Published online Aug 7, 2015. doi: 10.3748/wjg.v21.i29.8858
Peer-review started: February 23, 2015
First decision: March 26, 2015
Revised: May 1, 2015
Accepted: May 27, 2015
Article in press: May 27, 2015
Published online: August 7, 2015
Processing time: 168 Days and 16.1 Hours
AIM: To investigate pim-3 expression in hepatic stellate cells (HSCs) stimulated by lipopolysaccharide (LPS), and its protective effect on HSCs.
METHODS: Rat HSC-T6 cells were stimulated by LPS. The effect of LPS on proliferation and apoptosis of HSC-T6 cells was investigated by methyl thiazoyltetrazolium (MTT) assay and flow cytometry after annexin V-fluorescein isothiocyanate/propidium iodide double staining. pim-3 mRNA and protein were detected by reverse transcriptase polymerase chain reaction and Western blotting at 48 h when HSC-T6 cells were stimulated with 1 μg/mL LPS for 0, 3, 6, 12, 24 and 48 h. The cells without stimulation served as controls. To study the effect of pim-3 kinase on HSC-T6 cells, si-pim3 (siRNA against pim-3) was transfected into HSC-T6 cells. HSC-T6 cells were subjected to different treatments, including LPS, si-pim3, or si-pim3 plus LPS, and control cells were untreated. Protein expression of pim-3 was detected at 48 h after treatment, and cell proliferation at 24 and 48 h by MTT assay. Apoptosis was detected by flow cytometry, and confirmed with caspase-3 activity assay.
RESULTS: LPS promoted HSC-T6 cell proliferation and protected against apoptosis. Significantly delayed upregulation of pim-3 expression induced by LPS occurred at 24 and 48 h for mRNA expression (pim-3/β-actin RNA, 24 or 48 h vs 0 h, 0.81 ± 0.20 or 0.78 ± 0.21 vs 0.42 ± 0.13, P < 0.05), and occurred at 12 h and peaked at 24 and 48 h for protein expression (pim-3/GAPDH protein, 12, or 24 or 48 h vs 0 h, 0.68 ± 0.12, 1.47 ± 0.25 or 1.51 ± 0.23 vs 0.34 ± 0.04, P < 0.01). pim-3 protein was ablated by si-pim3 and upregulated by LPS in HSC-T6 cells at 48 h after treatment (pim-3/GAPDH: si-pim3, si-pim3 plus LPS or LPS vs control, 0.11 ± 0.05, 0.12 ± 0.05 or 1.08 ± 0.02 vs 0.39 ± 0.03, P < 0.01). Ablation of pim-3 by si-pim3 in HSC-T6 cells partly abolished proliferation (OD at 24 h, si-pim3 group or si-pim3 plus LPS vs control, 0.2987 ± 0.050 or 0.4063 ± 0.051 vs 0.5267 ± 0.030, P < 0.01; at 48 h 0.4634 ± 0.056 or 0.5433 ± 0.031 vs 0.8435 ± 0.028, P < 0.01; si-pim3 group vs si-pim3 plus LPS, P < 0.01 at 24 h and P < 0.05 at 48 h), and overexpression of pim-3 in the LPS group increased cell proliferation (OD: LPS vs control, at 24 h, 0.7435 ± 0.028 vs 0.5267 ± 0.030, P < 0.01; at 48 h, 1.2136 ± 0.048 vs 0.8435 ± 0.028, P < 0.01). Ablation of pim3 with si-pim3 in HSC-T6 cells aggravated apoptosis (si-pim3 or si-pim3 plus LPS vs control, 42.3% ±1.1% or 40.6% ± 1.3% vs 16.8% ± 3.3%, P < 0.01; si-pim3 vs si-pim3 plus LPS, P > 0.05), and overexpression of pim-3 in the LPS group attenuated apoptosis (LPS vs control, 7.32% ± 2.1% vs 16.8% ± 3.3%, P < 0.05). These results were confirmed by caspase-3 activity assay.
CONCLUSION: Overexpression of pim-3 plays a protective role in LPS-stimulated HSC-T6 cells.
Core tip: Hepatic stellate cell (HSC)-T6 cells stimulated by lipopolysaccharide (LPS) showed overexpression of pim-3 kinase. Overexpression of pim-3 in LPS-stimulated HSC-T6 cells protected against apoptosis and promoted proliferation. Knockdown of pim3 gene abolished proliferation of HSC-T6 cells and led to apoptosis. Overexpression of pim-3 induced by LPS play a protective role in rat hepatic stellate cells.
- Citation: Liu LH, Lai QN, Chen JY, Zhang JX, Cheng B. Overexpression of pim-3 and protective role in lipopolysaccharide-stimulated hepatic stellate cells. World J Gastroenterol 2015; 21(29): 8858-8867
- URL: https://www.wjgnet.com/1007-9327/full/v21/i29/8858.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i29.8858
Fibrosis and cirrhosis of the liver cause serious morbidity and mortality worldwide. Nearly all patients with chronic liver diseases experience liver fibrosis and some develop cirrhosis. Hepatic stellate cells (HSCs) are of pathogenetic relevance during the development, progression and regression of hepatic fibrosis. When the liver is injured, quiescent HSCs in the normal liver are activated. Activated HSCs secrete extracellular cell matrix (ECM) and inhibit ECM decomposition to promote progression of hepatic fibrosis. Promotion of apoptosis of activated HSCs may be an effective way to reverse fibrosis[1,2].
Lipopolysaccharide (LPS), which is found on the outer membrane of Gram-negative bacteria, is increased in the portal vein as the severity of hepatic fibrosis increases[3], due to increased portal vein pressure and gut permeability. LPS stimulates activity of HSCs and activates interaction of HSCs with Kupffer and endothelial cells to promote liver fibrosis and adjust portal vein pressure[4-6]. The cellular activation induced by LPS is accompanied with altered expression of several genes. Previous studies have revealed that LPS upregulates the activity of nuclear factor (NF)-κB and mitogen-activated protein kinase (MAPK) in activated HSCs[7-11], however, pim-3 kinase expression and its role in LPS-stimulated HSCs has not been reported.
pim kinase belongs to a serine/threoine protein kinase family that consists of pim-1, pim-2 and pim-3 and has been implicated in cell proliferation and apoptosis[12]. pim-3 kinase is overexpressed in both solid cancer cells and hematological malignancies, which contributes to tumor development through its anti-apoptosis and pro-proliferation functions[12]. In several normal cells, pim-3 kinase is upregulated by stress such as anoxia/reoxygenation injury, ischemia/reperfusion injury, or LPS, and protects against tissue injury[13,14]. Here, we investigated pim-3 expression and its protective role in LPS-stimulated HSCs.
HSC-T6 is an immortal rat cell line transfected with SV40 T antigen vector containing sarcoma virus promoter[15]. The cell line was a generous gift from Scott L. Friedman. LPS (Sigma, St Louis, MO, United States) was used to stimulate HSC-T6 cells. siRNA (Biomics Biotechnologies, Shanghai, China) was used to study pim-3 function in HSC-T6 cells. Total protein extraction kits were purchased from Solarbio (Beijing, China). The primary antibody to pim-3 was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, United States). Caspase-3 Activity Assay kit was from Beyotime Institute of Biotechnology (Nantong, China).
HSC-T6 cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS) at 37 °C in a humidified 5% CO2 atmosphere. The cultures were passaged after 80% confluence every 3 d.
The determination of cell proliferation was based on methyl thiazoyltetrazolium (MTT) metabolism. HSC-T6 cells were seeded into 96-well plates at 104 cells/well with 0.75% FBS for 24 h, as described previously, with some modification[16]. At the designated time, 20 μL MTT (5 mg/mL) was added to each well and the medium was removed after 4 h following addition of 150 μL DMSO to dissolve the dye for 10 min. The absorbance of each well was read at 490 nm by a spectrophotometer (Thermo Fisher Scientific, Shanghai, China). The experiments were performed in triplicate.
Apoptosis was measured using AnnexinV-fluorescein isothiocyanate (FITC)/PI apoptosis detection kit I (BD, San Jose, CA, United States). After 48 h treatment with different stimuli, the cells were digested by trypsin without EDTA and collected by centrifugation at 300 ×g for 5 min. Cells were washed twice with cooled PBS and resuspended with 100 μL binding buffer per 105 cells. Following incubation with 5 μL Annexin V-FITC and 5 μL PI solution in a dark room at room temperature for 15 min, 400 μL binding buffer was added and shaken slightly. The samples were collected and 104 cells were analyzed by a FACSCalibur flow cytometer (BD).
Protein was extracted from the treated HSC-T6 cells and the concentration was determined by BCA protein assay kits (Thermo Fisher Scientific). Caspase-3 activity was measured using the Caspase-3 Activity Assay kit (Beyotime Institute of Biotechnology). Cell extracts were mixed with Ac-DEVD-pNA substrate for 2 h at 37 °C in 96-well plates prior to colorimetric measurement of p-nitroanilide product at 405 nm.
HSC-T6 cells were seeded in six-well plates. After culturing for 12 h, cells received different treatments following serum starvation with 0.75% FBS. After treatment, total RNA was extracted from the cells using an RNA simple Total RNA kit (Tiangen, Beijing, China). The first strand of cDNA was synthesized with the reverse-transcript kit (Takara, Dalian, China). The following primers were used: (1)pim-3: forward: 5’-CACTGACTTTGATGGCACCC-3’ reverse: 5’-ATGCCCAGACGAAGACCA-3’(product of 770bp) (2) β-actin: forward: TCAGGTCATCACTATCGGCAAT reverse: AAAGAAAGGGTGTAAAACGCA (product of 432 bp). PCR was performed in a 25-μL reaction mixture containing 1 μL cDNA, 0.5 μL each primer, 0.25 μL rTaq DNA polymerase, and 2.0 μL dNTP. The PCR was performed with the following thermal cycling conditions: (1) denaturation at 95 °C for 5 min; (2) 35 cycles of denaturation at 94 °C for 45 s; and (3) primer annealing at 55 °C for 45 s and primer extension at 72 °C for 60 s, with a final extension at 72 °C for 10 min. The PCR products were electrophoresed in a 1.5% agarose gel containing ethidium bromide and visualized with UV light. The bands in the gels were quantified with Quantity one 4.62 and the level of a particular cDNA was normalized to that of β-actin product.
HSC-T6 cells were seeded in six-well plates. After culturing for 12 h, cells received different treatments following serum starvation with 0.75% FBS. After treatment, the cell pellets were collected, washed three times with ice-cold PBS, and resuspended in lysis buffer to extract protein. Protein concentration was determined by BCA protein assay kits (Thermo Fisher Scientific). The protein solution was heat-denatured with an equal volume of 2 × SDS loading buffer for 5 min and separated on 12% SDS-PAGE. The protein was then electro-transferred onto PVDF membranes. After blocking with 5% skimmed milk in PBS at 4 °C overnight, the membrane was incubated with each primary antibody, followed by incubation with a horseradish-peroxidase-conjugated secondary antibody. The membrane was then exposed to X-ray film and the quantification of the bands was carried out by Quantity one 4.62. GAPDH was used as an internal control for loading.
HSC-T6 cells were subjected to LPS (Escherichia coli 055:B5) treatment at different concentrations (10 ng/mL, 100 ng/mL, 1 μg/mL or 5 μg/mL) for 24 or 48 h following starvation with 0.75% FBS. MTT assay was conducted to achieve the optimum concentration of LPS for promotion of HSC-T6 cell proliferation. Apoptosis was detected by flow cytometry at 48 h after treatment with 1 μg/mL LPS. Reverse transcriptase polymerase chain reaction (RT-PCR) and western blotting were performed to detect pim-3 expression at 48 h when HSC-T6 cells were stimulated with 1 μg/mL LPS by different time-course (0 , 3 , 6 , 12 , 24 and 48 h). The cells without stimulation served as controls.
Short interfering RNA (siRNA) was synthesized by Biomics Biotechnologies. siRNA duplexes were designed to target AA(19)UU sequences in the open reading frame of mRNA encoding pim-3. Three siRNA against pim-3 (si-pim3) and one scrambled siRNA were transiently transfected into HSC-T6 cells with Lipofectamine 2000 transfection regent (Invitrogen, Carlsbad, CA, United States). One day before transfection, HSC-T6 cells were cultured in DMEM with no antibiotics, then in medium with serum-free complexes containing siRNA and Lipofectamine 2000 (20 pmol siRNA to 1 μL Lipofectamine 2000) for 6 h, followed by DMEM with 10% FBS. Forty-eight hours later, the cells were harvested and lysed and pim-3 mRNA and protein expression was detected by RT-PCR and western blotting to select the perfect siRNA duplex. The selected siRNA duplex (sense chain: 5’-UUCUCCGAACGUGUCACGUdTdT-3’ antisense chain 5’-ACGUGACACGUUCGGAGAAdTdT-3’) and the scrambled siRNA duplex (sense chain: 5’-UUCUCCGAACGUGUCACGUdTdT-3’, antisense chain: 5’-ACGUGACACGUUCGGAGAAdTdT-3’) was further blasted to search against another rat genome sequence to ensure its targets specificity. Experiments were divided into the following groups. Control group: HSC-T6 cells incubated without treatment. Liposome group: HSC-T6 cells incubated with equivalent liposome. Scramble group: scrambled RNA transfected into HSC-T6 cells. si-pim3 group: si-pim3 was transfected into HSC-T6 cells. LPS group: HSC-T6 cells treated with 1 μg/mL LPS. si-pim3 plus LPS group: si-pim3 was transfected into HSC-T6 cells, then treated with 1 μg/mL LPS. HSC-T6 cells were harvested at the designated time and cell proliferation (at 24 or 48 h after treatment), protein expression (at 48 h), and apoptosis (at 48 h) were determined. Each experiment was repeated three times.
Values were expressed at mean ± SD from duplicate samples. The difference in the means between two groups was tested by the Students’t test (two tailed), and that between the groups (above three groups) was tested by one-way ANOVA followed by Student-Newman-Keul test; P < 0.05 was considered statistically significant.
Proliferation of HSC-T6 cells treated with LPS at 10 ng/mL, 100 ng/mL, 1 μg/mL and 5 μg/mL was 1.19-, 1.19-, 1.39- and 1.31-fold that of the control group at 24 h after treatment; and 1.10-, 1.25-, 1.41- and 1.37-fold that at 48 h after treatment (Table 1). Proliferation with 1 μg/mL LPS was the highest. Our results show that LPS promotes HSC-T6 cell proliferation and the optimum concentration is 1 μg/mL.
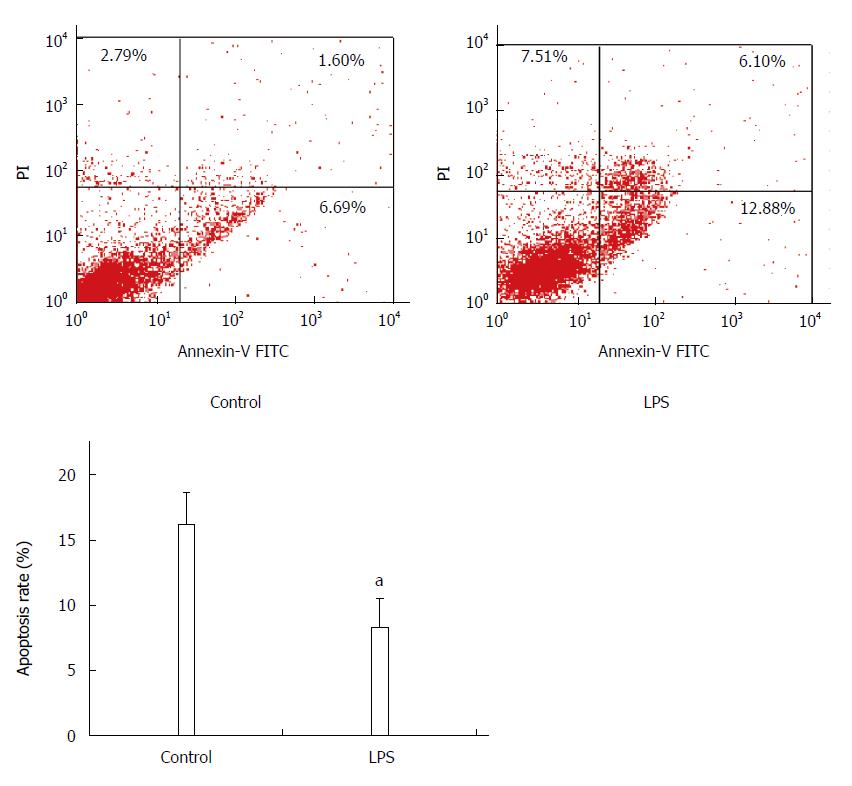
Apoptosis was detected by flow cytometry following Annexin-FITC/PI double staining, and FITC+PI- spots represented early apoptosis, while FITC+PI+ spots represented late apoptosis. Total apoptosis rate of HSC-T6 cells treated with LPS was lower than that of the control group (control vs LPS, 16.3% ± 2.4% vs 8.3% ± 2.3%, P < 0.05), which suggests that LPS has an inhibitory effect on HSC-T6 apoptosis (Figure 1).
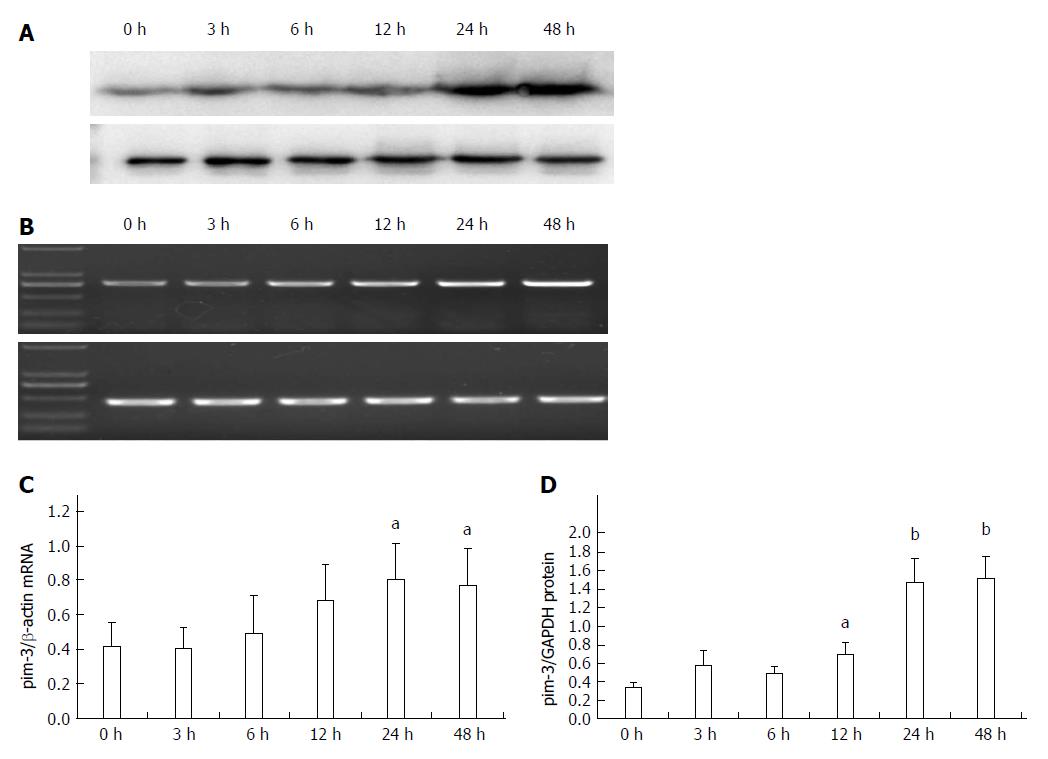
Expression of pim-3 mRNA in LPS-stimulated HSC-T6 cells at 3, 6, 12, 24 and 48 h after treatment was 0.95-, 1.28-, 1.63-, 1.94- and 1.84-fold that of the control group, respectively (Figure 2). Expression was significantly increased at 24 and 48 h after treatment (P < 0.05, compared to that at 0 and 3 h). Expression of pim-3 protein in LPS-treated cells at 3, 6, 12, 24 and 48 h after treatment was 1.67-, 1.42-, 2.01-, 4.30- and 4.42-fold that of the control group, respectively. Expression increased significantly at 12 h (P < 0.05, compared with 0 h), reached a plateau at 24 h, and was sustained at a high level at 48 h (P < 0.01, compared with 0 h). The results clearly demonstrated that LPS significantly increases expression of pim-3 in HSC-T6 cells at transcriptional and translational levels and delays overexpression of pim-3.
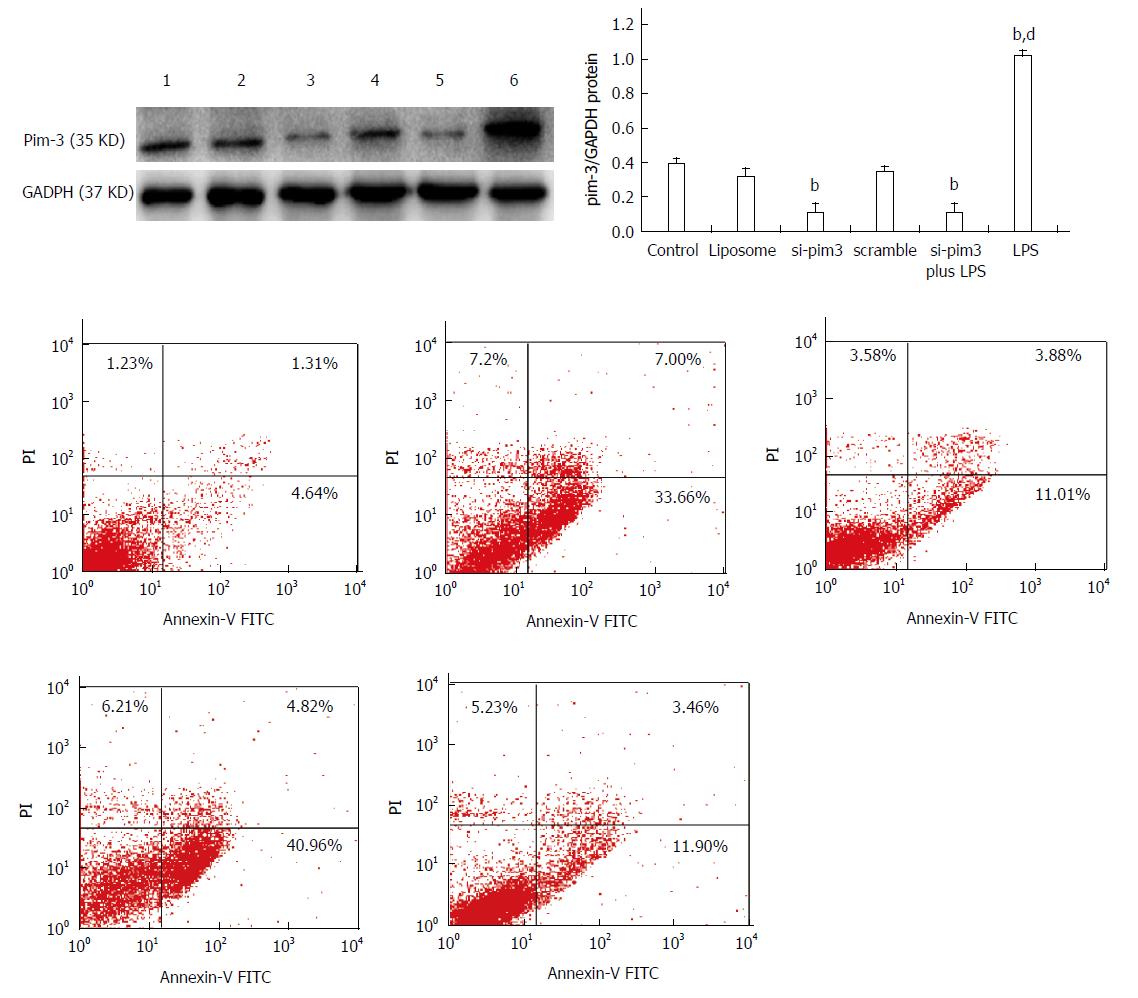
In order to clarify the role of endogenous pim-3 in HSC-T6 cells, we tested cell proliferation and apoptosis under the condition of pim-3 ablation by RNA interference (RNAi) and overexpression of pim-3 induced by LPS. Three si-pim3 duplexes were used and pim-3 mRNA and protein levels were measured in HSC-T6 cells at 48 h after si-pim3 transfection. si-pim3 was selected depending on the experiment for its strongest inhibition. The selected si-pim3 duplex (sense chain: 5’-UUCUCCGAACGUGUCACGUdTdT-3’ antisense chain 5’-ACGUGACACGUUCGGAGAAdTdT-3’) could reduce about 70% expression of pim-3 protein. pim-3 expression was confirmed by western blotting, and pim-3 protein was ablated by si-pim3 and upregulated by LPS at 48 h after treatment (Figure 3) (pim-3/GAPDH: si-pim3 or si-pim3 plus LPS or LPS vs control, 0.11 ± 0.05 or 0.12 ± 0.05 or 1.08 ± 0.02 vs 0.39 ± 0.03, P < 0.01).
To determine the effect of pim-3 kinase on HSC-T6 cell proliferation, we compared proliferation among different groups treated with si-pim3 transfection (ablation of pim-3), LPS (overexpression of pim-3), or LPS combined si-pim3 transfection (ablation of pim-3). HSC-T6 cell proliferation in the si-pim3 group and si-pim3 plus LPS group was significantly decreased, compared with the control group (OD: si-pim3 or si-pim3 plus LPS vs control, at 24 h, 0.2987 ± 0.050 or 0.4063 ± 0.051 vs 0.5267 ± 0.030, P < 0.05; at 48 h, 0.4634 ± 0.056 or 0.5434 ± 0.031 vs 0.8435 ± 0.028, P < 0.05), whereas proliferation in the LPS group was significantly increased, compared with the control group (OD: LPS vs control, at 24 h, 0.7435 ± 0.028 vs 0.5267 ± 0.030, P < 0.01; at 48 h, 1.2136 ± 0.048 vs 0.8435 ± 0.028, P < 0.01) (Table 2). These results indicated that ablation of pim-3 in the si-pim3 group inhibited HSC-T6 cell proliferation, and overexpression of pim-3 in the LPS group promoted HSC-T6 proliferation, which suggests that endogenous pim-3 has potential pro-proliferative activity in HSC-T6 cells. HSC-T6 cell proliferation in the si-pim3 plus LPS group was significantly increased, compared with the si-pim3 group (P < 0.05), however, they had similar expression of pim-3, which means that LPS has another approach to promote cell proliferation independent of pim-3 kinase.
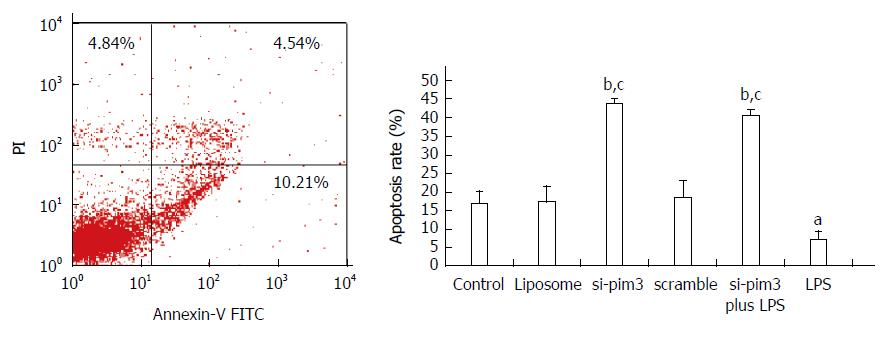
The apoptosis rate of HSC-T6 cells in the LPS group with overexpression of pim-3 was significantly lower than that of the control group (LPS vs control, 7.32% ± 2.1% vs 16.8% ± 3.3%, P < 0.05) (Figure 4). In contrast, the apoptosis rate of the si-pim3 group with ablation of pim-3 was remarkably higher than that of the control group (si-pim3 vs control, 42.3% ± 1.1% vs 16.8% ± 3.3%, P < 0.01), but similar to that of the si-pim3 plus LPS group (si-pim3 vs si-pim3 plus LPS, 42.3% ± 1.1% vs 40.6% ± 1.3%, P > 0.05). These results demonstrated that ablation of pim-3 can aggregate HSC-T6 cell apoptosis and endogenous pim-3 kinase may protect HSC-T6 cells from apoptosis.
Caspase-3 is a cell death executioner, which can be activated by various apoptosis signals. To confirm the results from flow cytometry following Annexin V-FITC/PI staining, we quantified the activity of caspase-3. Caspase-3 activity in the LPS group was lower than in the control group (Table 3). In contrast, the activity in the si-pim3 group was significantly higher than in the control group (P < 0.05), and similar to that in the si-pim3 plus LPS group. All the results were consistent with those from flow cytometry following Annexin-FITC/PI double staining.
To the best of our knowledge, the present work is the first report about the expression of pim-3, as well as its role in HSCs treated with LPS. In this study, HSC-T6 cells stimulated by LPS showed overexpression of pim-3 kinase. Overexpression of pim-3 in LPS-stimulated HSC-T6 cells protected against apoptosis and promoted proliferation, however, knockdown of pim3 gene by si-pim3 abolished proliferation of HSC-T6 cells and led to apoptosis. These results suggest that overexpression of pim-3 induced by LPS has a protective role in rat HSCs.
Endotoxin in the portal vein and circulating blood is increased by aggravation of chronic liver disease and hepatic fibrosis[3], and contributes to hepatic fibrosis[5,17]. LPS could indirectly activate HSCs and protect against apoptosis by soluble mediators from Kupffer cells[18-20], or apoptotic bodies from damaged hepatocytes[21,22]. Activated HSCs have several phenotypes, including proliferation, fibrogenesis, contractility, inflammatory signaling, and chemotaxis[23]. LPS also directly activates HSCs, and the cells show the inflammatory phenotype and secrete cytokines and chemokines[7,24]. Here, we studied the effect of LPS on proliferation and apoptosis of HSCs. The proliferation of primary activated HSCs in response to LPS, assessed by [3H]-thymidine incorporation, is unchanged[7], while that from HSC lines assessed by MTT is increased[25]. The difference may be due to differently derived cells and detection assays. In our study, LPS has a beneficial effect on proliferation of HSC-T6 cells and protects cells from apoptosis.
Antibiotic use is a way to eliminate gut-derived endotoxin, and animal experiments have demonstrated its efficacy in hepatic fibrosis[5]. However, long-term use of antibiotics increases the possibility of drug resistance, and presently no antibiotics are approved for use against hepatic fibrosis. Thus, it may be necessary to explore alterations in survival gene expression in LPS-treated HSCs, to help find new ways to avoid the effect of LPS on hepatic fibrosis. Previous studies have demonstrated that LPS upregulates secretion of tissue inhibitor of metalloproteinase (TIMP-)1[9] (necessary for prevention of HSC apoptosis[26]) and interleukin-6 (necessary for HSC trans-differentiation[27]), and upregulates expression of intracellular survival signal molecules, such as NF-κB, extracellular signal-regulated kinase (ERK) and C-Jun N-terminal kinase[11,16].
pim kinase is one of the serine/threoine protein kinase family, which at least includes pim-1, pim-2 and pim-3. The three members are well conserved in vertebrates and show structural similarity and functional overlap. They all have a role in promoting cell growth and inhibiting apoptosis. Many tumor cells overexpress pim-3, such as solid cancers and hematological malignancies[12]. Normal cells induced by special stimuli can also upregulate expression of pim-3 kinase, such as cardiomyocytes with anoxia/reoxgenation injury[13] , endothelial cell with tumor necrosis factor-α[28], and intestinal mucosa with LPS[14]. Our study clearly demonstrated that pim-3 expression is upregulated in HSCs treated with LPS. Unlike other survival signal molecules such as NF-κB and MAPK, which are characteristic of rapid and transient upregulation in LPS-stimulated HSCs[11], pim-3 expression shows late and persistent upregulation, which suggests that overexpression of pim-3 depends on other upstream signaling molecules. pim-3 kinase is involved in accelerating the cell cycle and protecting against apoptosis. pim-3 kinase can phosphorylate p27kipl, inducing 14-3-3 binding and proteasome-dependent degradation, thus relieving the inhibition of the cell cycle and promoting proliferation[29]. pim-3 also phosphorylates BCL-xL/BCL-2-associated death promoter (BAD), inducing 14-3-3 binding and degradation, leading to release of Bcl-XL and Bcl-2[30,31], thus promoting cell survival[32]. Meanwhile, pim-3 can phosphorylate signal transducer and activator of transcription (STAT)3[33], promote antiapoptotic protein synthesis of Bcl-XL and survivin, leading to cell survival. pim-3 kinase is constitutively active and does not require post-translational modifications for the induction of kinase activity. Ablation of pim-3 by RNAi was used to explore its function in HSCs treated with LPS. Our results show that endogenous pim-3 could play a protective role in LPS-stimulated HSC-T6 cells, according to the results from ablation of pim-3 by si-pim3 and overexpression of pim-3 induced by LPS. The apoptosis rate of si-pim3-treated cells was similar to that of cells treated by si-pim3 plus LPS, which means that the antiapoptotic effect of LPS is completely inhibited by si-pim3. However, proliferation of si-pim3-treated cells was lower than that of the cells treated by si-pim3 plus LPS, although they had similar expression of pim-3, which suggests that cell proliferation induced by LPS has another mechanism independent of pim-3. LPS upregulates pim3 kinase and stimulates several other survival kinases, such as NF-κB and ERK. The results suggest that pim-3 kinase is downstream of other survival genes that are dependent on LPS, which coincides with the timing of pim-3 expression. Further study is needed to ascertain the relationship of pim-3 and other survival kinases.
In conclusion, the present study provides evidence that LPS can upregulate pim-3 expression in activated HSCs, and pim-3 expression can promote cell proliferation and inhibit apoptosis. Thus, pim-3 kinase may be an antifibrotic candidate target and blocker of pim-3 kinase, and an effective way to reverse liver fibrosis.
We appreciate the assistance from the staff in Jiangxi Provincial Key Laboratory of Molecular Medicine, China.
pim-3 kinase is implicated in cell proliferation and anti-apoptosis. Upregulation of pim-3 expression often occurs in cancer and normal cells upon stress. Hepatic stellate cells (HSCs) are often challenged with gut-derived endotoxin.
pim-3 kinase often plays a protective role when cells, such as cardiomyocytes, are challenged with anoxia or ischemic injury, or intestinal mucosa with lipopolysaccharide (LPS). Promotion of HSC apoptosis may be an effective way to reverse fibrosis. However, there is no research about pim-3 expression in HSCs induced by LPS, and its role in activated HSCs when challenged with LPS.
This is the first report about the expression of pim-3 as well as its role in HSCs treated with LPS. We demonstrated that overexpression of pim-3 is induced by LPS and plays a protective role in rat HSCs.
This study provided us with a new candidate target to relieve hepatic fibrosis.
This study is meaningful and the findings that pim-3 might be involved in LPS-stimulated hepatic stellate cells are interesting.
| 1. | Elsharkawy AM, Oakley F, Mann DA. The role and regulation of hepatic stellate cell apoptosis in reversal of liver fibrosis. Apoptosis. 2005;10:927-939. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 309] [Cited by in RCA: 327] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 2. | Kong D, Zhang F, Zhang Z, Lu Y, Zheng S. Clearance of activated stellate cells for hepatic fibrosis regression: molecular basis and translational potential. Biomed Pharmacother. 2013;67:246-250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 40] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 3. | Lin RS, Lee FY, Lee SD, Tsai YT, Lin HC, Lu RH, Hsu WC, Huang CC, Wang SS, Lo KJ. Endotoxemia in patients with chronic liver diseases: relationship to severity of liver diseases, presence of esophageal varices, and hyperdynamic circulation. J Hepatol. 1995;22:165-172. [PubMed] |
| 4. | Pradere JP, Troeger JS, Dapito DH, Mencin AA, Schwabe RF. Toll-like receptor 4 and hepatic fibrogenesis. Semin Liver Dis. 2010;30:232-244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 123] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 5. | Zhu Q, Zou L, Jagavelu K, Simonetto DA, Huebert RC, Jiang ZD, DuPont HL, Shah VH. Intestinal decontamination inhibits TLR4 dependent fibronectin-mediated cross-talk between stellate cells and endothelial cells in liver fibrosis in mice. J Hepatol. 2012;56:893-899. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 123] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 6. | Seki E, De Minicis S, Osterreicher CH, Kluwe J, Osawa Y, Brenner DA, Schwabe RF. TLR4 enhances TGF-beta signaling and hepatic fibrosis. Nat Med. 2007;13:1324-1332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1361] [Cited by in RCA: 1604] [Article Influence: 84.4] [Reference Citation Analysis (1)] |
| 7. | Brun P, Castagliuolo I, Pinzani M, Palù G, Martines D. Exposure to bacterial cell wall products triggers an inflammatory phenotype in hepatic stellate cells. Am J Physiol Gastrointest Liver Physiol. 2005;289:G571-G578. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 115] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 8. | Guo J, Loke J, Zheng F, Hong F, Yea S, Fukata M, Tarocchi M, Abar OT, Huang H, Sninsky JJ. Functional linkage of cirrhosis-predictive single nucleotide polymorphisms of Toll-like receptor 4 to hepatic stellate cell responses. Hepatology. 2009;49:960-968. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 175] [Cited by in RCA: 184] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 9. | De Minicis S, Seki E, Uchinami H, Kluwe J, Zhang Y, Brenner DA, Schwabe RF. Gene expression profiles during hepatic stellate cell activation in culture and in vivo. Gastroenterology. 2007;132:1937-1946. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 328] [Cited by in RCA: 364] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 10. | Paik YH, Schwabe RF, Bataller R, Russo MP, Jobin C, Brenner DA. Toll-like receptor 4 mediates inflammatory signaling by bacterial lipopolysaccharide in human hepatic stellate cells. Hepatology. 2003;37:1043-1055. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 492] [Cited by in RCA: 513] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 11. | Thirunavukkarasu C, Watkins SC, Gandhi CR. Mechanisms of endotoxin-induced NO, IL-6, and TNF-alpha production in activated rat hepatic stellate cells: role of p38 MAPK. Hepatology. 2006;44:389-398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 83] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 12. | Narlik-Grassow M, Blanco-Aparicio C, Carnero A. The PIM family of serine/threonine kinases in cancer. Med Res Rev. 2014;34:136-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 186] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 13. | Liu D, He M, Yi B, Guo WH, Que AL, Zhang JX. Pim-3 protects against cardiomyocyte apoptosis in anoxia/reoxygenation injury via p38-mediated signal pathway. Int J Biochem Cell Biol. 2009;41:2315-2322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 14. | Chen JY, Shen XW, Zhang JX. [Protective role of Pim-3 gene in intestinal mucosa damaged by burn or lipopolysaccharide]. Zhonghua Yixue Zazhi. 2007;87:2960-2964. [PubMed] |
| 15. | Vogel S, Piantedosi R, Frank J, Lalazar A, Rockey DC, Friedman SL, Blaner WS. An immortalized rat liver stellate cell line (HSC-T6): a new cell model for the study of retinoid metabolism in vitro. J Lipid Res. 2000;41:882-893. [PubMed] |
| 16. | Liu YW, Huang YT. Inhibitory effect of tanshinone IIA on rat hepatic stellate cells. PLoS One. 2014;9:e103229. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 28] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 17. | Isayama F, Hines IN, Kremer M, Milton RJ, Byrd CL, Perry AW, McKim SE, Parsons C, Rippe RA, Wheeler MD. LPS signaling enhances hepatic fibrogenesis caused by experimental cholestasis in mice. Am J Physiol Gastrointest Liver Physiol. 2006;290:G1318-G1328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 105] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 18. | Zhang X, Yu WP, Gao L, Wei KB, Ju JL, Xu JZ. Effects of lipopolysaccharides stimulated Kupffer cells on activation of rat hepatic stellate cells. World J Gastroenterol. 2004;10:610-613. [PubMed] |
| 19. | Tomita K, Tamiya G, Ando S, Ohsumi K, Chiyo T, Mizutani A, Kitamura N, Toda K, Kaneko T, Horie Y. Tumour necrosis factor alpha signalling through activation of Kupffer cells plays an essential role in liver fibrosis of non-alcoholic steatohepatitis in mice. Gut. 2006;55:415-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 318] [Cited by in RCA: 358] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 20. | Saile B, Matthes N, Knittel T, Ramadori G. Transforming growth factor beta and tumor necrosis factor alpha inhibit both apoptosis and proliferation of activated rat hepatic stellate cells. Hepatology. 1999;30:196-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 126] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 21. | Jiang JX, Mikami K, Venugopal S, Li Y, Török NJ. Apoptotic body engulfment by hepatic stellate cells promotes their survival by the JAK/STAT and Akt/NF-kappaB-dependent pathways. J Hepatol. 2009;51:139-148. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 135] [Cited by in RCA: 134] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 22. | Wang K, Lin B, Brems JJ, Gamelli RL. Hepatic apoptosis can modulate liver fibrosis through TIMP1 pathway. Apoptosis. 2013;18:566-577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 51] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 23. | Friedman SL. Mechanisms of hepatic fibrogenesis. Gastroenterology. 2008;134:1655-1669. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2139] [Cited by in RCA: 2191] [Article Influence: 121.7] [Reference Citation Analysis (1)] |
| 24. | Harvey SA, Dangi A, Tandon A, Gandhi CR. The transcriptomic response of rat hepatic stellate cells to endotoxin: implications for hepatic inflammation and immune regulation. PLoS One. 2013;8:e82159. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 36] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 25. | Bai T, Lian LH, Wu YL, Wan Y, Nan JX. Thymoquinone attenuates liver fibrosis via PI3K and TLR4 signaling pathways in activated hepatic stellate cells. Int Immunopharmacol. 2013;15:275-281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 87] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 26. | Murphy FR, Issa R, Zhou X, Ratnarajah S, Nagase H, Arthur MJ, Benyon C, Iredale JP. Inhibition of apoptosis of activated hepatic stellate cells by tissue inhibitor of metalloproteinase-1 is mediated via effects on matrix metalloproteinase inhibition: implications for reversibility of liver fibrosis. J Biol Chem. 2002;277:11069-11076. [RCA] [DOI] [Full Text] [Cited by in Crossref: 328] [Cited by in RCA: 347] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 27. | Lakner AM, Moore CC, Gulledge AA, Schrum LW. Daily genetic profiling indicates JAK/STAT signaling promotes early hepatic stellate cell transdifferentiation. World J Gastroenterol. 2010;16:5047-5056. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 34] [Cited by in RCA: 39] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 28. | Yang H, Wang Y, Qian H, Zhang P, Huang C. Pim protein kinase-3 is regulated by TNF-α and promotes endothelial cell sprouting. Mol Cells. 2011;32:235-241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 29. | Morishita D, Katayama R, Sekimizu K, Tsuruo T, Fujita N. Pim kinases promote cell cycle progression by phosphorylating and down-regulating p27Kip1 at the transcriptional and posttranscriptional levels. Cancer Res. 2008;68:5076-5085. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 243] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 30. | Popivanova BK, Li YY, Zheng H, Omura K, Fujii C, Tsuneyama K, Mukaida N. Proto-oncogene, Pim-3 with serine/threonine kinase activity, is aberrantly expressed in human colon cancer cells and can prevent Bad-mediated apoptosis. Cancer Sci. 2007;98:321-328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 106] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 31. | Li YY, Popivanova BK, Nagai Y, Ishikura H, Fujii C, Mukaida N. Pim-3, a proto-oncogene with serine/threonine kinase activity, is aberrantly expressed in human pancreatic cancer and phosphorylates bad to block bad-mediated apoptosis in human pancreatic cancer cell lines. Cancer Res. 2006;66:6741-6747. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 147] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 32. | Nawijn MC, Alendar A, Berns A. For better or for worse: the role of Pim oncogenes in tumorigenesis. Nat Rev Cancer. 2011;11:23-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 360] [Cited by in RCA: 398] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 33. | Chang M, Kanwar N, Feng E, Siu A, Liu X, Ma D, Jongstra J. PIM kinase inhibitors downregulate STAT3(Tyr705) phosphorylation. Mol Cancer Ther. 2010;9:2478-2487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 47] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
P- Reviewer: Al-Gayyar MMH, Chen CJ, Zhang YP S- Editor: Ma YJ L- Editor: Kerr C E- Editor: Liu XM