Published online Jan 21, 2015. doi: 10.3748/wjg.v21.i3.854
Peer-review started: May 13, 2014
First decision: July 9, 2014
Revised: July 30, 2014
Accepted: September 18, 2014
Article in press: September 19, 2014
Published online: January 21, 2015
Processing time: 252 Days and 8.6 Hours
AIM: To investigate the antiproliferative activity of cinobufacini on human hepatocellular carcinoma HepG2 cells and the possible mechanism of its action.
METHODS: HepG2 cells were treated with different concentrations of cinobufacini. Cell viability was measured by methylthiazolyl tetrazolium (MTT) assay. Cell cycle distribution was analyzed by flow cytometry (FCM). Cytoskeletal and nuclear alterations were observed by fluorescein isothiocyanate-phalloidin and DAPI staining under a laser scanning confocal microscope. Changes in morphology and ultrastructure of cells were detected by atomic force microscopy (AFM) at the nanoscale level.
RESULTS: MTT assay indicated that cinobufacini significantly inhibited the viability of HepG2 cells in a dose-dependent manner. With the concentration of cinobufacini increasing from 0 to 0.10 mg/mL, the cell viability decreased from 74.9% ± 2.7% to 49.41% ± 2.2% and 39.24% ± 2.1% (P < 0.05). FCM analysis demonstrated cell cycle arrest at S phase induced by cinobufacini. The immunofluorescence studies of cytoskeletal and nuclear morphology showed that after cinobufacini treatment, the regular reorganization of actin filaments in HepG2 cells become chaotic, while the nuclei were not damaged seriously. Additionally, high-resolution AFM imaging revealed that cell morphology and ultrastructure changed a lot after treatment with cinobufacini. It appeared as significant shrinkage and deep pores in the cell membrane, with larger particles and a rougher cell surface.
CONCLUSION: Cinobufacini inhibits the viability of HepG2 cells via cytoskeletal destruction and cell membrane toxicity.
Core tip: Cinobufacini is effective against hepatocarcinoma. However, its mechanism of action has not been determined. The present study investigated the effect of cinobufacini on HepG2 cells and the alterations in cell morphology and membrane ultrastructure. We used atomic force microscopy (AFM) to study the changes in cell membrane ultrastructure induced by cinobufacini. We demonstrated that AFM is a useful tool in verifying cell response to cinobufacini treatment. We observed nuclear morphology and actin filaments in the cytoskeleton by laser scanning confocal microscopy. The cellular changes allowed us to understand better the biophysical functions of HepG2 cells induced by cinobufacini.
- Citation: Wu Q, Lin WD, Liao GQ, Zhang LG, Wen SQ, Lin JY. Antiproliferative effects of cinobufacini on human hepatocellular carcinoma HepG2 cells detected by atomic force microscopy. World J Gastroenterol 2015; 21(3): 854-861
- URL: https://www.wjgnet.com/1007-9327/full/v21/i3/854.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i3.854
Cinobufacini is a water-soluble Chinese medicine that is extracted from the skin of Bufo bufo gargarizans Cantor[1]. It has been proven to be effective against a variety of malignant tumor cells, such as breast cancer[2], lung cancer[3] and hepatocellular carcinoma[1,4] cells. In recent years, it has also shown satisfactory therapeutic effects against cancer in clinical studies[5-7]. Although cinobufacini is widely used clinically, little is known about its anti-tumor mechanisms. In particular, there are no detailed data on the changes it induces in cell membrane morphology. The present study sought to investigate the effect of cinobufacini on human hepatoma cell line HepG2 and the alterations in cell morphology and cell membrane ultrastructure.
Atomic force microscopy (AFM) is a powerful tool for nanoscale imaging of cells[8-10], and an important diagnostic instrument[11]. In this study, AFM was used to visualize cell morphology and membrane ultrastructure, which can provide information about the surface topography of the cell at the nanometric level. We used AFM to image the changes in HepG2 cell membrane ultrastructure induced by cinobufacini. We also demonstrated that AFM is a useful tool in discerning and verifying cell response to cinobufacini. In addition, we also analyzed the cell cycle by flow cytometry (FCM), and observed the nuclear morphology and actin filaments in the cytoskeleton by laser scanning confocal microscopy (LSCM). The changes observed in the cells allow us to understand better the biophysical functions of HepG2 cells treated by cinobufacini.
All reagents used in the experiments were of analytical grade. Fetal bovine serum (FBS), 2.5% trypsin, RPMI-1640 medium, methylthiazolyl tetrazolium (MTT) and DMSO were purchased from Gibco (Carlsbad, CA, United States). Glutamine, penicillin and streptomycin were purchased from Hyclone (Logan, UT, United States). Triton X-100 and 4% paraformaldehyde were purchased from Sigma (St Louis, MO, United States). Fluorescein isothiocyanate (FITC)-phalloidin and DAPI were purchased from Biyuntian Biological (Shanghai, China). Cell cycle phase determination kit was bought from Keygen Biotechnology (Nanjing, China). Cinobufacini was provided by Jinchan Biochemistry Company Ltd. (Anhui, China). Human hepatoma cell line HepG2 was donated by the First Affiliated Hospital of Jinan University.
HepG2 cells were cultured in RPMI-1640 medium supplemented with 10% FBS at 37 °C in a humidified atmosphere containing 5% CO2, and the medium was refreshed every 2-3 d. The cinobufacini was diluted to appropriate concentrations with free medium. Cells were harvested with 0.25% trypsin when needed.
The effect of cinobufacini on cell viability was detected by MTT assay. HepG2 cells were plated at a density of 5000 cells/well in 96-well plates. After 24 h of culture, the cells were treated with cinobufacini at a final concentrations of 0, 0.01, 0.05 or 0.1 mg/mL. After incubation for 48 h, 20 μL MTT dye solution (5 mg/mL) was added to each well and incubated at 37 °C for 4 h. The medium was removed and formazan was dissolved in 150 μL DMSO. A570 of each group was then measured with a spectrophotometer (Tecan, Switzerland). Cell viability was expressed by the following formula: Viability (%) = (Atreated/Acontrol) × 100%. Experiments were repeated three times.
The effect of cinobufacini on the cell cycle of HepG2 cells was analyzed by FCM (Becton Dickinson, CA, United States). HepG2 cells were seeded at a density of 1 × 106 cells/mL in six-well plates, and treated with different concentrations of cinobufacini (0, 0.05 or 0.1 mg/mL) for 48 h. Cells were harvested and fixed in 70% ethanol and stored at 4 °C overnight. The fixed cells were centrifuged at 1000 g for 5 min and washed with cold PBS three times. At last, cells were incubated with 50 μg/mL propidium iodide (PI) containing 8 μg/mL RNase in the dark at 37 °C for 30 min. The DNA content of cells was quantified by FCM.
HepG2 cells grown on coverslips were treated with 0.1 mg/mL cinobufacini or free medium for 48 h. The actin filaments in HepG2 cells were visualized by staining with FITC-phalloidin. Cells in each group were fixed with 4% paraformaldehyde for 15 min, rinsed three times with PBS, then treated with 0.2% Triton X-100 for 20 min at room temperature, and rinsed with PBS three times again. The cells were incubated with 1 mmol/L FITC-phalloidin for 1 h in the dark at room temperature. Subsequently, we added 50 mmol/L DAPI to label the nuclei, for 15 min in the dark at room temperature. The cells were washed in PBS to remove the unbound FITC-phalloidin and DAPI. Finally, the cytoskeletal and nuclear morphology was imaged by LSCM (LSCM510 Meta Duo Scan; Carl Zeiss, Jena, Germany).
HepG2 cells were seeded on the slide and treated with different concentrations of cinobufacini (0, 0.05 or 0.1 mg/mL) for 48 h at 37 °C in 5% CO2. The cells were fixed with 4% paraformaldehyde for 15 min, washed three times with PBS, and air dried at room temperature. An atomic force microscope (Autoprobe CP Research, Veeco, CA, United States) was used in the contact mode to obtain topographic images. The silicon nitride tips (UL20B; Park Scientific Instruments) used in all AFM measurements were irradiated with UV in air for 15 min, to remove any organic contaminants prior to use. The curvature radius of the tips was < 10 nm, with a force constant set at 2.8 N/m and oscillation frequency set at 255 kHz. The prepared samples were placed on the XY scanning station of AFM, and > 5 cells were measured. The acquired images were only processed with the instrument-equipped software (Image Processing Software Version 2.1, IP 2.1) to eliminate low-frequency background noise in the scanning direction.
All data are presented as mean ± SD from at least three independent experiments. Statistical analyses were performed with SPSS version 13.0 software. Comparisons between groups were performed using two independent samples t-test; comparisons between multiple groups were tested by one-way analysis of variance; and comparison between any group using the Student-Newman-Keuls test. A P-value < 0.05 was considered statistically significant.
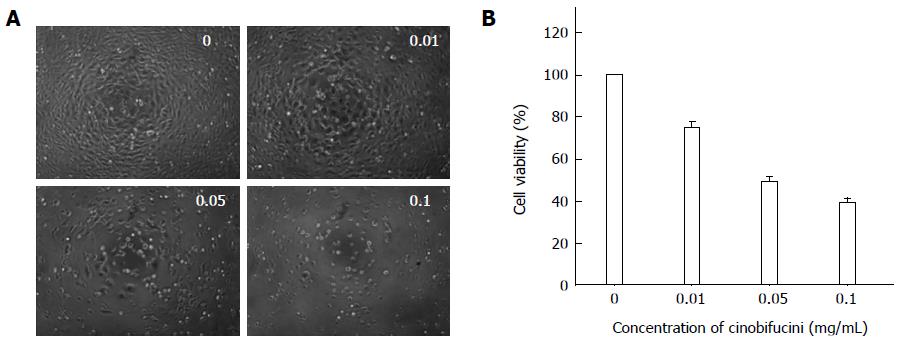
HepG2 cells were treated with different concentration of cinobufacini for 48 h. Under the inverted microscope (Figure 1A), the cells in the control group were closely arranged and well adherent in large numbers. After treatment with different concentrations of cinobufacini (0, 0.01, 0.05 or 0.10 mg/mL), the cells became round and had poor adherence, and were fewer in number, especially at a high concentration of cinobufacini.
As shown in Figure 1B, MTT assay showed that proliferation of HepG2 cells was significantly inhibited. With the concentration of cinobufacini increasing, the cell viability decreased from 74.9% ± 2.7% to 49.41% ± 2.2% and 39.24% ± 2.1%, which suggested that the inhibitory effect of cinobufacini on HepG2 cell viability was dose-dependent (P < 0.05).
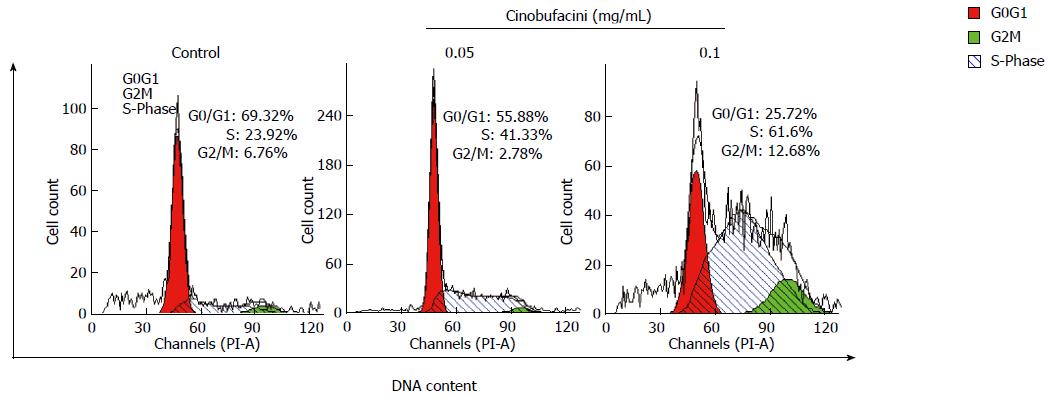
After treatment with cinobufacini at different concentrations for 48 h, cell cycle distribution of HepG2 cells was analyzed by FCM through PI staining. Figure 2 shows that the percentage of HepG2 cells in S phase increased when the concentration of cinobufacini increased. After treatment with cinobufacini at a concentration of 0.10 mg/mL, the percentage of cells in S phase increased to 61.60%, which was significantly higher than that in the control group (23.92%). These results suggested that cinobufacini caused cell cycle arrest in S phase.
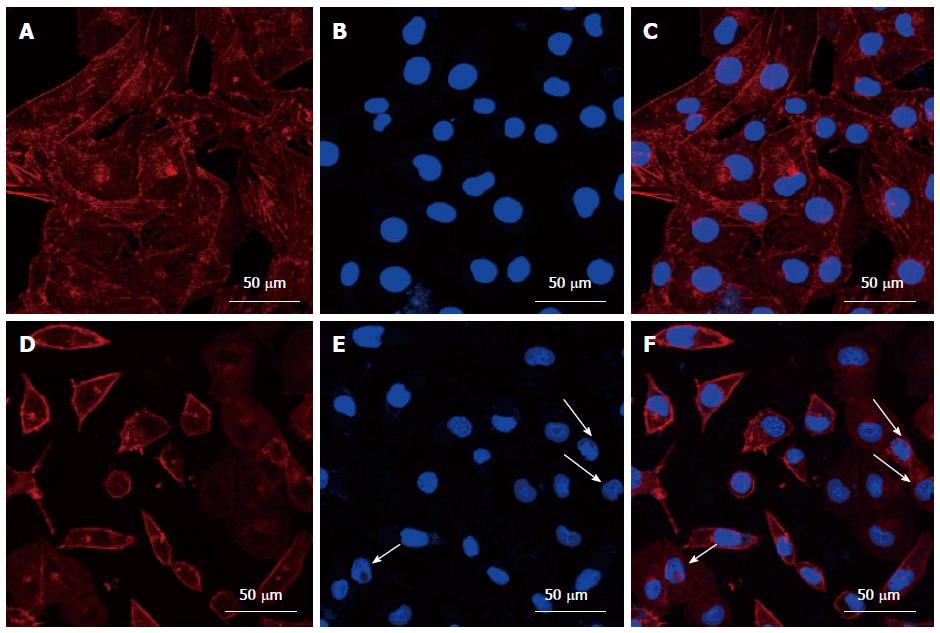
The cytoskeleton plays an important role in cell morphology, movement, and even apoptosis[2]. Actin microfilaments are important constituents of the cytoskeleton. To observe the organization of the cytoskeleton, actin was stained by FITC-phalloidin and imaged by LSCM. The cells in the control group were rich in actin with regular, parallel organization (Figure 3A). After treatment with cinobufacini, actin assembly changed dramatically. It became disordered with fewer filaments in the cells (Figure 3D). We used DAPI to stain the cell nuclei. The nuclear morphology of control HepG2 cells was intact and plump (Figure 3B). After treatment with cinobufacini for 48 h, most of the nuclei shrank, and some of the nuclei were condensed, but few were fragmented (Figure 3E, white arrows). Figure 3C and Figure 3F were merged images of (Figure 3A, B) and (Figure 3D, E).
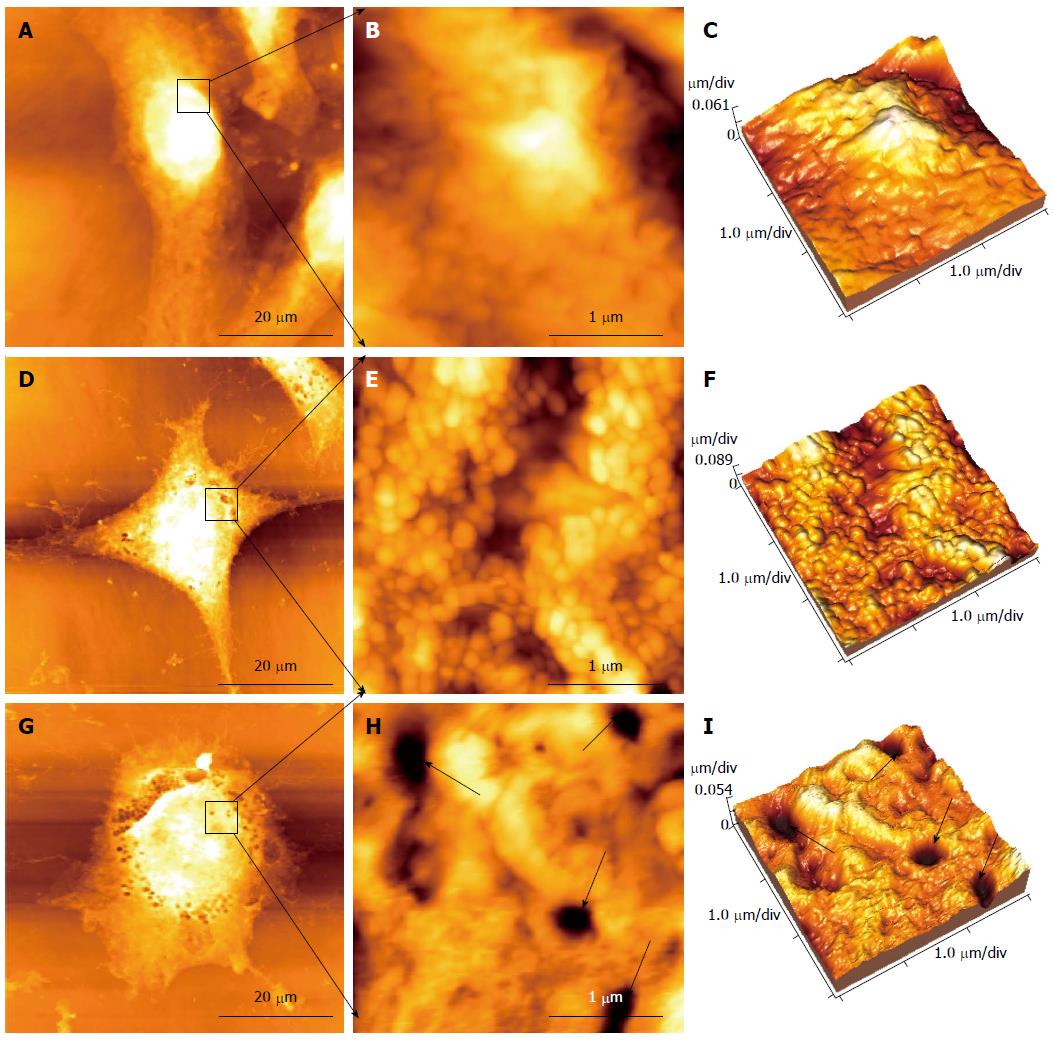
AFM was used for cell imaging and observing various changes in surface morphology and ultrastructure of HepG2 cells after treatment with different concentrations of cinobufacini. HepG2 cells in the control group were fusiform in shape (Figure 4A). The ultrastructure of the cell surface was homogeneous and displayed granular morphology, with uniform particles, and the cell membrane was relatively smooth with intact and plump cells (Figure 4B, C). After treatment with 0.05 mg/mL cinobufacini for 48 h, cell morphology changed to polygonal (Figure 4D), and the cell surface also became rougher with particles of different size (Figure 4E, F). As the concentration increased to 0.1 mg/mL, further changes in cell morphology were seen, and they appeared to be irregular and even became rounded (Figure 4G), with some pores in the cell membrane (Figure 4H, I, black arrows). These results could help us to acquire more detailed information about the toxic effect of cinobufacini on the cell membrane.
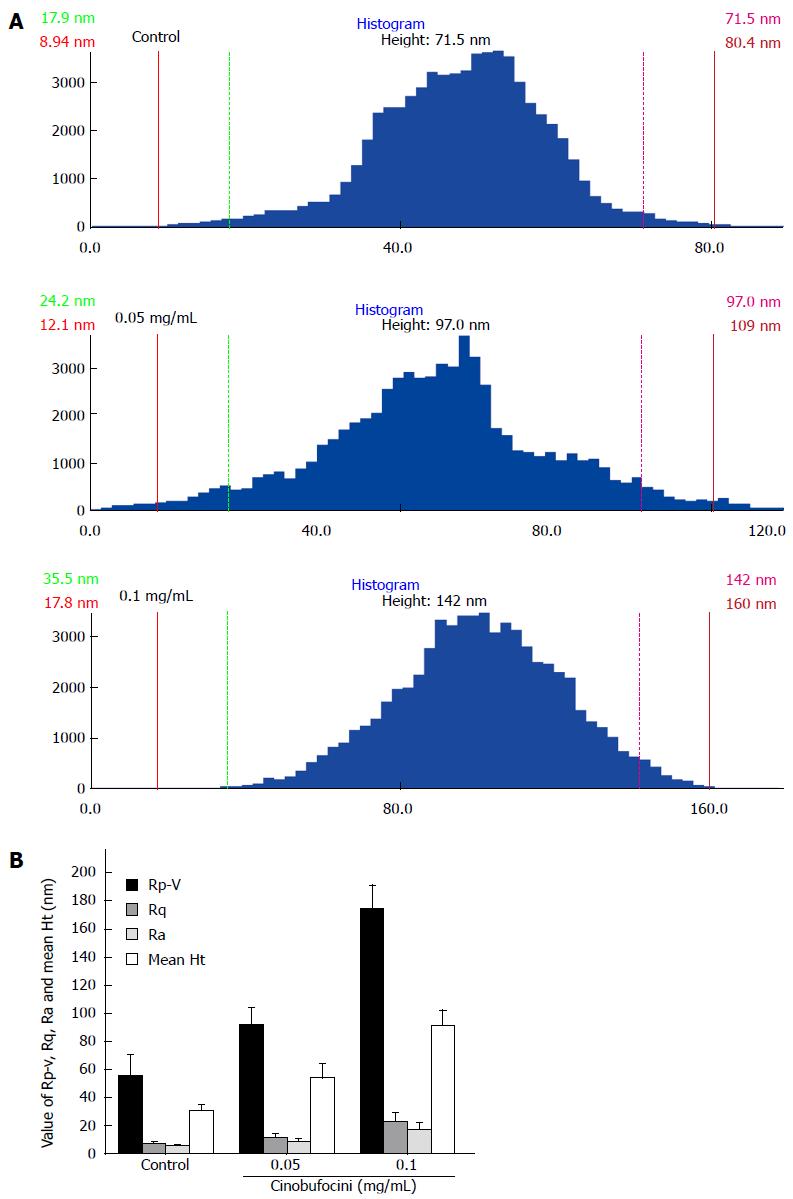
By measuring and comparing different areas of cellular ultrastructure, we observed that the average size of surface particles on control HepG2 cells was about 71.5 nm, which increased to 97.0 nm and 142.0 nm for cells treated with 0.05 mg/mL and 0.1 mg/mL cinobufacini, respectively (Figure 5A). We detected that peak to valley roughness (Rp-v), root-mean-square roughness (Rq), average roughness (Ra) and mean height (mean Ht) of the cell membrane surface increased with cinobufacini concentration. As shown in Figure 5B, Rp-v was 55.70 ± 14.74, 92.03 ± 12.03 and 174.53 ± 16.56 nm (F = 56.580, P = 0.000); Rq was 7.32 ± 1.69, 11.66 ± 2.79 and 23.35 ± 6.04 nm (F = 14.136, P = 0.000); Ra was 5.72 ± 1.07, 9.02 ± 2.15 and 17.82 ± 4.66 nm (F = 13.683, P = 0.000). Mean Ht was 30.46 ± 4.82, 54.29 ± 9.99 and 91.35 ± 11.05 nm (F = 23.895, P = 0.000). All these data showed that cinobufacini treatment induced much rougher cell membranes.
As a traditional Chinese antineoplastic drug, cinobufacini has been proved to be effective against hepatocarcinoma, and is extensively used clinically[4,7]. Clinical data indicate that total effective rate of cinobufacini against hepatocarcinoma is 44.4%, and cinobufacini can also improve quality of life[12]. However, the mechanism of cinobufacini against hepatocarcinoma has not yet been identified, and especially little is known about its effect on membrane morphology and the mechanism involved. In the present investigation, human HepG2 cells were treated with different concentrations of cinobufacini for 48 h. AFM was used to visualize cell morphology and membrane ultrastructure, which can show the surface topography of the cell at the nanometer scale. Moreover, MTT assay and FCM were used to analyze cell viability and cell cycle distribution, and LSCM was used to observe the alterations in the cytoskeleton and cell nuclei. Our results demonstrated that cinobufacini could induce cytotoxicity, growth inhibition and S phase arrest in HepG2 cells, which were associated with prominent alterations in cell membrane ultrastructure and cytoskeleton.
MTT assay showed that cinobufacini significantly inhibited the viability of HepG2 cells in a dose-dependent manner. Interruption of the tumor cell cycle so as to inhibit cell growth is the main mechanism of current antineoplastic drugs[13]. Cells are damaged by anti-cancer drugs at different phases of the cell cycle: G1 (growth and preparation of the chromosomes), S (synthesis of DNA), G2 (preparation to divide), and M (cell division). Our study showed that cinobufacini could induce cell cycle arrest at S phase. This observation is consistent with previous studies that showed cinobufacini-induced arrest of several types of human cancer cells in S phase[2,13]. S phase arrest implies that the effects of cinobufacini involve disruption of DNA synthesis and replication, which lead to in inhibition of cell proliferation.
For further investigation of the inhibitory effect of cinobufacini, we observed the alterations in cytoskeleton by staining with FITC-phalloidin and LSCM, and detected the changes in cell membrane ultrastructure by AFM. Immunofluorescence images showed the regular organization of actin filaments in HepG2 cells became chaotic and significantly reduced after treatment with cinobufacini, suggesting that cinobufacini disrupts polymerization of the actin filaments network. It is well known that the cytoskeleton contains three major constitutes: actin microfilaments, intermediate filaments, and microtubules. The cytoskeleton can be remodeled during the cellular processes, including motility, migration, adhesion, and proliferation[14]. Besides, the cytoskeletal network plays vital roles in cell morphology maintenance[2]. It was found that the course of the cell cycle depends on correct cytoskeletal arrangement[15]. As a result, cinobufacini can induce cytoskeletal rearrangement. Moreover, to detect the morphology of cell nuclei, DAPI was used to stain the cells by binding to double-stranded DNA in the nuclei. After treatment with cinobufacini for 48 h, cell nuclei showed shrinkage and chromatin condensation, but not fragmentation, which implied that the nuclear damage was less serious than cytoskeletal damage induced by cinobufacini. Taken together, these data suggest that cinobufacini inhibits the proliferation of HepG2 cells via disorganization of the cytoskeleton.
As a nondestructive surface imaging tool, AFM can obtain images of the cell surface at the nanoscale, which can provide us with qualitative and quantitative information on the architecture of cell membranes[10,16]. AFM was widely used in cell imaging, especially in cancer detection[17,18]. It can probe modifications of cell morphology induced by drugs in the imaging mode[19]. The tapping mode of AFM was used to detect the variety of changes in surface morphology and ultrastructure of HepG2 cells treated with different concentrations of cinobufacini. AFM images revealed that cinobufacini could damage the cell membrane in a dose-dependent manner. It appeared to be significant shrinkage and deep pores in the cell membrane, which were similar to the signs of apoptosis[20], especially after treatment with 0.1 mg/mL cinobufacini. With AFM at the subcellular level, we visualized that after treatment with cinobufacini, particles on the cell membrane were bigger than in the control group. It has been reported that the visible protruding particles are clusters of membrane proteins[21], which means that some biological events have occurred, such as opening/closing of ion channels, structural disruption, or changes in the chemical composition of the outer membrane proteins[22]. Changes in cell morphology and ultrastructure are closely related to cell function[23-25]. AFM images provided evidence for the toxic effects of cinobufacini on HepG2 cell membranes, which help us to achieve a better understanding of the mechanism of action of cinobufacini.
In conclusion, the present study demonstrated that cinobufacini inhibited the viability of HepG2 cells and arrested the cell cycle at S phase, which were due to the cytoskeletal destruction and membrane toxicity induced by cinobufacini. These would be the therapeutic targets of cinobufacini.
Cinobufacini is a traditional Chinese antineoplastic medicine and widely used in clinical cancer therapy in China. As an anti-tumor drug, it has been used in clinical cancer research in recent years. Atomic force microscopy (AFM) is a nondestructive surface imaging tool, which can provide the qualitative and quantitative information on cancer research.
Cinobufacini has been proven to be effective against a variety of malignant tumor cells, such as breast cancer, lung cancer and hepatocellular carcinoma cells. It has also achieved satisfactory therapeutic results in cancer. Little is known about its anti-tumor mechanisms in the cancer cells. In particular, there are no detailed data on changes in cell membrane morphology and the mechanism involved.
This is believed to be the first study to use AFM to image the changes in cell membrane ultrastructure in HepG2 cells induced by cinobufacini. These AFM images provide evidence for the toxic effects of cinobufacini on the HepG2 cell membrane, which help us to achieve a better understanding of the mechanism of action of cinobufacini.
This study demonstrated that AFM is a useful tool in discerning and verifying cell response to cinobufacini treatment.
This is an original molecular research investigating the possible anti-cancer effect of cinobufacini on a hepatocellular cancer cell line. Previous studies of this traditional Chinese medicine have shown the possible involvement of apoptotic pathways in its anti-cancer effect. Here, the authors’ study revealed the morphological changes in cancer cells by AFM after treatment with different concentrations of cinobufacini. These findings also suggest apoptotic cell death.
| 1. | Qi FH, Li AY, Lv H, Zhao L, Li JJ, Gao B, Tang W. Apoptosis-inducing effect of cinobufacini, Bufo bufo gargarizans Cantor skin extract, on human hepatoma cell line BEL-7402. Drug Discov Ther. 2008;2:339-343. [PubMed] |
| 2. | Ma L, Song B, Jin H, Pi J, Liu L, Jiang J, Cai J. Cinobufacini induced MDA-MB-231 cell apoptosis-associated cell cycle arrest and cytoskeleton function. Bioorg Med Chem Lett. 2012;22:1459-1463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 31] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 3. | Wang JY, Chen L, Zheng Z, Wang Q, Guo J, Xu L. Cinobufocini inhibits NF-κB and COX-2 activation induced by TNF-α in lung adenocarcinoma cells. Oncol Rep. 2012;27:1619-1624. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 4. | Xie RF, Li ZC, Gao B, Shi ZN, Zhou X. Bufothionine, a possible effective component in cinobufocini injection for hepatocellular carcinoma. J Ethnopharmacol. 2012;141:692-700. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 37] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 5. | Qin TJ, Zhao XH, Yun J, Zhang LX, Ruan ZP, Pan BR. Efficacy and safety of gemcitabine-oxaliplatin combined with huachansu in patients with advanced gallbladder carcinoma. World J Gastroenterol. 2008;14:5210-5216. [PubMed] |
| 6. | Qi F, Li A, Zhao L, Xu H, Inagaki Y, Wang D, Cui X, Gao B, Kokudo N, Nakata M. Cinobufacini, an aqueous extract from Bufo bufo gargarizans Cantor, induces apoptosis through a mitochondria-mediated pathway in human hepatocellular carcinoma cells. J Ethnopharmacol. 2010;128:654-661. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 107] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 7. | Meng Z, Yang P, Shen Y, Bei W, Zhang Y, Ge Y, Newman RA, Cohen L, Liu L, Thornton B. Pilot study of huachansu in patients with hepatocellular carcinoma, nonsmall-cell lung cancer, or pancreatic cancer. Cancer. 2009;115:5309-5318. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 238] [Cited by in RCA: 231] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 8. | Müller DJ, Dufrêne YF. Atomic force microscopy: a nanoscopic window on the cell surface. Trends Cell Biol. 2011;21:461-469. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 283] [Cited by in RCA: 249] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 9. | Kim KS, Cho CH, Park EK, Jung MH, Yoon KS, Park HK. AFM-detected apoptotic changes in morphology and biophysical property caused by paclitaxel in Ishikawa and HeLa cells. PLoS One. 2012;7:e30066. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 105] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 10. | Pi J, Jin H, Liu R, Song B, Wu Q, Liu L, Jiang J, Yang F, Cai H, Cai J. Pathway of cytotoxicity induced by folic acid modified selenium nanoparticles in MCF-7 cells. Appl Microbiol Biotechnol. 2013;97:1051-1062. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 65] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 11. | Ji XL, Ma YM, Yin T, Shen MS, Xu X, Guan W. Application of atomic force microscopy in blood research. World J Gastroenterol. 2005;11:1709-1711. [PubMed] |
| 12. | Sun Y, Lu XX, Liang XM, Cui XN. The impact of Cinobufacini injection on proliferation and apoptosis of human hepatoma HepG-2 cells. Zhongde Linchuang Zhongliu Zazhi. 2011;10:27-30. |
| 13. | Sun Y, Lu XX, Liang XM, Cui XN. Impact of Cinobufacini injection on proliferation and cell cycle of human hepatoma HepG-2 cells. Zhongde Linchuang Zhongliu Zazhi. 2011;10:321-324. |
| 14. | Jin H, Pi J, Huang X, Huang F, Shao W, Li S, Chen Y, Cai J. BMP2 promotes migration and invasion of breast cancer cells via cytoskeletal reorganization and adhesion decrease: an AFM investigation. Appl Microbiol Biotechnol. 2012;93:1715-1723. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 70] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 15. | Jiang J, Jin H, Liu L, Pi J, Yang F, Cai J. Curcumin disturbed cell-cycle distribution of HepG2 cells via cytoskeletal arrangement. Scanning. 2013;35:253-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 16. | Shao W, Jin H, Huang J, Qiu B, Xia R, Deng Z, Cai J, Chen Y. AFM investigation on Ox-LDL-induced changes in cell spreading and cell-surface adhesion property of endothelial cells. Scanning. 2013;35:119-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 17. | Wang J, Wan Z, Liu W, Li L, Ren L, Wang X, Sun P, Ren L, Zhao H, Tu Q. Atomic force microscope study of tumor cell membranes following treatment with anti-cancer drugs. Biosens Bioelectron. 2009;25:721-727. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 72] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 18. | Lekka M, Laidler P. Applicability of AFM in cancer detection. Nat Nanotechnol. 2009;4:72; author reply 72-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 119] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 19. | Pillet F, Chopinet L, Formosa C, Dague E. Atomic Force Microscopy and pharmacology: from microbiology to cancerology. Biochim Biophys Acta. 2014;1840:1028-1050. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 20. | Jin H, Zhong X, Wang Z, Huang X, Ye H, Ma S, Chen Y, Cai J. Sonodynamic effects of hematoporphyrin monomethyl ether on CNE-2 cells detected by atomic force microscopy. J Cell Biochem. 2011;112:169-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 39] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 21. | Le Grimellec C, Lesniewska E, Giocondi MC, Finot E, Vié V, Goudonnet JP. Imaging of the surface of living cells by low-force contact-mode atomic force microscopy. Biophys J. 1998;75:695-703. [PubMed] |
| 22. | Wang M, Ruan Y, Chen Q, Li S, Wang Q, Cai J. Curcumin induced HepG2 cell apoptosis-associated mitochondrial membrane potential and intracellular free Ca(2+) concentration. Eur J Pharmacol. 2011;650:41-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 101] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 23. | Ke C, Jin H, Cai J. AFM studied the effect of celastrol on β1 integrin-mediated HUVEC adhesion and migration. Scanning. 2013;35:316-326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 24. | Pan Y, Wu Q, Liu R, Shao M, Pi J, Zhao X, Qin L. Inhibition effects of gold nanoparticles on proliferation and migration in hepatic carcinoma-conditioned HUVECs. Bioorg Med Chem Lett. 2014;24:679-684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 25. | Pan Y, Wu Q, Qin L, Cai J, Du B. Gold nanoparticles inhibit VEGF165-induced migration and tube formation of endothelial cells via the Akt pathway. Biomed Res Int. 2014;2014:418624. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 76] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
P- Reviewer: Doganay L, Li ZF S- Editor: Qi Y L- Editor: Wang TQ E- Editor: Liu XM