PDAC (P = 0.009) (Table 2).
| Variable | Univariate | Multivariate | ||
| P value | HR | 95%CI | P value | |
| T stage | 0.031 | 1.812 | 1.008-3.258 | 0.047 |
| N stage | 0.026 | 2.016 | 1.112-3.657 | 0.021 |
| Perineural invasion | 0.016 | 2.611 | 1.246-5.471 | 0.011 |
| Linc00675 expression | 0.013 | 4.620 | 1.233-4.336 | 0.009 |
Because linc00675 showed a significant correlation with lymph node metastasis and perineural invasion, we went on to assess the value of linc00675 in predicting tumor progression after surgery and conducted a ROC (Receiver Operating Characteristic) curve analysis. Results showed that for predicting tumor progression within one year, the area under the ROC curve was 0.893 (P < 0.0001, Figure 2C); and for predicting progression within six months, the area under the ROC curve was 0.928 (P < 0.0001, Figure 2D). These findings suggested that linc00675 has potential diagnostic value in predicting recurrence in PDAC patients after radical surgical resection.
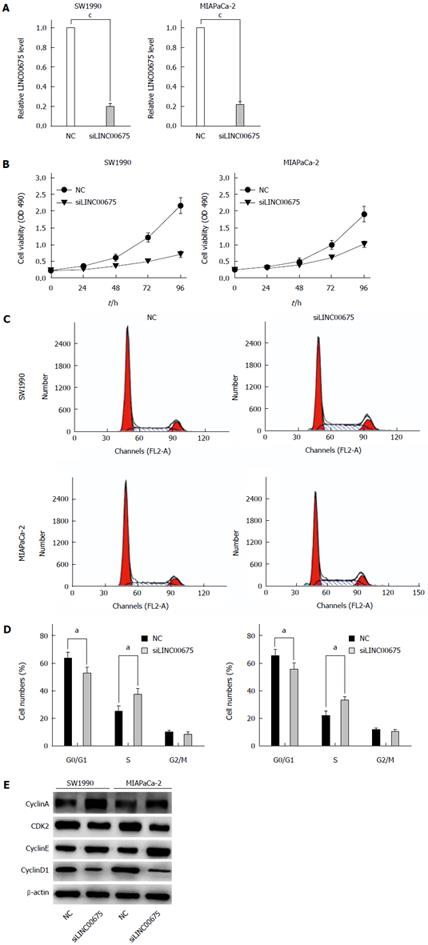
To further examine whether linc00675 functions in pancreatic cancer progression, in vitro studies were conducted. We knocked down linc00675 expression in SW1990 and MIA PaCa-2 cells using small interfering RNAs; the most effective siRNA that showed more than 70% knockout efficiency was selected for the following test (Figure 3A). Linc00675 depletion resulted in decreased tumor cell proliferation in both the pancreatic cancer cell lines SW1990 and MIA PaCa-2, as determined by MTT assay (Figure 3B). Further flow cytometry analysis showed that linc00675 knockdown induced S phase arrest in both SW1990 and MIA PaCa-2 cells (Figure 3C and D). The expression of Cyclin E, CyclinA, Cyclin D1 and CDK2, which are markers of S phase arrest, was analyzed by Western blot. In both linc00675 knockdown-treated SW1990 and Miapaca-2 cells, Cyclin E and Cyclin A, which are responsible for G1/S transition and S phase progression, respectively, were significantly upregulated, whereas the levels of Cyclin D1 and CDK2, which are suppressed in the S phase, were found to be significantly reduced (Figure 3E). The latter findings are consistent with S phase arrest via reduced expression of linc00675 in PDAC cell lines.
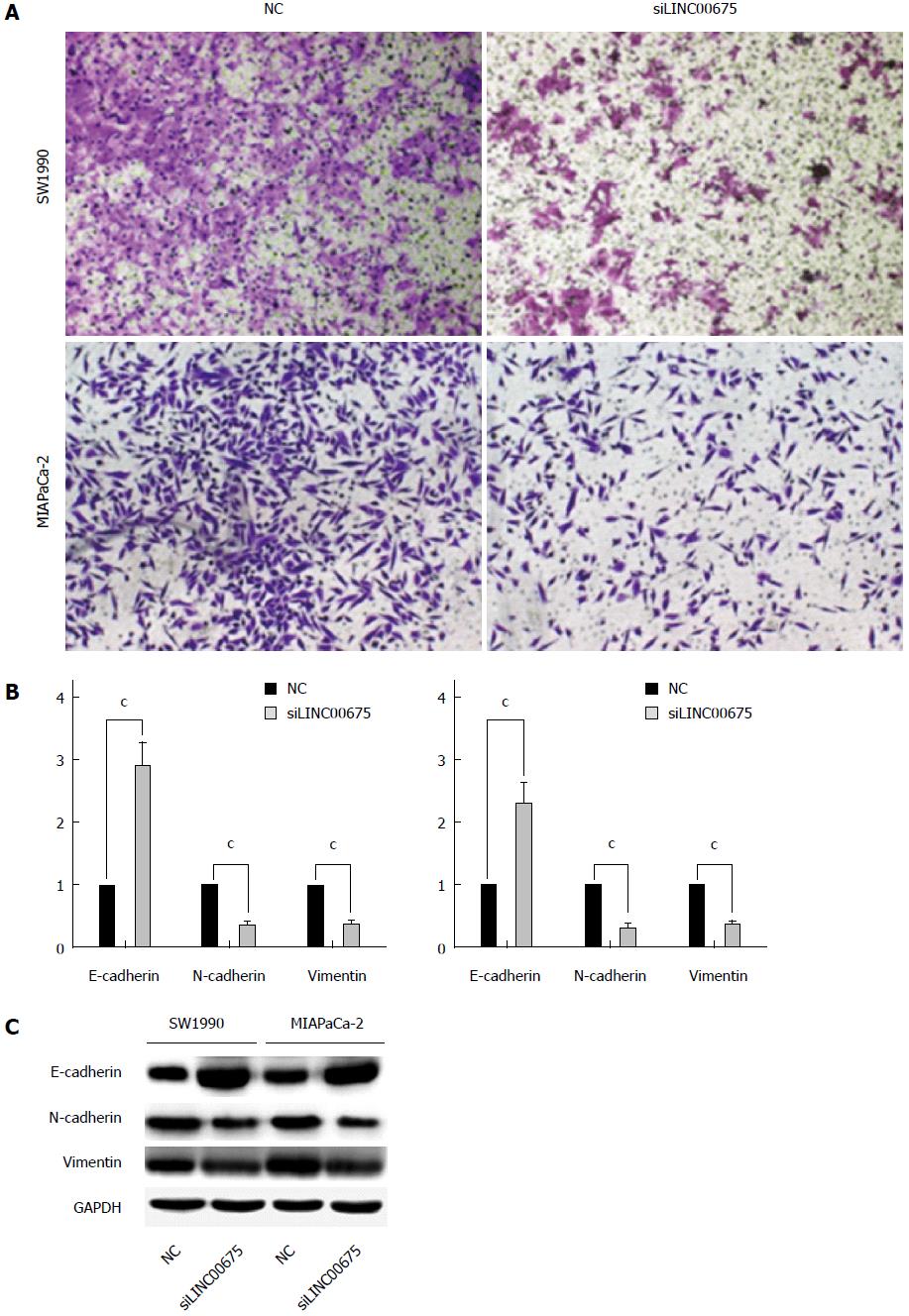
Cell invasiveness is closely correlated with cancer metastasis. We therefore examined whether linc00675 knockdown affects invasiveness of pancreatic cancer cells. A Matrigel invasion assay showed that linc00675 knockdown significantly inhibited invasiveness of SW1990 and MIA PaCa-2 cells (Figure 4A). Since EMT is closely related to cell invasiveness, we also examined whether the suppression of linc00675 can affect the expression of epithelial-mesenchymal transition (EMT)-related genes. Both PCR (Figure 4B) and Western blot analyses (Figure 4C) showed that suppression of linc00675 in pancreatic cells resulted in decreased expression of mesenchymal markers N-cadherin and vimentin, and upregulation of epithelial marker E-cadherin.
To our knowledge, the findings of the present study provide the first evidence of the potential clinical utility of linc00675 expression as a prognostic factor in PDAC, and support a role for this lncRNA in cancer cell behavior. We showed that linc00675 is frequently highly expressed in PDAC tissues as well as in PDAC cell lines, and its overexpression positively correlates with lymph node metastasis, perineural invasion and poor prognosis in PDAC patients, suggesting that linc00675 might to be a useful biomarker. Supporting this observation, functional studies revealed a strong correlation between linc00675 level and malignant behavior of pancreatic carcinoma cells.
LncRNAs are non-protein coding transcripts longer than 200 nucleotides which can regulate gene transcription indirectly through the targeting and recruitment of chromatin-modifying complexes as well as directly at the transcriptional or posttranscriptional levels[14,15]. An increasing number of studies have revealed that lncRNAs have important functions and implicated them in a wide range of diseases. Moreover, lncRNA biology is attracting great attention in cancer research because dysregulated lncRNAs occur in a variety of cancers, including pancreatic cancer[10,16]. Recently, several lncRNAs have been identified as oncogenes or tumor suppressors during cancer progression. In pancreatic cancer, the lncRNA HOTAIR[17], MALAT1[18] and HULC[19] have been found to be associated with either clinical characteristics of PDAC patients or cancer cell behavior. Abnormally expressed lncRNAs may play a causal role in pancreatic cancer initiation, development and progression by regulating cell proliferation, migration, invasion, and EMT[20-22]. However, research investigating functional lncRNAs in PDAC is still limited. In this study, based on microarray screening, we found that linc00675 expression in PDAC tissues was 672 times that in CP tissues. Notably, the level of linc00675 in pancreatic cancer tissues was markedly higher than other lncRNAs already found to recurrently upregulated and of potential prognostic use, further supporting the value of linc00675 as a biomarker and therapeutic target.
Identification of biomarkers that accurately predict survival, disease recurrence and response to chemotherapy would help to assess individual risk and treatment selection. Besides protein coding genes, noncoding RNAs have also been shown to possess potential diagnostic and prognostic value. Efforts have been made to identify molecular predictive factors for lymph node metastasis, perineural invasion, and the survival of cancer patients[23,24]. Here we found that linc00675 expression correlated with the prognosis of PDAC patients. PDAC patients with a high linc00675 level in tumor tissue had a very poor outcome. Importantly, this subset of patients showed a higher recurrence rate with increased linc00675 expression. By ROC curve analysis, the AUC using linc00675 as a predictor for tumor recurrence was around 0.9, which is higher than that reported for most prognostic tumor markers, such as CA19-9. Linc00675 may therefore prove to be a more useful prognostic factor in PDAC patients than biomarkers reported to date.
The prognostic value of linc00675 in patients with PDAC is supported by functional experiments. We modulated its expression in SW1990 and Miacapa-2 cell lines and found that suppression of linc00675 could reduce cell proliferation and invasion ability, which was consistent with clinical findings. Interestingly, we found that knockdown of linc00675 resulted in S phase arrest in pancreatic cancer cells. Gemcitabine, a chemotherapy agent, exerts its cytotoxic effect mainly by targeting tumor cells in S phase, which remains a standard therapy in PDAC patients. Some tumor suppressor genes and molecules were identified to increase gemcitabine sensitivity in pancreatic cancer cells through S-phase arrest[25]. Because the patients analyzed in this study were receiving gemcitabine-based chemotherapy after surgery, and linc00675 showed S phase arrest in pancreatic cancer cells, it will be interesting to explore whether linc00675 contributes to increased gemcitabine sensitivity.
In summary, we found strong expression of linc00675 in patients with PDAC, and suggest that linc00675 may serve as an oncogenic lincRNA that promotes pancreatic cancer cell growth and progression. Further study is required to completely define the function of linc00675, its utility in guiding patient management and its potential as a therapeutic target.
Pancreatic ductal adenocarcinoma (PDAC) is a highly malignant digestive tumor with extremely poor prognosis. Long intergenic noncoding RNAs (lincRNAs) have key roles in the regulation of multiple biological processes, including development, differentiation and carcinogenesis. There is, therefore, a need to explore the potential of lincRNAs as markers of diagnosis and prognosis and to investigate their biological functions to improve the outcome of PDAC patients.
Recently, lincRNAs have been found to play critical roles in cancer initiation, progression and metastasis. LincRNA expression has been associated with patient survival and may be useful in outcome prediction and the design of anticancer treatments. Several lincRNAs have been implicated in pancreatic tumorigenesis; however, the role of linc00675 in PDAC is still unknown. In this study, the authors demonstrate that linc00675 was highly expressed in PDAC tissues compared with adjacent normal tissues. Increased expression of linc00675 in PDAC tissues positively correlated with poor survival and tumor progression. These results indicate that linc00675 could be a potential prognostic factor for PDAC patients.
This is the first study to report that linc00675 is overexpressed in PDAC tissue compared with adjacent normal tissue. The overexpression of linc00675 positively correlates with poor survival and short-term recurrence in patients with PDAC and in functional experiments was shown to promote pancreatic cancer cell growth and progression.
This study showed that the linc00675 expression level may be useful as a predictor of prognosis in pancreatic cancer.
Linc00675 serves as an oncogenic lincRNA that promotes pancreatic cancer cell growth and progression. Since linc00675 is associated with the malignancy phenotypes of pancreatic cancer, further study is required to determine the potential roles of linc00675 as a candidate therapeutic target.
This is an interesting study with valuable information regarding the expression and clinical impact of linc00675 in pancreatic ductal adenocarcinoma.
| 1. | Reichert M, Rustgi AK. Pancreatic ductal cells in development, regeneration, and neoplasia. J Clin Invest. 2011;121:4572-4578. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 182] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 2. | Michl P, Gress TM. Current concepts and novel targets in advanced pancreatic cancer. Gut. 2013;62:317-326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 125] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 3. | Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3924] [Cited by in RCA: 4503] [Article Influence: 264.9] [Reference Citation Analysis (0)] |
| 4. | Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494-1504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1668] [Cited by in RCA: 1887] [Article Influence: 111.0] [Reference Citation Analysis (0)] |
| 5. | Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629-641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4004] [Cited by in RCA: 3982] [Article Influence: 234.2] [Reference Citation Analysis (0)] |
| 6. | Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651-669. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1443] [Cited by in RCA: 1488] [Article Influence: 114.5] [Reference Citation Analysis (0)] |
| 7. | Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. Br J Cancer. 2013;108:2419-2425. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 582] [Cited by in RCA: 612] [Article Influence: 47.1] [Reference Citation Analysis (0)] |
| 8. | Brunner AL, Beck AH, Edris B, Sweeney RT, Zhu SX, Li R, Montgomery K, Varma S, Gilks T, Guo X. Transcriptional profiling of long non-coding RNAs and novel transcribed regions across a diverse panel of archived human cancers. Genome Biol. 2012;13:R75. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 197] [Cited by in RCA: 195] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 9. | Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354-361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1424] [Cited by in RCA: 1574] [Article Influence: 104.9] [Reference Citation Analysis (0)] |
| 10. | Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391-407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1266] [Cited by in RCA: 1481] [Article Influence: 105.8] [Reference Citation Analysis (0)] |
| 11. | Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703-719. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1254] [Cited by in RCA: 1519] [Article Influence: 108.5] [Reference Citation Analysis (0)] |
| 12. | Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071-1076. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4202] [Cited by in RCA: 4303] [Article Influence: 268.9] [Reference Citation Analysis (0)] |
| 13. | Deng Q, He B, Gao T, Pan Y, Sun H, Xu Y, Li R, Ying H, Wang F, Liu X. Up-regulation of 91H promotes tumor metastasis and predicts poor prognosis for patients with colorectal cancer. PLoS One. 2014;9:e103022. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 69] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 14. | Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 537] [Cited by in RCA: 571] [Article Influence: 33.6] [Reference Citation Analysis (0)] |
| 15. | Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15 Spec No 1:R17-R29. [PubMed] |
| 16. | Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1127] [Cited by in RCA: 1352] [Article Influence: 90.1] [Reference Citation Analysis (0)] |
| 17. | Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616-1625. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 631] [Cited by in RCA: 690] [Article Influence: 53.1] [Reference Citation Analysis (0)] |
| 18. | Jiao F, Hu H, Yuan C, Wang L, Jiang W, Jin Z, Guo Z, Wang L. Elevated expression level of long noncoding RNA MALAT-1 facilitates cell growth, migration and invasion in pancreatic cancer. Oncol Rep. 2014;32:2485-2492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 132] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 19. | Peng W, Gao W, Feng J. Long noncoding RNA HULC is a novel biomarker of poor prognosis in patients with pancreatic cancer. Med Oncol. 2014;31:346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 127] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 20. | Sun YW, Chen YF, Li J, Huo YM, Liu DJ, Hua R, Zhang JF, Liu W, Yang JY, Fu XL. A novel long non-coding RNA ENST00000480739 suppresses tumour cell invasion by regulating OS-9 and HIF-1α in pancreatic ductal adenocarcinoma. Br J Cancer. 2014;111:2131-2141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 98] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 21. | Ma C, Nong K, Zhu H, Wang W, Huang X, Yuan Z, Ai K. H19 promotes pancreatic cancer metastasis by derepressing let-7’s suppression on its target HMGA2-mediated EMT. Tumour Biol. 2014;35:9163-9169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 237] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 22. | Pang EJ, Yang R, Fu XB, Liu YF. Overexpression of long non-coding RNA MALAT1 is correlated with clinical progression and unfavorable prognosis in pancreatic cancer. Tumour Biol. 2015;36:2403-2407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 160] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 23. | Xiao Z, Luo G, Liu C, Wu C, Liu L, Liu Z, Ni Q, Long J, Yu X. Molecular mechanism underlying lymphatic metastasis in pancreatic cancer. Biomed Res Int. 2014;2014:925845. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 24. | Bapat AA, Hostetter G, Von Hoff DD, Han H. Perineural invasion and associated pain in pancreatic cancer. Nat Rev Cancer. 2011;11:695-707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 264] [Cited by in RCA: 362] [Article Influence: 24.1] [Reference Citation Analysis (1)] |
| 25. | Guo Y, Ziesch A, Hocke S, Kampmann E, Ochs S, De Toni EN, Göke B, Gallmeier E. Overexpression of heat shock protein 27 (HSP27) increases gemcitabine sensitivity in pancreatic cancer cells through S-phase arrest and apoptosis. J Cell Mol Med. 2015;19:340-350. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
P- Reviewer: Park JY, Ritchie S, Stanojevic GZ, Wongkham S S- Editor: Wang JL L- Editor: Wang TQ E- Editor: Liu XM