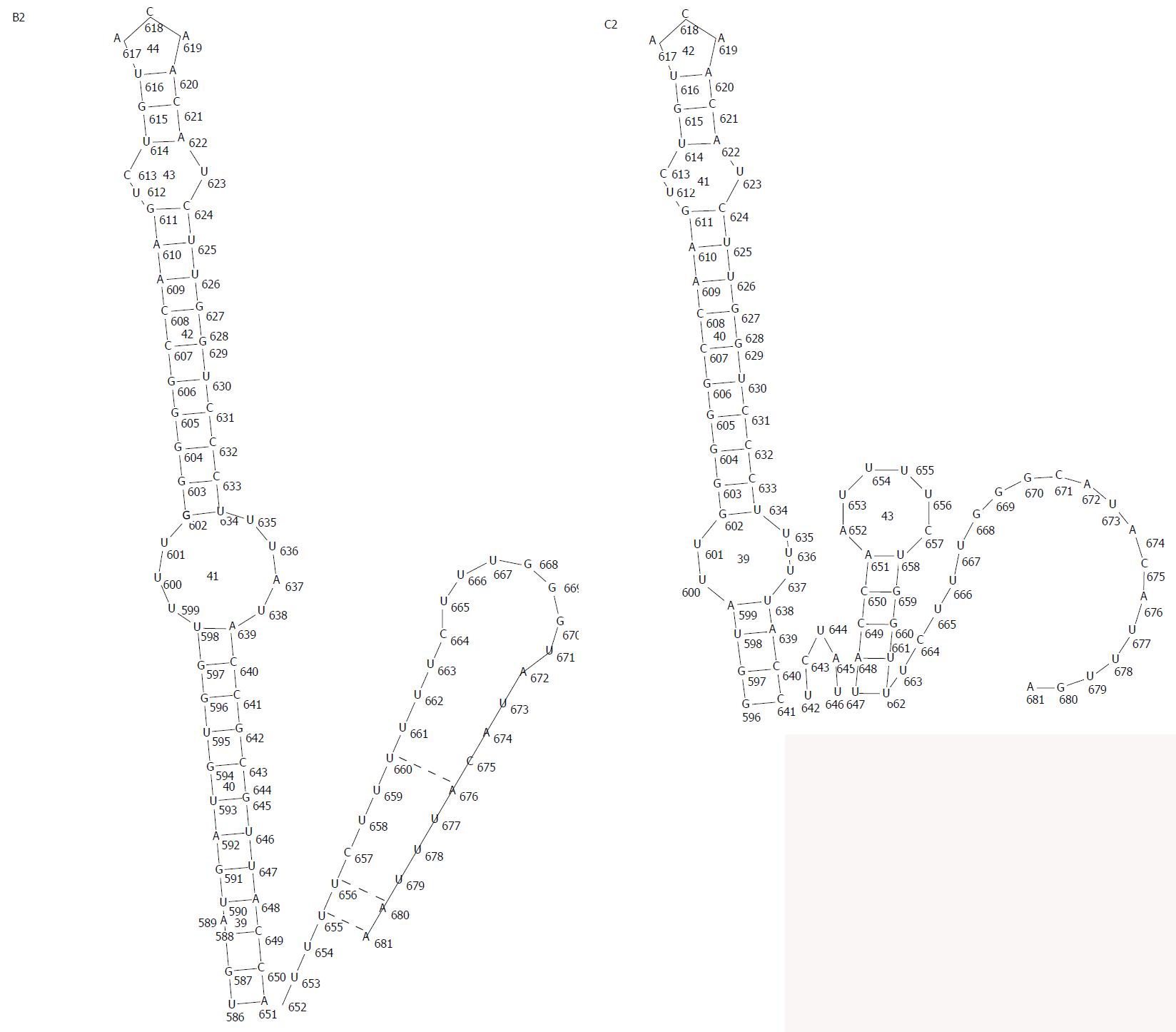Published online Jun 7, 2015. doi: 10.3748/wjg.v21.i21.6684
Peer-review started: December 26, 2014
First decision: January 22, 2015
Revised: March 3, 2015
Accepted: April 3, 2015
Article in press: April 3, 2015
Published online: June 7, 2015
Processing time: 168 Days and 18.5 Hours
AIM: To analyze the hepatitis B virus (HBV) characters in China, as well as the correlation between several HBV mutation and hepatitis symptoms.
METHODS: A total of 1148 HBV genome sequences from patients throughout China were collected via the National Center For Biotechnology Information database (information including: genotype, territory and clinical status). HBV genotypes were classified by a direct reference from the Genbank sequence annotation, phylogenetic tree and online software analysis (http://www.ncbi.nlm.nih.gov/projects/genotyping/formpage.cgi). The phylogenetic tree was constructed based on the neighbor-joining method by MEGA5.0 software. HBV sequences were grouped based on phylogenetic tree and the distance between the groups was calculated by using the computer between group mean distance methods. Seven hundred and twelve HBV sequences with clear annotation of clinical symptoms were selected to analyses the correlation of mutation and clinical symptoms. Characteristics of sequences were analyzed by using DNAStar and BioEdit software packages. The codon usage bias and RNA secondary structures analysis were performed by RNAdraw software. Recombination analysis was performed by using Simplot software.
RESULTS: In China, HBV genotype C was the predominant in Northeastern, genotype B was predominant in Central Southern areas, genotype B and C were both dominant in Southwestern areas, and the recombinant genotype C/D was predominant in Northwestern areas. C2 and B2 were identified as the two major sub-genotypes, FJ386674 might be a putative sub-genotype as B10. The basal core promoter double mutation and pre-C mutation showed various significant differences between hepatitis symptoms. In addition to ATG, many other HBV initiation codons also exist. HBV has codon usage bias; the termination codon of X, C and P open reading frames (ORF) were TAA, TAG, and TGA, respectively. The major stop codons of S-ORF were TAA (96.45%) and TGA (83.60%) in B2 and C2 subtype, respectively.
CONCLUSION: This study recapitulated the epidemiology of HBV in China, and the information might be meaningful critical for the future prevention and therapy of HBV infections.
Core tip: This study recapitulated the epidemiology of hepatitis B virus (HBV) in China. Genotype C was the predominant HBV genotype in Northeastern, genotype B was predominant in Central Southern areas, genotype B and C were both dominant in Southwestern areas, and the recombinant genotype C/D was predominant in Northwestern areas. C2 and B2 were identified as the two major subgenotypes, FJ386674 might be a putative sub-genotype as B10. Moreover, the termination codon usage bias of B2 (TAA) and C2 (TGA) subtype and the correlation between HBV sequence mutations and clinical symptoms were also determined.
- Citation: Li HM, Wang JQ, Wang R, Zhao Q, Li L, Zhang JP, Shen T. Hepatitis B virus genotypes and genome characteristics in China. World J Gastroenterol 2015; 21(21): 6684-6697
- URL: https://www.wjgnet.com/1007-9327/full/v21/i21/6684.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i21.6684
Hepatitis B virus (HBV) belongs to the family Hepadnaviridae, it is an enveloped virus with a circular, partially double-stranded DNA genome of 3.2 kb. It contains four partially overlapping open-reading frames (ORF) preS1/S2/S, preC/C, P and X. HBV infection can induce diseases, such as acute hepatitis, chronic hepatitis, hepatocirrhosis, and hepatocellular carcinoma (HCC), and thus severely threats global human health. HBV is one of the most successful human pathogens, with an estimated 2 billion people have serological evidence of past or present infected with HBV worldwide, of whom 250 million have chronic hepatitis B (CHB) infection[1]. More than 75% of patients with hepatitis B virus live in the western Pacific and Southeast Asia[2]. China is a country that has a high incidence of HBV, with more than 120 million hepatitis B patients. Approximately 15%-40% of the hepatitis B virus carriers eventually developed HBV-related cirrhosis or HCC[3]. Each year about 600000 people die from liver disease caused by HBV infection[4].
Okamoto et al[5] firstly proposed the concept of HBV genotypes in 1988 and assigned each newly identified genotype based on the criterion of ≥ 8% of the whole HBV genome difference. HBV genotype A has been shown to be primarily distributed in Northern Europe and Africa; genotype B and C in Southeastern Asia; genotype D in the Middle East, North Africa, and Europe. With technological development, more HBV genotypes have been found, genotype E in Africa; genotype F in South America; genotype G in United States and France; and genotype H in Europe and North America[6]. Recently, genotype I and J were reported in Vietnam and in Japan[7,8], respectively, making a total genotype count to date of 10. The distribution of these HBV genotypes has obviously geographic-associated features[9]. Previous studies have indicated that HBV genotypes might be associated with serotype, liver disease progression and mutations in the BCP and pre-C region[10]. Therefore, in this study, the distribution of genotypes and subgenotypes in China were analyzed firstly. Then, the characters of HBV subgenotype B2 and C2 were analyzed. Finally, the correlation between BCP double mutation/pre-C mutation and clinical symptoms was also determined.
A total of 1148 HBV genome sequences from patients throughout China were collected via the NCBI database (information including: genotype, territory and clinical status). Sequences were divided into 6 groups based on the administrative territory of Chinese mainland. The reference sequences used in this study were listed[11-24].
HBV genotypes were classified by a direct reference from the Genbank sequence annotation, phylogenetic tree and online software analysis (http: //http://www.ncbi.nlm.nih.gov/projects/genotyping/formpage.cgi)[25,26].
Phylogenetic tree analysis was performed by MEGA5.0 software. Three reference sequences of each HBV genotype and one to five reference sequences of the B, C and I sub-genotypes were selected based on the previous reports[27,28]. The phylogenetic tree was constructed based on the neighbor-joining method. HBV sequences were grouped based on phylogenetic tree. The distance between the groups was then calculated by using the computer between-group mean distance methods. Sub-genotype clustering was accomplished based on both the phylogenetic tree and the distances between the groups. Characteristics of sequences were analyzed by using DNAStar and BioEdit software packages[29]. The codon usage bias and RNA secondary structures analysis were performed by RNAdraw software[30]. Recombination analysis was performed by using Simplot software[31].
The HBV-associated clinical symptoms of 1148 sequences included asymptomatic (ASC), CHB, acute-on-chronic liver failure (ACLF), acute hepatitis B (AHB), liver cirrhosis (LC), HCC and HBsAg positive (HBsAg+). Incomplete sequences (< 3215 bp) and clinical symptoms with only HBsAg positive were excluded, while 712 HBV sequences were selected for further analysis of the correlation between BCP double mutation/pre-C mutation and clinical symptoms.
Statistical analysis and plotting of the data were accomplished by Excel software. SPSS software was used for significance analysis t-test. P < 0.05 was regarded as statistically significance.
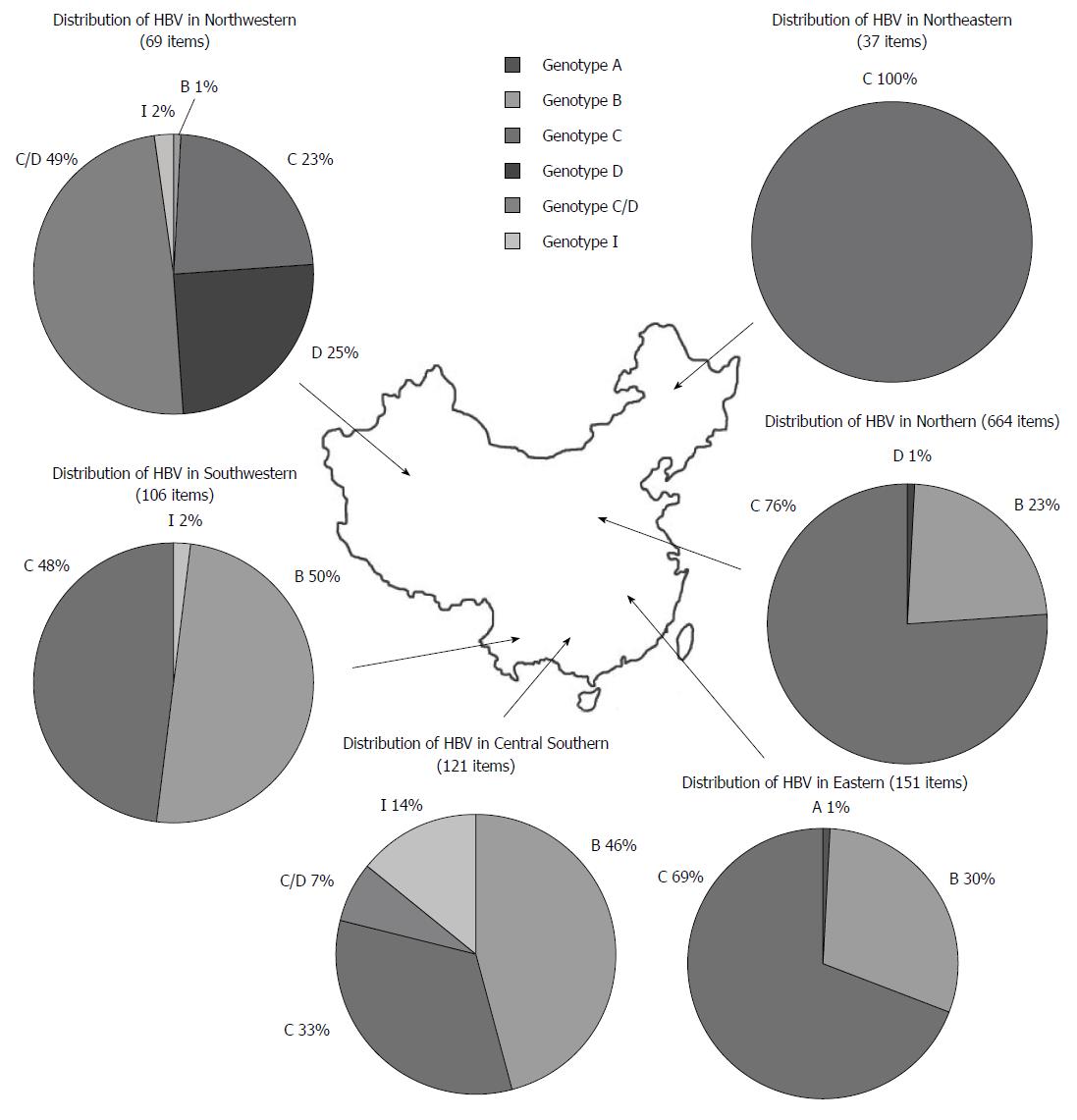
In this study, a total of 1148 HBV sequences were analyzed, including 320 genotype B, 739 genotype C, 26 genotype D, 42 C/D recombinant genotype, 20 genotype I and 1 genotype A sequences. In Northeastern, Northern, and Eastern regions, genotype C was the predominant HBV genotype, followed by genotype B. Genotype A and D were only present in very small numbers. In Central Southern areas, genotype B was predominant, followed by genotype C, I, and recombinant genotype C/D. In Southwestern areas, genotype B and C were both dominant, with few genotype I. Finally, in Northwestern areas, the recombinant genotype C/D was predominant, followed by genotype D, C, I, and B (Figure 1).
Based on the specificity of HBV distribution in Chinese territories, and removal of the incomplete sequences (< 3215 bp), 12, 478, 83, 21, 60 and 14 complete genome sequences of genotype B and C were selected from Chinese Northeastern, Northern, Eastern, Central Southern, Southeastern, and Northwestern areas, respectively. Results suggested that subgenotype B2 was the major HBV genotype B (197/208, 94.71%) in the Chinese territory, followed by subgenotype B6’ (11/208, 5.29%)[13,15,20]. Meanwhile, C2 (439/460, 95.44%) was the major subgenotype identified for genotype C, followed by subgenotype C1 (15/460, 3.62%) and C12 (6/460, 1.3%). In addition, I1 subgenotype was the major subgenotype of Chinese genotype I (20/20,100%).
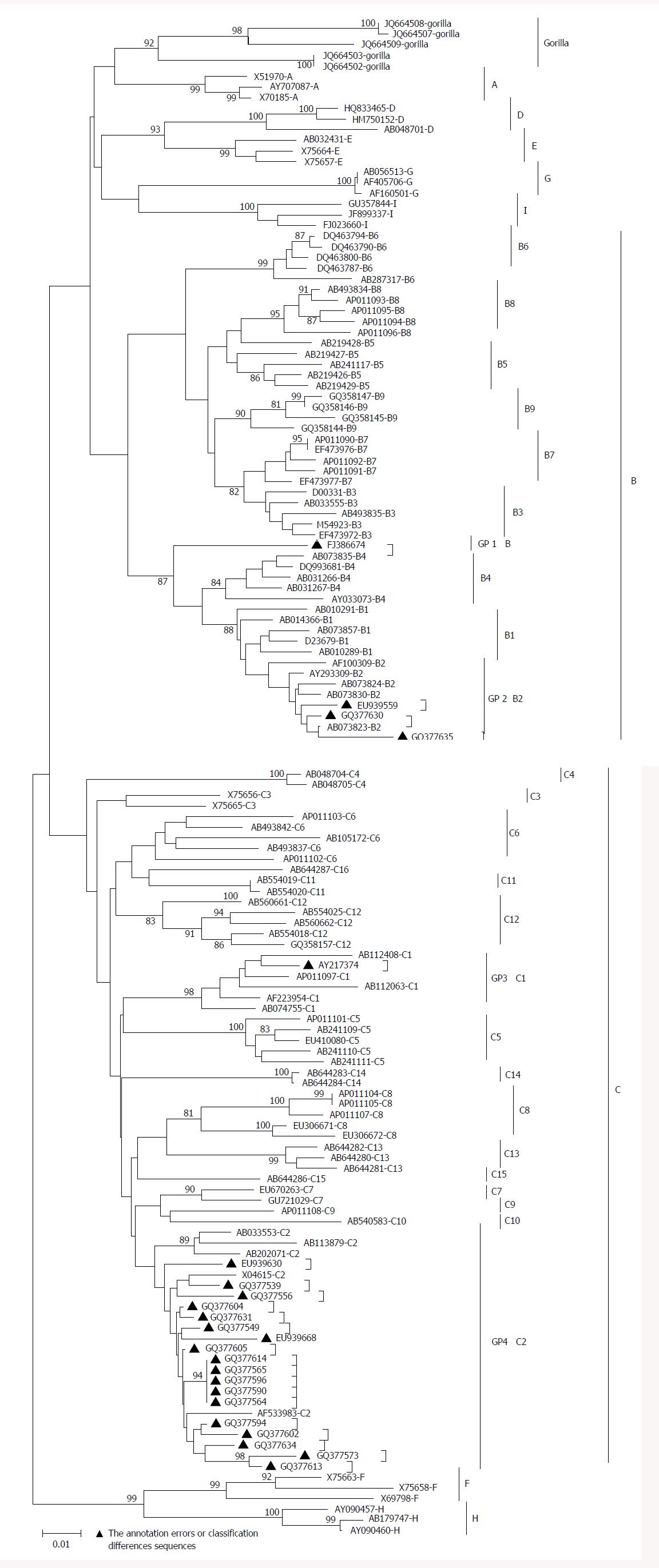
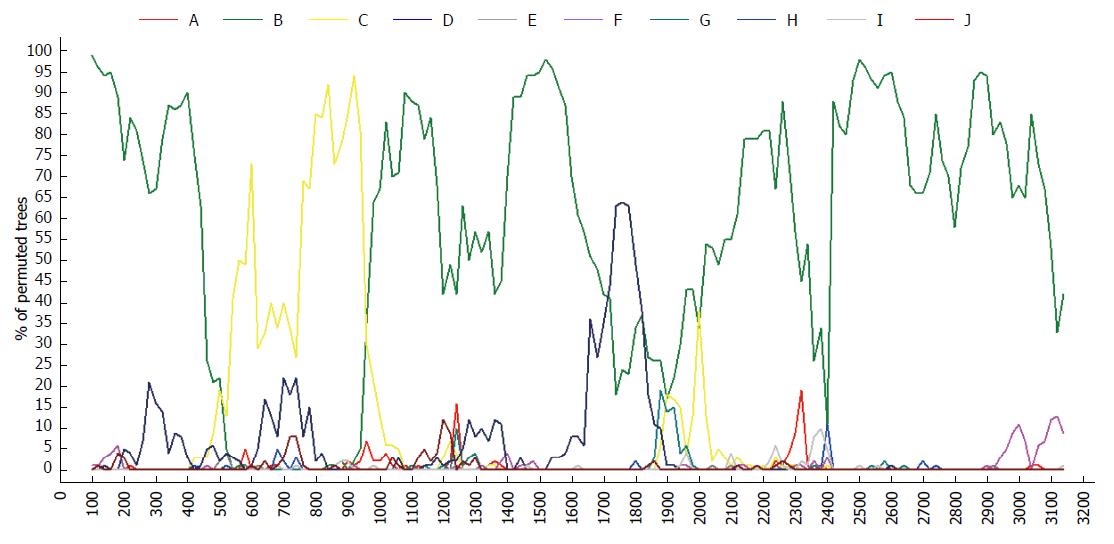
Due to differential classification methods, 23 sequences were classified with controversial genotyping and/or sub-genotyping results[32,33] (Table 1). Among these sequences, the phylogenetic tree revealed that FJ386674 had a new clad separating from the major trunk of genotype B with a 87% bootstrap value (Figure 2), the genetic distance between FJ386674 and genotypes (A, C-J) were more than 8%, but the genetic distance between FJ386674 and other B subgenotypes was more than 4% (0.05 ± 0.00 to 0.07 ± 0.01) and less than 8% (Table 2). Then we further analyzed FJ386674 by Simplot software, and results showed that FJ386674 was possibly the B/C/H recombination genotype (Figure 3).
| Sequence ID | NCBI annotation | Online software Genotyping | MEGA5.0 softwarePhylogenetic tree |
| FJ386674 | C | B | B10 |
| EU939559 | C | B | B2 |
| GQ377630 | C4 | B | B2 |
| GQ377635 | C4 | B | B2 |
| AY217374 | B | C | C1 |
| EU939668 | B2 | C | C2 |
| GQ377556 | B | C | C2 |
| GQ377539 | B | C | C2 |
| GQ377590 | B | C | C2 |
| EU939630 | B2 | C | C2 |
| GQ377596 | B | C | C2 |
| GQ377614 | B | C | C2 |
| GQ377634 | B | C | C2 |
| GQ377604 | B | C | C2 |
| GQ377594 | B | C | C2 |
| GQ377602 | B | C | C2 |
| GQ377565 | B | C | C2 |
| GQ377631 | C1 | C | C2 |
| GQ377605 | C1 | C | C2 |
| GQ377573 | B | B | C2 |
| GQ377613 | B | B | C2 |
| GQ377549 | B | B | C2 |
| GQ377564 | B | B | C2 |
| A | B | C | D | E | F | G | H | I | J | |
| B | 0.10 ± 0.01 | |||||||||
| C | 0.10 ± 0.01 | 0.12 ± 0.00 | ||||||||
| D | 0.12 ± 0.01 | 0.13 ± 0.01 | 0.13 ± 0.01 | |||||||
| E | 0.11 ± 0.01 | 0.12 ± 0.01 | 0.13 ± 0.01 | 0.09 ± 0.01 | ||||||
| F | 0.15 ± 0.01 | 0.16 ± 0.01 | 0.15 ± 0.01 | 0.16 ± 0.01 | 0.15 ± 0.01 | |||||
| G | 0.14 ± 0.01 | 0.15 ± 0.01 | 0.15 ± 0.01 | 0.14 ± 0.01 | 0.13 ± 0.01 | 0.17 ± 0.02 | ||||
| H | 0.16 ± 0.02 | 0.16 ± 0.01 | 0.15 ± 0.01 | 0.16 ± 0.01 | 0.16 ± 0.01 | 0.10 ± 0.01 | 0.17 ± 0.02 | |||
| I | 0.09 ± 0.01 | 0.11 ± 0.01 | 0.08 ± 0.01 | 0.13 ± 0.01 | 0.12 ± 0.01 | 0.15 ± 0.01 | 0.14 ± 0.01 | 0.16 ± 0.01 | ||
| J | 0.14 ± 0.02 | 0.13 ± 0.01 | 0.13 ± 0.01 | 0.15 ± 0.02 | 0.14 ± 0.02 | 0.16 ± 0.02 | 0.16 ± 0.02 | 0.17 ± 0.02 | 0.13 ± 0.01 | |
| FJ386674 | 0.11 ± 0.01 | 0.06 ± 0.01 | 0.10 ± 0.01 | 0.14 ± 0.01 | 0.13 ± 0.01 | 0.16 ± 0.01 | 0.15 ± 0.01 | 0.17 ± 0.02 | 0.11 ± 0.01 | 0.13 ± 0.01 |
| B1 | B2 | B3 | B4 | B5 | B6 | B7 | B8 | B9 | ||
| B2 | 0.03 ± 0.00 | |||||||||
| B3 | 0.05 ± 0.01 | 0.05 ± 0.01 | ||||||||
| B4 | 0.04 ± 0.00 | 0.04 ± 0.00 | 0.05 ± 0.01 | |||||||
| B5 | 0.05 ± 0.01 | 0.05 ± 0.00 | 0.03 ± 0.00 | 0.05 ± 0.01 | ||||||
| B6 | 0.05 ± 0.01 | 0.05 ± 0.01 | 0.04 ± 0.00 | 0.06 ± 0.01 | 0.05 ± 0.01 | |||||
| B7 | 0.05 ± 0.01 | 0.05 ± 0.01 | 0.03 ± 0.00 | 0.05 ± 0.01 | 0.03 ± 0.00 | 0.04 ± 0.00 | ||||
| B8 | 0.05 ± 0.00 | 0.05 ± 0.00 | 0.03 ± 0.00 | 0.05 ± 0.00 | 0.03 ± 0.00 | 0.04 ± 0.01 | 0.03 ± 0.00 | |||
| B9 | 0.05 ± 0.01 | 0.05 ± 0.00 | 0.03 ± 0.00 | 0.05 ± 0.00 | 0.03 ± 0.00 | 0.05 ± 0.01 | 0.03 ± 0.00 | 0.03 ± 0.00 | ||
| FJ386674 | 0.05 ± 0.01 | 0.05 ± 0.00 | 0.07 ± 0.01 | 0.06 ± 0.01 | 0.07 ± 0.01 | 0.07 ± 0.01 | 0.07 ± 0.01 | 0.06 ± 0.01 | 0.06 ± 0.01 |
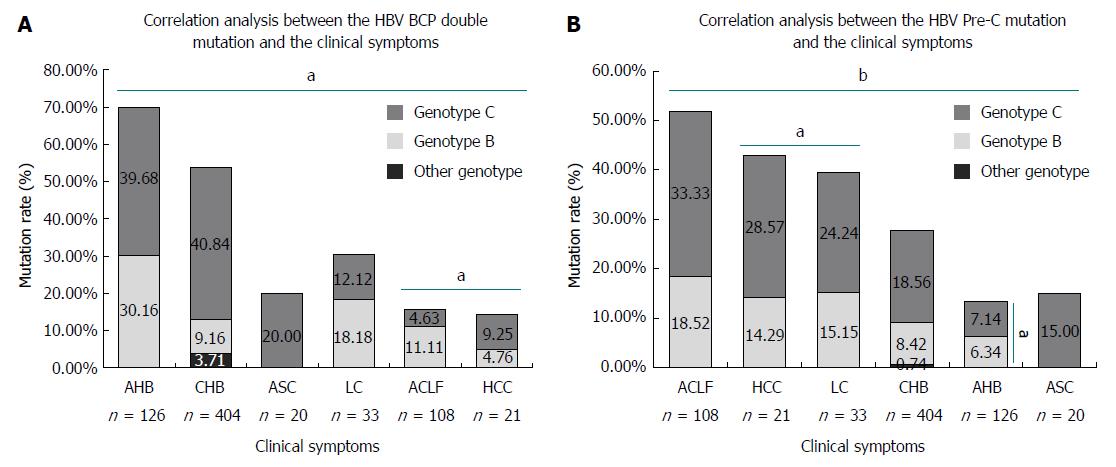
Results suggested a significant BCP double mutation (A1762T, G1764A) and pre-C mutation (G1896A) among the various hepatitis symptoms (t = 3.646, P = 0.015; t = 4.981, P = 0.004, respectively, Figure 4). A BCP double mutation was observed between ACLF and HCC and was statistically significant (t = 20.562, P = 0.031). The mutation difference in pre-C was significant between HCC and LC (t = 23.703, P = 0.027). In addition, the BCP double mutation was more frequently present in AHB (88/126, 69.84%), CHB (217/404, 53.71%), and LC (10/33, 30.30%). Pre-C showed more frequent mutations in ACLF (56/108, 51.85%), HCC (9/21, 42.86%), and LC (13/33, 39.39%). The above sequences were then subjected to analyses to determine the relationship between genotype and HBV sequence mutation. The results showed no significant differences for both the BCP double mutation (t = 2.382, P = 0.253) and the pre-C mutation (t = 3.089, P = 0.199) between genotype B and C. For AHB, a significant difference was observed between genotype B and C in the pre-C (t = 16.850, P = 0.038).
After excluding the incomplete sequences (< 3215 bp), the start and stop codons from 197 B2 genotype sequences and 439 C2 subgenotype sequences were analyzed. Results suggested that start codon mutations were observed in the ORFs. In addition to ATG, many other initiation codons of HBV also exist. The termination codon of X, C and P ORF were TAA, TAG, and TGA, respectively. Meanwhile, stop codon usage bias of S-ORF from the sub-genotypes B2 and C2 HBV genomes was observed. The dominant S-ORF stop codon in sub-genotype B2 was TAA (96.45%), followed by TGA (3.55%), while the S-ORF stop codon in sub-genotype C2 preferentially used TGA (83.6%), followed by TAA (16.4%). Based on the RNAdraw results, RNA secondary structure of subgenotype B2 with TAA termination codon was similar to a hairpin loop, due to 681 (A), 680 (A) and 676 (A) might be paired with 655 (U), 656 (U) and 660 (U), respectively. However, there were no such base pairs existing in the C2 subgenotype with TGA termination codon except possibly base-pairs of 681 (A) and 655 (U) (Figure 5).
HBV genotypes and sub-genotypes had obvious geographic features according to previous reports[34-36]. The current study also suggested a differential distribution of HBV genotypes in China. Within the northern areas of the Qinling Mountains-Huaihe River Line, genotype C (75.3%) was predominant, followed by a smaller percentage of type B (23.4%) and D (1.3%). While Sunbul[37] reported that genotype B was the major genotype in southern China, our results showed that the ratios of genotype B and C in the southern areas of China were 41% and 57%, respectively. This inconsistency may be due to differences in the selection of subjects and quantity of tested samples. Meanwhile, Northwestern China was dominated by recombinant genotype C/D and genotype D, with percentages of 49.3% and 24.6%, respectively. This result is consistent with a study by Yin et al[38]. Additionally, our investigation indicated that genotype I (originally reported as recombinant genotype A/C/G) was mainly located in the Guangxi[39], Shaanxi[40], Yunnan[41], and Sichuan Provinces[42], and I1 was the major sub-genotype in China.
From a geological perspective, many of the identified provinces were located on the Silk Route. For instance, the Guangdong Province was adjacent to Hong Kong and Macao; the Hainan and Taiwan regions were separated by the strait; Hong Kong, Macao, and Taiwan were once European colonies, where genotype A and D were dominant. Thus, we postulated that genotype I and recombinant genotype C/D was the result of a mixed genotype infection since it has already been discovered that recombination can occur in different genotypes of parental HBV strains[40,43]. Moreover, some studies proposed that genotype I may have existed for a long time in Shaanxi Province without being recognized, creating the question of how genotype I arose historically[40]. We hypothesized that a mixed genotype infection in patients from these areas may have occurred at first and subsequently resulted in recombinants. Furthermore, multiple factors, including extreme environmental effects[44] and special religious influences[42,45], may have helped preserved the resultant recombinant by natural selection.
To date, the definition of a new sub-genotype has been classified utilizing several major instructions. Firstly, a novel sub-genotype should be different from the known sub-genotype by 4% over the complete sequences. Secondly, a new sub-genotype should be an independent branch in the phylogenetic tree. Finally, a novel sub-genotype should have a bootstrap value over 75%[46].
In this study, Simplot results showed that FJ386674 represented a recombinant of genotypes B, C and H, with its two recombination breakpoints: one between nucleotides 500 and 960, and another from nucleotides 1700 to 1820 (Figure 3). Considering the results of phylogenetic tree and genetic distance, we designated FJ386674 as a putative sub-genotype B10.
However, despite the development of several new criteria for new sub-genotype classification, a number of controversial results still exist[32,33]. For instance, in this study, we found that 23 sequences had different genotyping and/or sub-genotyping due to different classification methods, and evolutionary distance among B3 and B5, B7-B9 (0.03 ± 0.00) was < 4%, a result consistent with a previous report indicating that B5, B7-B9 should be classified as a quasi-strain of B3[20,46]. Thus, the systematic approach of HBV putative sub-genotype classification needs to be further improved.
Many HBV mutations might be tightly associated with liver disease progression[47-49]. This study showed BCP double mutations were significantly different in ACLF and HCC. The results suggested that HCC had a lower mutation rate in the BCP region as compared with that of ACLF, which is consistent with a previous report[32]. However, other studies indicated that the BCP double mutation was associated with liver disease progression[47,48]. This inconsistency between current studies might be affected by many factors, like the genetic background of selected patients, the numbers of samples, and/or genotypes[49]. Some studies suggested a synergenistic action of the BCP double mutation and HBV genotype C in liver disease progression[50]. Although no statistically significant difference was observed between genotype B and C (P = 0.253) for all other hepatitis symptoms, the ratio of the BCP double mutation in genotype C has a tendency to be higher than that in genotype B (Figure 4), which is consistent with a previous study[51]. This might explain the intriguing relationship between genotype C and liver disease progression. Thus, the investigation of liver disease progression should not only look at the genotypes, but should also consider BCP mutations.
It has been suggested that the pre-C mutation is also tightly associated with liver disease progression[48,52]. In this study, we revealed that the pre-C mutation showed significant differences in HCC and LC, the mutation rate of the pre-C in ACLF, LC and HCC were higher than that of CHB (Figure 4), which is consistent with previous studies[48,52-55]. Thus, an HBV pre-C mutation might be tightly associated with liver disease progression.
In addition, some studies have reported the hepatitis B virus genotype C is associated with the process of liver disease[51,56]. This study also suggested no statistically significant difference in the HBV pre-C mutation between types B and C (P = 0.199). The detailed analysis indicated that only AHB was significantly different between types B and C (P = 0.038), while the mutation ratio in the same region was higher in type C without statistical significance for the other symptoms. This may explain why patients infected with genotype C HBV are more susceptible to the development of ACLF LC and HCC. We also found that the mutation rate of pre-C in ACLF was as high as 49.66% (Figure 4), which may be supported by a report that identified this mutation as a potential biomarker for ACLF onset[32]. Thus, the investigation of liver disease progression should always consider multiple factors, including the HBV genotype and associated mutations in order to achieve the most comprehensive understanding of the disease.
Generally, the start codon of a nucleic acid in living organisms is ATG. Results of this study showed that the P-ORF start codon is highly conserved in a mutation-free manner. However, several start codon mutations were observed in the other ORFs. The preS2 region of HBV nucleic acid sequence had the highest mutation rate, resulting in the most variable amino acid mutation, ATG turn into AAA (lysine), AAG (lysine), ACG (threonine), AGG (arginine), AGT (serine), ATA (isoleucine), ATT(isoleucine), CCA(proline), CCG (proline), GTG (valine), GTT (valine), TTG (leucine). These results were consistent with previous reports from Vietnam, Korea, China, and Thailand[57,58]. In fact, the same mutation also exists in other species[59-63]. This study showed that the start codon mutation rate in genotype C2 (81/439, 18.45%) was higher than that of genotype B2 (26/197, 13.20%), which is consistent with a previous study[64]. Furthermore, one study suggested that the PreS2 start codon mutation might be related with liver cancer progression or active DNA replication[65]. We revealed that the start codon mutation rate of genotype C2 HBV (63/439, 14.35%) was also higher than that of subtype B2 (15/197, 7.61%), although it was not statistically significant (P = 0.19). This result indicated that genotype C HBV might be more susceptible for developing liver disease when compared with genotype B[65,66]. Thus, a mutation of the HBV PreS2 start codon is a likely cause for liver disease progression. Unfortunately, it is still unclear what the mechanisms are and whether this alteration affects HBV survival, replication, and expression in host cells. Therefore, further studies are needed to elucidate this mechanism.
In all organisms, the stop codons of biological nucleic acids are TAA, TAG, or TGA. Previous reports indicate that many viruses including foot-and-mouth disease viruses, influenza A virus subtype H5N1 and human bocavirus, have codon usage bias[67]. HBV was no exception; in this study we found that all stop codons of P-ORF were TGA, while the majority stop codons for X-ORF and C-ORF were TAA and TAG, respectively. The S-ORF stop codon in sub-genotype B2 preferentially used TAA, followed by TGA; on the contrary, the dominant S-ORF stop codon for sub-genotype C2 was TGA, followed by TAA, which was consistent with a previous report[68]. We further analyzed the termination codon of subgenotype B2 and C2 by RNAdraw software to predict RNA secondary structure of the HBsAg protein. Results showed that subgenotype B2 with TAA termination codon was similar to a stem-loop structure, and subgenotype C2 with TGA termination codon was similar to a single-stranded structure. Previous studies have shown that specific nucleotide sequences in the stem-loop structure are critical for RNA stability, alternative splicing, packaging and encapsidation[69-71]. The RNA secondary structure of subgenotype B2 with TAA termination codon might be more stable than subgenotype C2 with TGA termination codon because of base pairing. Therefore, we suggest that termination codon usage bias might be a reason for genotype C producing more serious clinical symptoms compared with genotype B. This still requires further investigation.
In conclusion, the samples collected in this study showed territory-associated features that recapitulate the epidemiology of HBV in China. C2 and B2 were identified as the two major subgenotypes in China. FJ386674 might be a putative sub-genotype of B10. The major stop codons of S-ORF were TAA (92.2%) and TGA (79.65%) in B2 and C2 subtypes, respectively. These data will facilitate researcher ability to connect sequence mutations with liver disease progression and to investigate the genetic heterogeneity of HBV genomes. This information might be meaningful for the future prevention and therapy of HBV infections.
Hepatitis B virus (HBV) infection can induce diseases, such as acute hepatitis, chronic hepatitis, hepatocirrhosis, and hepatocellular carcinoma, and thus severely threats global human health.
Previous studies suggested that liver disease progression might be associated with HBV genotypes, serotypes, basal core promoter (BCP) double mutation and pre-C mutation.
This study recapitulated the epidemiology of HBV in China. C2 and B2 were identified as the two major sub-genotypes in China. FJ386674 might be a new sub-genotype as B10. The BCP double mutation and pre-C mutation showed various significant differences between hepatitis symptoms. In addition to ATG, many other HBV initiation codons also exist. HBV has codon usage bias, the termination codons of X, C and P open reading frame (ORF) were TAA, TAG, and TGA, respectively. The major stop codons of S-ORF were TAA (92.2%) and TGA (79.65%) in B2 and C2 subtypes, respectively.
The study results recapitulated the epidemiology of HBV in China, and these data will facilitate researcher ability to connect sequence mutations with liver disease progression and to investigate the genetic heterogeneity of HBV genomes. This information might be meaningful for the future prevention and therapy of HBV infections.
Codon usage bias is the disequilibrium phenomenon of synonymous codon usage, which encodes the same kinds of amino acids in biological matter. Because this phenomenon is associated with the carrier of genetic information molecules of DNA and the biological function of proteins, it has important biological significance.
This study provides important information on HBV genotype distribution and genome characteristics in China. New information provided in this manuscript is also of some clinical value.
| 1. | Li L, Shen H, Li A, Zhang Z, Wang B, Wang J, Zheng X, Wu J, Yang D, Lu M. Inhibition of hepatitis B virus (HBV) gene expression and replication by HBx gene silencing in a hydrodynamic injection mouse model with a new clone of HBV genotype B. Virol J. 2013;10:214. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 14] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 2. | Gust ID. Epidemiology of hepatitis B infection in the Western Pacific and South East Asia. Gut. 1996;38 Suppl 2:S18-S23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 3. | Fattovich G, Bortolotti F, Donato F. Natural history of chronic hepatitis B: special emphasis on disease progression and prognostic factors. J Hepatol. 2008;48:335-352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 909] [Cited by in RCA: 988] [Article Influence: 54.9] [Reference Citation Analysis (1)] |
| 4. | Kao JH, Chen DS. Global control of hepatitis B virus infection. Lancet Infect Dis. 2002;2:395-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 596] [Cited by in RCA: 615] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 5. | Okamoto H, Tsuda F, Sakugawa H, Sastrosoewignjo RI, Imai M, Miyakawa Y, Mayumi M. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J Gen Virol. 1988;69:2575-2583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 725] [Cited by in RCA: 773] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 6. | Arauz-Ruiz P, Norder H, Robertson BH, Magnius LO. Genotype H: a new Amerindian genotype of hepatitis B virus revealed in Central America. J Gen Virol. 2002;83:2059-2073. [PubMed] |
| 7. | Tran TT, Trinh TN, Abe K. New complex recombinant genotype of hepatitis B virus identified in Vietnam. J Virol. 2008;82:5657-5663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 195] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 8. | Tatematsu K, Tanaka Y, Kurbanov F, Sugauchi F, Mano S, Maeshiro T, Nakayoshi T, Wakuta M, Miyakawa Y, Mizokami M. A genetic variant of hepatitis B virus divergent from known human and ape genotypes isolated from a Japanese patient and provisionally assigned to new genotype J. J Virol. 2009;83:10538-10547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 325] [Cited by in RCA: 338] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 9. | Huy TT, Sall AA, Reynes JM, Abe K. Complete genomic sequence and phylogenetic relatedness of hepatitis B virus isolates in Cambodia. Virus Genes. 2008;36:299-305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 10. | Lin CL, Kao JH. The clinical implications of hepatitis B virus genotype: Recent advances. J Gastroenterol Hepatol. 2011;26 Suppl 1:123-130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 256] [Cited by in RCA: 283] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 11. | Huy TT, Ushijima H, Quang VX, Win KM, Luengrojanakul P, Kikuchi K, Sata T, Abe K. Genotype C of hepatitis B virus can be classified into at least two subgroups. J Gen Virol. 2004;85:283-292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 106] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 12. | Osiowy C, Giles E, Tanaka Y, Mizokami M, Minuk GY. Molecular evolution of hepatitis B virus over 25 years. J Virol. 2006;80:10307-10314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 124] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 13. | Sakamoto T, Tanaka Y, Simonetti J, Osiowy C, Borresen ML, Koch A, Kurbanov F, Sugiyama M, Minuk GY, McMahon BJ. Classification of hepatitis B virus genotype B into 2 major types based on characterization of a novel subgenotype in Arctic indigenous populations. J Infect Dis. 2007;196:1487-1492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 60] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 14. | Nagasaki F, Niitsuma H, Cervantes JG, Chiba M, Hong S, Ojima T, Ueno Y, Bondoc E, Kobayashi K, Ishii M. Analysis of the entire nucleotide sequence of hepatitis B virus genotype B in the Philippines reveals a new subgenotype of genotype B. J Gen Virol. 2006;87:1175-1180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 15. | Shen T, Gao JM, Zou YL, Dong H, Yan XM. Novel hepatitis B virus subgenotype in the southern Yunnan Province of China. Intervirology. 2009;52:340-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 16. | Mulyanto SN, Surayah K, Tsuda F, Ichiyama K, Takahashi M, Okamoto H. A nationwide molecular epidemiological study on hepatitis B virus in Indonesia: identification of two novel subgenotypes, B8 and C7. Arch Virol. 2009;154:1047-1059. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 55] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 17. | Mulyanto SN, Surayah K, Tjahyono AA, Jirintai S, Takahashi M, Okamoto H. Identification and characterization of novel hepatitis B virus subgenotype C10 in Nusa Tenggara, Indonesia. Arch Virol. 2010;155:705-715. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 31] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 18. | Mulyanto SN, Wahyono A, Jirintai S, Takahashi M, Okamoto H. Analysis of the full-length genomes of novel hepatitis B virus subgenotypes C11 and C12 in Papua, Indonesia. J Med Virol. 2011;83:54-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 19. | Mulyanto P, Depamede SN, Wahyono A, Jirintai S, Nagashima S, Takahashi M, Nishizawa T, Okamoto H. Identification of four novel subgenotypes (C13-C16) and two inter-genotypic recombinants (C12/G and C13/B3) of hepatitis B virus in Papua province, Indonesia. Virus Res. 2012;163:129-140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 40] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 20. | Shi W, Zhu C, Zheng W, Carr MJ, Higgins DG, Zhang Z. Subgenotype reclassification of genotype B hepatitis B virus. BMC Gastroenterol. 2012;12:116. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 21. | Shi W, Zhu C, Zheng W, Zheng W, Ling C, Carr MJ, Higgins DG, Zhang Z. Subgenotyping of genotype C hepatitis B virus: correcting misclassifications and identifying a novel subgenotype. PLoS One. 2012;7:e47271. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 22. | Thedja MD, Muljono DH, Nurainy N, Sukowati CH, Verhoef J, Marzuki S. Ethnogeographical structure of hepatitis B virus genotype distribution in Indonesia and discovery of a new subgenotype, B9. Arch Virol. 2011;156:855-868. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 45] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 23. | Olinger CM, Jutavijittum P, Hübschen JM, Yousukh A, Samountry B, Thammavong T, Toriyama K, Muller CP. Possible new hepatitis B virus genotype, southeast Asia. Emerg Infect Dis. 2008;14:1777-1780. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 139] [Cited by in RCA: 145] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 24. | Nguyen CH, Ishizaki A, Chung PT, Hoang HT, Nguyen TV, Tanimoto T, Lihana R, Matsushita K, Bi X, Pham TV. Prevalence of HBV infection among different HIV-risk groups in Hai Phong, Vietnam. J Med Virol. 2011;83:399-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 25. | Sugauchi F, Orito E, Ichida T, Kato H, Sakugawa H, Kakumu S, Ishida T, Chutaputti A, Lai CL, Ueda R. Hepatitis B virus of genotype B with or without recombination with genotype C over the precore region plus the core gene. J Virol. 2002;76:5985-5992. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 218] [Cited by in RCA: 233] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 26. | Sugauchi F, Kumada H, Sakugawa H, Komatsu M, Niitsuma H, Watanabe H, Akahane Y, Tokita H, Kato T, Tanaka Y. Two subtypes of genotype B (Ba and Bj) of hepatitis B virus in Japan. Clin Infect Dis. 2004;38:1222-1228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 63] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 27. | Norder H, Couroucé AM, Coursaget P, Echevarria JM, Lee SD, Mushahwar IK, Robertson BH, Locarnini S, Magnius LO. Genetic diversity of hepatitis B virus strains derived worldwide: genotypes, subgenotypes, and HBsAg subtypes. Intervirology. 2004;47:289-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 630] [Cited by in RCA: 651] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 28. | Veazjalali M, Norder H, Magnius L, Jazayeri SM, Alavian SM, Mokhtari-Azad T. A new core promoter mutation and premature stop codon in the S gene in HBV strains from Iranian patients with cirrhosis. J Viral Hepat. 2009;16:259-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 18] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 29. | Harari A, Ooms M, Mulder LC, Simon V. Polymorphisms and splice variants influence the antiretroviral activity of human APOBEC3H. J Virol. 2009;83:295-303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 134] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 30. | Matzura O, Wennborg A. RNAdraw: an integrated program for RNA secondary structure calculation and analysis under 32-bit Microsoft Windows. Comput Appl Biosci. 1996;12:247-249. [PubMed] |
| 31. | Mamadou S, Vidal N, Montavon C, Ben A, Djibo A, Rabiou S, Soga G, Delaporte E, Mboup S, Peeters M. Emergence of complex and diverse CRF02-AG/CRF06-cpx recombinant HIV type 1 strains in Niger, West Africa. AIDS Res Hum Retroviruses. 2003;19:77-82. [PubMed] |
| 32. | Xu Z, Ren X, Liu Y, Li X, Bai S, Zhong Y, Wang L, Mao P, Wang H, Xin S. Association of hepatitis B virus mutations in basal core promoter and precore regions with severity of liver disease: an investigation of 793 Chinese patients with mild and severe chronic hepatitis B and acute-on-chronic liver failure. J Gastroenterol. 2011;46:391-400. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 60] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 33. | Xu Z, Liu Y, Xu T, Chen L, Si L, Wang Y, Ren X, Zhong Y, Zhao J, Xu D. Acute hepatitis B infection associated with drug-resistant hepatitis B virus. J Clin Virol. 2010;48:270-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 38] [Article Influence: 2.4] [Reference Citation Analysis (1)] |
| 34. | Kao JH, Chen PJ, Lai MY, Chen DS. Hepatitis B genotypes correlate with clinical outcomes in patients with chronic hepatitis B. Gastroenterology. 2000;118:554-559. [PubMed] |
| 35. | Schaefer S, Magnius L, Norder H. Under construction: classification of hepatitis B virus genotypes and subgenotypes. Intervirology. 2009;52:323-325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 43] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 36. | Dunford L, Carr MJ, Dean J, Nguyen LT, Ta Thi TH, Nguyen BT, Connell J, Coughlan S, Nguyen HT, Hall WW. A multicentre molecular analysis of hepatitis B and blood-borne virus coinfections in Viet Nam. PLoS One. 2012;7:e39027. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 56] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 37. | Sunbul M. Hepatitis B virus genotypes: global distribution and clinical importance. World J Gastroenterol. 2014;20:5427-5434. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 264] [Cited by in RCA: 304] [Article Influence: 25.3] [Reference Citation Analysis (8)] |
| 38. | Yin J, Zhang H, He Y, Xie J, Liu S, Chang W, Tan X, Gu C, Lu W, Wang H. Distribution and hepatocellular carcinoma-related viral properties of hepatitis B virus genotypes in Mainland China: a community-based study. Cancer Epidemiol Biomarkers Prev. 2010;19:777-786. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 81] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 39. | Fang ZL, Hué S, Sabin CA, Li GJ, Yang JY, Chen QY, Fang KX, Huang J, Wang XY, Harrison TJ. A complex hepatitis B virus (X/C) recombinant is common in Long An county, Guangxi and may have originated in southern China. J Gen Virol. 2011;92:402-411. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 28] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 40. | Yu H, Yuan Q, Ge SX, Wang HY, Zhang YL, Chen QR, Zhang J, Chen PJ, Xia NS. Molecular and phylogenetic analyses suggest an additional hepatitis B virus genotype “I”. PLoS One. 2010;5:e9297. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 111] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 41. | Kang WY, Bi SL, Ding ZR, Tian BJ, Zhao ZX, Li H. [Molecular identification of hepatitis B virus genotype I in Yunnan Province, the People’s Republic of China]. Bing Du Xue Bao. 2011;27:215-217. [PubMed] |
| 42. | Tong W, He J, Sun L, He S, Qi Q. Hepatitis B virus with a proposed genotype I was found in Sichuan Province, China. J Med Virol. 2012;84:866-870. [PubMed] |
| 43. | Zhou B, Wang Z, Yang J, Sun J, Li H, Tanaka Y, Mizokami M, Hou J. Novel evidence of HBV recombination in family cluster infections in western China. PLoS One. 2012;7:e38241. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 44. | Zhou B, Xiao L, Wang Z, Chang ET, Chen J, Hou J. Geographical and ethnic distribution of the HBV C/D recombinant on the Qinghai-Tibet Plateau. PLoS One. 2011;6:e18708. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 38] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 45. | Shen T, Yan XM, Liu HX, Zhang BX, Li L, Zhang JP, Wang JL, Xiao CJ. Genotype I of hepatitis B virus was found in east Xishuangbanna, China and molecular dynamics of HBV/I. J Viral Hepat. 2015;22:37-45. [PubMed] |
| 46. | Shi W, Zhang Z, Ling C, Zheng W, Zhu C, Carr MJ, Higgins DG. Hepatitis B virus subgenotyping: history, effects of recombination, misclassifications, and corrections. Infect Genet Evol. 2013;16:355-361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 81] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 47. | Liu CJ, Chen BF, Chen PJ, Lai MY, Huang WL, Kao JH, Chen DS. Role of hepatitis B virus precore/core promoter mutations and serum viral load on noncirrhotic hepatocellular carcinoma: a case-control study. J Infect Dis. 2006;194:594-599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 98] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 48. | Liu S, Zhang H, Gu C, Yin J, He Y, Xie J, Cao G. Associations between hepatitis B virus mutations and the risk of hepatocellular carcinoma: a meta-analysis. J Natl Cancer Inst. 2009;101:1066-1082. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 279] [Cited by in RCA: 332] [Article Influence: 19.5] [Reference Citation Analysis (1)] |
| 49. | Yu MW, Yeh SH, Chen PJ, Liaw YF, Lin CL, Liu CJ, Shih WL, Kao JH, Chen DS, Chen CJ. Hepatitis B virus genotype and DNA level and hepatocellular carcinoma: a prospective study in men. J Natl Cancer Inst. 2005;97:265-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 417] [Cited by in RCA: 423] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 50. | Chen QY, Harrison TJ, Sabin CA, Li GJ, Huang GM, Yang JY, Wang XY, Li H, Liu MH, Fang ZL. The Effect of HBV Genotype C on the Development of HCC Differs Between Wild-Type Viruses and Those With BCP Double Mutations (T(1762)A(1764)). Hepat Mon. 2014;14:e16214. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 51. | Yuen MF, Tanaka Y, Shinkai N, Poon RT, But DY, Fong DY, Fung J, Wong DK, Yuen JC, Mizokami M. Risk for hepatocellular carcinoma with respect to hepatitis B virus genotypes B/C, specific mutations of enhancer II/core promoter/precore regions and HBV DNA levels. Gut. 2008;57:98-102. [PubMed] |
| 52. | Rezende RE, Fonseca BA, Ramalho LN, Zucoloto S, Pinho JR, Bertolini DA, Martinelli AL. The precore mutation is associated with severity of liver damage in Brazilian patients with chronic hepatitis B. J Clin Virol. 2005;32:53-59. [PubMed] |
| 53. | Jammeh S, Tavner F, Watson R, Thomas HC, Karayiannis P. Effect of basal core promoter and pre-core mutations on hepatitis B virus replication. J Gen Virol. 2008;89:901-909. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 102] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 54. | Ou JH. Molecular biology of hepatitis B virus e antigen. J Gastroenterol Hepatol. 1997;12:S178-S187. [PubMed] |
| 55. | Scaglioni PP, Melegari M, Wands JR. Posttranscriptional regulation of hepatitis B virus replication by the precore protein. J Virol. 1997;71:345-353. [PubMed] |
| 56. | Ochwoto M, Chauhan R, Gopalakrishnan D, Chen CY, Ng’ang’a Z, Okoth F, Kioko H, Kimotho J, Kaiguri P, Kramvis A. Genotyping and molecular characterization of hepatitis B virus in liver disease patients in Kenya. Infect Genet Evol. 2013;20:103-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 57. | Huy TT, Ushijima H, Win KM, Luengrojanakul P, Shrestha PK, Zhong ZH, Smirnov AV, Taltavull TC, Sata T, Abe K. High prevalence of hepatitis B virus pre-s mutant in countries where it is endemic and its relationship with genotype and chronicity. J Clin Microbiol. 2003;41:5449-5455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 85] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 58. | Suwannakarn K, Tangkijvanich P, Thawornsuk N, Theamboonlers A, Tharmaphornpilas P, Yoocharoen P, Chongsrisawat V, Poovorawan Y. Molecular epidemiological study of hepatitis B virus in Thailand based on the analysis of pre-S and S genes. Hepatol Res. 2008;38:244-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 23] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 59. | Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453-1462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5555] [Cited by in RCA: 5453] [Article Influence: 188.0] [Reference Citation Analysis (0)] |
| 60. | Lusic H, Gustilo EM, Vendeix FA, Kaiser R, Delaney MO, Graham WD, Moye VA, Cantara WA, Agris PF, Deiters A. Synthesis and investigation of the 5-formylcytidine modified, anticodon stem and loop of the human mitochondrial tRNAMet. Nucleic Acids Res. 2008;36:6548-6557. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 50] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 61. | Sakamoto W, Tan SH, Murata M, Motoyoshi F. An unusual mitochondrial atp9-rpl16 cotranscript found in the maternal distorted leaf mutant of Arabidopsis thaliana: implication of GUG as an initiation codon in plant mitochondria. Plant Cell Physiol. 1997;38:975-979. [PubMed] |
| 62. | Takemoto C, Spremulli LL, Benkowski LA, Ueda T, Yokogawa T, Watanabe K. Unconventional decoding of the AUA codon as methionine by mitochondrial tRNAMet with the anticodon f5CAU as revealed with a mitochondrial in vitro translation system. Nucleic Acids Res. 2009;37:1616-1627. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 94] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 63. | Kadowaki K, Ozawa K, Kazama S, Kubo N, Akihama T. Creation of an initiation codon by RNA editing in the coxI transcript from tomato mitochondria. Curr Genet. 1995;28:415-422. [PubMed] |
| 64. | Choi MS, Kim DY, Lee DH, Lee JH, Koh KC, Paik SW, Rhee JC, Yoo BC. Clinical significance of pre-S mutations in patients with genotype C hepatitis B virus infection. J Viral Hepat. 2007;14:161-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 73] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 65. | Lu J, Gong W, Cheng H, Wu Z, Li D, Wang X, Liang P, Zhang J. Detection of HBV genotypes of tumor tissues and serum by a fluorescence polarization assay in north-western China’s hepatocellular carcinoma patients. Virol J. 2011;8:362. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 66. | Kao JH. Hepatitis B virus genotypes and hepatocellular carcinoma in Taiwan. Intervirology. 2003;46:400-407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 110] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 67. | Kessler MD, Dean MD. Effective population size does not predict codon usage bias in mammals. Ecol Evol. 2014;4:3887-3900. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 15] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 68. | Li JQ, Tian JH, Liu F, Du SC. [Hepatitis B surface antigen terminates codon bias selection]. Beijing Da Xue Xue Bao. 2008;40:270-272. [PubMed] |
| 69. | Pollack JR, Ganem D. An RNA stem-loop structure directs hepatitis B virus genomic RNA encapsidation. J Virol. 1993;67:3254-3263. [PubMed] |
| 70. | Grover A, Houlden H, Baker M, Adamson J, Lewis J, Prihar G, Pickering-Brown S, Duff K, Hutton M. 5’ splice site mutations in tau associated with the inherited dementia FTDP-17 affect a stem-loop structure that regulates alternative splicing of exon 10. J Biol Chem. 1999;274:15134-15143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 228] [Cited by in RCA: 221] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 71. | Jambor H, Mueller S, Bullock SL, Ephrussi A. A stem-loop structure directs oskar mRNA to microtubule minus ends. RNA. 2014;20:429-439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 50] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
P- Reviewer: Lazarevic I, Sagnelli E S- Editor: Yu J L- Editor: O’Neill M E- Editor: Ma S