Published online Apr 7, 2015. doi: 10.3748/wjg.v21.i13.3960
Peer-review started: September 4, 2014
First decision: September 27, 2014
Revised: November 7, 2014
Accepted: December 14, 2014
Article in press: December 16, 2014
Published online: April 7, 2015
Processing time: 216 Days and 3.7 Hours
AIM: To examine the methylation levels of interleukin-1 receptor-associated kinase 3 (IRAK3) and GLOXD1 and their potential clinical applications in hepatocellular carcinoma (HCC).
METHODS: mRNA expression and promoter methylation of IRAK3 and GLOXD1 in HCC cells were analyzed by reverse transcription-polymerase chain reaction (RT-PCR) and methylation-specific PCR (MSP), respectively. Using pyrosequencing results, we further established a quantitative MSP (Q-MSP) system for the evaluation of IRAK3 and GLOXD1 methylation in 29 normal controls and 160 paired HCC tissues and their adjacent nontumor tissues. We also calculated Kaplan-Meier survival curves to determine the applications of gene methylation in the prognosis of HCC.
RESULTS: IRAK3 and GLOXD1 expression was partially restored in several HCC cell lines after treatment with 5-aza-2′-deoxycytidine (DNA methyltransferase inhibitor; 5DAC). A partial decrease in the methylated band was also observed in the HCC cell lines after 5DAC treatment. Using GLOXD1 as an example, we found a significant correlation between the data obtained from the methylation array and from pyrosequencing. The methylation frequency of IRAK3 and GLOXD1 in HCC tissues was 46.9% and 63.8%, respectively. Methylation of IRAK3 was statistically associated with tumor stage. Moreover, HCC patients with IRAK3 methylation had a trend toward poor 3-year disease-free survival (P < 0.05).
CONCLUSION: IRAK3 and GLOXD1 were frequently methylated in HCC tissues compared to normal controls and nontumor tissues. IRAK3 methylation was associated with tumor stage and poor prognosis of patients. These data suggest that IRAK3 methylation is a novel prognostic marker in HCC.
Core tip: The methylation biomarker is relatively stable in tissue samples and body fluids, suggesting that it is a good tool for the detection, diagnosis, prognosis, and even therapy of hepatocellular carcinoma (HCC). Our study not only demonstrated frequent methylation of interleukin-1 receptor-associated kinase 3 (IRAK3) and GLOXD1 in HCC but also found that IRAK3 methylation was positively associated with poor 3-year disease-free survival of patients. This indicates that IRAK3 methylation could be used as a potential biomarker for prediction of prognosis in HCC.
-
Citation: Kuo CC, Shih YL, Su HY, Yan MD, Hsieh CB, Liu CY, Huang WT, Yu MH, Lin YW. Methylation of
IRAK3 is a novel prognostic marker in hepatocellular carcinoma. World J Gastroenterol 2015; 21(13): 3960-3969 - URL: https://www.wjgnet.com/1007-9327/full/v21/i13/3960.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i13.3960
Hepatocellular carcinoma (HCC) is one of the most common causes of cancer deaths in the world[1]. HCC is a serious disease because it is difficult to detect in its early stages; this leads to a very poor prognosis and high mortality. It is believed that studying the molecular mechanisms of HCC development can help us to design better strategies for disease detection or prognosis prediction[2].
Aberrant changes in DNA methylation patterns, which alter gene expression and subsequently drive malignant transformation, are recognized as a common event during carcinogenesis[3] and are also found during the development of HCC[4,5]. Identification of these events not only allows for a detailed understanding of the hepatocarcinogenesis but also provides potential clinical applications in the diagnosis or prognosis of HCC[6]. Recently, technical advances in array systems have led to the development of higher-resolution genome-wide methods for DNA methylation analysis, such as Infinium assay[7]. It has also been successfully used in the study of HCC[8-17]. By using array-based platforms, researchers can simultaneously profile the DNA methylation of a large number of genes or the entire genome. Furthermore, by validating the results from the high-throughput screening approach, researchers can effectively discover more novel genes that may have potential applications in clinical practice.
In our recent study[18], we found several aberrantly methylated genes in HCC by using the Infinium HumanMethylation27 BeadChip and then verified 34 genes by methylation-specific PCR (MSP). Of these genes, we further showed that frequent methylation of homeobox A9 (HOXA9) in HCC tissues and plasma samples from patients could be a helpful biomarker to assist in HCC detection. However, several novel genes in our array data were not further validated by quantitative MSP (QMSP), such as interleukin-1 receptor-associated kinase 3 (IRAK3) and 4-hydroxyphenylpyruvate dioxygenase-like (HPDL, also known as GLOXD1). IRAK3 plays an important role in alcohol-induced liver injury[19], and HPDL is an important enzyme in the catabolic pathway of tyrosine in the liver[20]. Moreover, there are no quantitative data about the methylation levels of IRAK3 and GLOXD1 in HCC. In this study, we aimed to examine the methylation levels of IRAK3 and GLOXD1 in HCC by QMSP and to further test whether these two genes have potential clinical applications in the diagnosis or prognosis of HCC.
A normal liver cell line (THLE-3) and 6 HCC cell lines (HepG2, SK-HEP1, TONG, Mahlavu, PLC/PRF/5, and HuH6) were used in this study. THLE-3, HepG2, and SK-HEP1 cells were purchased from American Type Culture Collection. TONG, Mahlavu, PLC/PRF/5, and HuH6 cells were provided by Professor K.H. Lin (Chuang-Gung University, Taiwan). For 5-aza-2′-deoxycytidine (5DAC) treatment, HCC cells were prepared as previously described and harvested directly for reverse transcription-polymerase chain reaction (RT-PCR) and MSP[18]. THLE-3, 3 HCC cell lines (PLC/PRF/5, HepG2, and HuH6), and 7 types of pooled tissues (each type was independently pooled with an equal amount of DNA from 5 tissues) were used as the samples for methylation analysis by pyrosequencing. The primer sequences of RT-PCR and MSP are summarized in Table 1.
| Primer | Sequence (5'→3') | Amplicon (bp) |
| RT-PCR | ||
| IRAK3-Forward | ATGCAGTGTAAGAAGCATTGGA | 247 |
| IRAK3-Reverse | GCAGGTAGTGAATGGCTTTGG | |
| GLOXD1-Forward | CCCTTCCTACCCGGCTTCA | 122 |
| GLOXD1-Reverse | TGGAACCAGCGCAAAAGTGT | |
| Pyrosequencing | ||
| GLOXD1-Forward | GAAGGGAGGTTTAGTGTTTAAGGA | 242 |
| GLOXD1-Reverse | AGCTGGACATCACCTCCCACAACGCCACCCCAACCAAAAACA | |
| Universal primer | AGCTGGACATCACCTCCCACAACG-Biotin | |
| Sequencing primer | AGGTTTAGTGTTTAAGGAT | |
| MSP/Q-MSP | ||
| IRAK3-Forward | AGGAGATCGTTTAGTCGTGGGGTAC | 110 |
| IRAK3-Reverse | ACCTCTACGATAAAAACGAAACTACCG | |
| IRAK3-Probe | CTACCGAAACAAACAAAATA | |
| GLOXD1-Forward | AGGATGTGATTAGGCGTGAGGTTC | 122 |
| GLOXD1-Reverse | AAAAAAACGAAACCCGTAACTCCG | |
| GLOXD1-Probe | FAM-CGCTACTCTTTCCCC |
The Taiwan Liver Cancer Network (TLCN) is funded by the National Science Council to provide researchers in Taiwan with primary liver cancer tissues and their associated clinical information. With the approval by the TLCN User Committee and the Institutional Review Board of the Tri-Service General Hospital (TSGH), 29 normal parts of liver hemangiomas (as normal controls) and a total of 160 HCC tissues and their paired adjacent nontumor tissues were used in this study. Among these samples, 40 HCC tissues and their paired adjacent nontumor tissues were obtained from TSGH; the others were obtained from TLCN. These specimens were obtained during surgery, frozen immediately in liquid nitrogen and preserved at -80 °C until DNA extraction. The diagnosis of HCC samples was confirmed by histology. The clinicopathological characteristics of the patients are summarized in Table 2.
| Characteristic | Cases |
| Age, yr | 59 ± 14 |
| Mean ± SD | |
| Gender | |
| Female | 94 |
| Male | 66 |
| Hepatitis | |
| HBV-positive | 68 |
| HCV-positive | 62 |
| Double-negative | 30 |
| Cirrhosis | |
| No | 77 |
| Yes | 80 |
| Unknown | 3 |
| Tumor size, cm | |
| ≤ 3 | 52 |
| > 3 | 108 |
| Nodule | |
| Solitary | 98 |
| Multiple | 62 |
| AFP level, ng/mL | |
| ≤ 10 | 45 |
| > 10 | 113 |
| Unknown | 2 |
| Stage | |
| I | 60 |
| II | 46 |
| III | 47 |
| IV | 7 |
| Invasion | |
| No | 85 |
| Yes | 75 |
| Recurrence | |
| No | 58 |
| Yes | 36 |
| Unknown | 66 |
| Survival | |
| No | 71 |
| Yes | 27 |
| Unknown | 62 |
Genomic DNA from tissue samples was extracted and prepared for sodium bisulfite treatment and methylation analysis as previously described[21]. Pyrosequencing for the methylation levels of 11 CpG sites in a GLOXD1 promoter was carried out using PCR and sequencing primers, as previously described[22]. The primers for pyrosequencing were designed with PyroMark Assay Design 2.0 software (Qiagen, Hilden, Germany) to amplify and sequence bisulfite-treated DNA. PCR was carried out in a 20 μL reaction mix containing 1 μL bisulfite-converted DNA, 2 × RBC SensiZyme HotStart Taq Mastermix (RBC Bioscience Corp., Taipei, Taiwan), and primers using the following program: 95 °C for 15 min, then 49 cycles of 95 °C for 30 s, 62 °C for 30 s and 72 °C for 30 s, with a final extension at 72 °C for 10 min. The biotinylated PCR product was purified by binding to streptavidin-sepharose beads, washed, and denatured. The sequencing primer was then added to the PCR products, and pyrosequencing was performed using the PyroMark Q24 system (Qiagen). Q-MSP was performed in the TaqMan probe system using the LightCycler 480 system (Roche Applied Science, Mannheim, Germany) and prepared as previously described[18]. The COL2A gene was used as an internal reference by amplifying non-CpG sequences. Results with cycle threshold values (Cq values) of COL2A > 38 were defined as detection failures. The DNA methylation level was determined as a methylation index using the following formula: 100 × 2 [(Cq of COL2A) - (Cq of target genes)][23]. Each set of amplifications included a positive control, a negative control, and a non-template control. The primer and probe sequences of pyrosequencing and Q-MSP are summarized in Table 1.
The prism software (version 4.03; Graphpad Software Inc, La Jolla, CA) was used for statistical analyses. The unpaired t-test and paired t-test were used to determine the difference of the methylation index between tissues with different disease status. Fisher’s exact test, χ2 test, and χ2 test for trend were used to evaluate the association between gene methylation and clinical parameters. Pearson correlation was used to compare the consistency of different techniques. Receiver operating characteristic (ROC) curves were generated to determine the optimal cut-off point of gene methylation for discriminating tumors and normal controls. Kaplan-Meier curves were used to estimate survival fraction of patients for 3 years after treatment. Log-rank tests were used to compare the survival of patients with or without gene methylation.
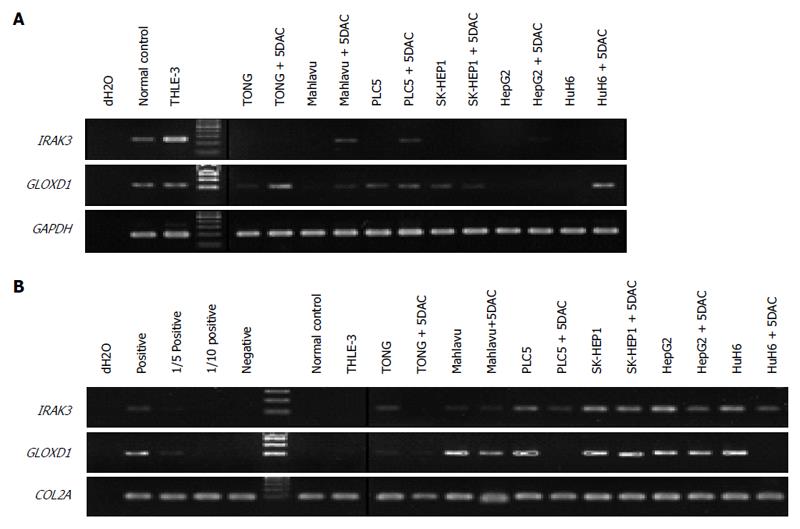
To confirm the results from the methylation array, we first analyzed the correlation between gene expression and promoter methylation of IRAK3 and GLOXD1 in cell lines by RT-PCR and MSP (Figure 1). Expression analysis showed that IRAK3 and GLOXD1 were expressed in normal control and THLE-3 cells but down-regulated in several HCC cell lines (Figure 1A). In addition, the expression of IRAK3 and GLOXD1 was partially restored after treatment with 5DAC (a DNA methyltransferase inhibitor). Methylation analysis revealed that IRAK3 and GLOXD1 methylation was detected mainly in HCC cell lines, and a partial decrease in the methylated band was also observed in the HCC cell lines after 5DAC treatment (Figure 1B). These results implied that IRAK3 and GLOXD1 were down-regulated in HCC cell lines through promoter methylation.
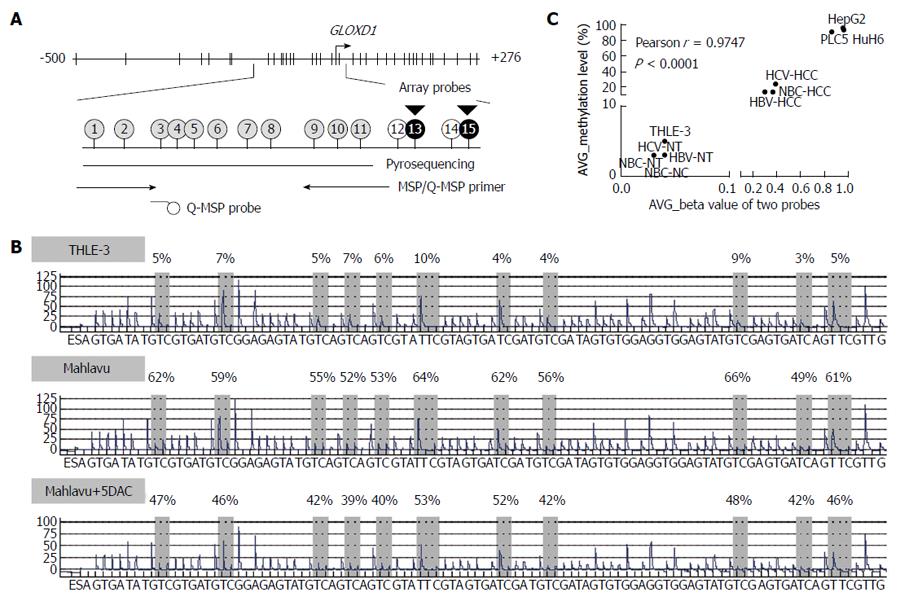
We then confirmed the methylation levels of IRAK3 and GLOXD1 in cell lines and pooled samples by pyrosequencing (Figure 2; GLOXD1 as an example). Methylation levels of 11 CpG sites in GLOXD1 promoter that is close to the two probe sites on array were examined (Figure 2A). It revealed that the GLOXD1 methylation level was 3%-10% in THLE-3 cells and 49%-66% in Mahlavu cells (Figure 2B). Consistent with MSP, a partial decrease in the methylation level of the GLOXD1 promoter was observed in Mahlavu cells after 5DAC treatment (39%-53%). Furthermore, the average β value for different array probes was significantly correlated to the average methylation level of the 11 CpG sites in the samples used in the methylation array (r = 0.9747, Figure 2C). In addition, GLOXD1 methylation was much lower in THLE-3 cells, the pooled normal controls, and each type of pooled nontumor tissues compared to HCC cell lines and all types of pooled tumor tissues. Finally, we designed a primer and probe set based on the CpG methylation results of pyrosequencing to carry out Q-MSP analysis in larger clinical samples.
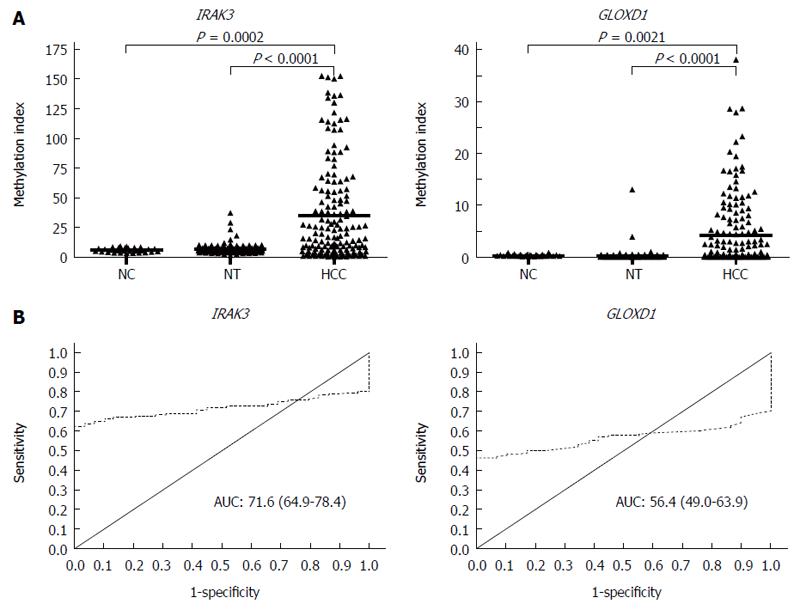
To examine the methylation levels of IRAK3 and GLOXD1 in HCC, we analyzed 29 normal controls, 160 paired HCC tissues, and their adjacent nontumor tissues using Q-MSP (Figure 3). Promoter methylation of IRAK3 and GLOXD1 was both significantly increased in HCC tissues compared to normal controls and nontumor tissues (Figure 3A). Furthermore, to find a best cut-off value for defining methylated cases, ROC curve analysis of each gene was performed to discriminate normal controls and HCC tissues (Figure 3B). As summarized in Table 3, IRAK3 and GLOXD1 methylated cases were mainly present in HCC tissues (102/160, 63.8%; 75/160, 46.9%) compared to normal controls (1/29, 3.4%; 2/29, 6.9%).
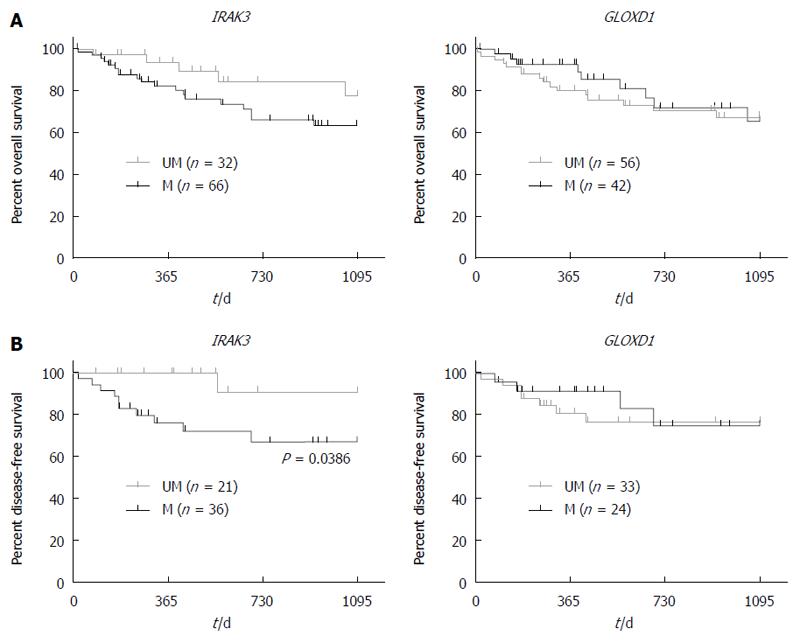
To evaluate the association of gene methylation with clinicopathological characteristics, we analyzed a total of 160 HCC patients (Table 4 and Figure 4). As shown in Table 4, there was a statistically significant correlation between IRAK3 methylation and tumor stage (P = 0.03), but no significant association was shown between GLOXD1 methylation and clinicopathological parameters. As shown in Figure 4, HCC patients with IRAK3 methylation was found to have a trend toward poor 3-year disease-free survival (P = 0.0386, log-rank test) but not in patients with or without GLOXD1 methylation.
| Methylation status | IRAK3, M-Index | P value | GLOXD1, M-Index | P value | ||
| Characteristic | ≤8.90 | > 8.90 | ≤0.60 | > 0.60 | ||
| Cases | 58 | 102 | 85 | 75 | ||
| Age, yr | ||||||
| ≤ 59 | 27 | 47 | 1.00 | 45 | 29 | 0.08 |
| > 59 | 31 | 55 | 40 | 46 | ||
| Gender | ||||||
| Female | 25 | 41 | 0.74 | 39 | 27 | 0.26 |
| Male | 33 | 61 | 46 | 48 | ||
| Hepatitis | ||||||
| HBV-positive | 21 | 47 | 0.32 | 38 | 30 | 0.22 |
| HCV-positive | 23 | 39 | 28 | 34 | ||
| Double-negative | 14 | 16 | 19 | 11 | ||
| Cirrhosis | ||||||
| No | 33 | 44 | 0.07 | 46 | 31 | 0.15 |
| Yes | 23 | 57 | 38 | 42 | ||
| Unknown | 2 | 1 | 1 | 2 | ||
| Tumor size, cm | ||||||
| ≤ 3 | 16 | 36 | 0.38 | 25 | 27 | 0.40 |
| > 3 | 42 | 66 | 60 | 48 | ||
| Nodule | ||||||
| Solitary | 37 | 61 | 0.74 | 53 | 45 | 0.87 |
| Multiple | 21 | 41 | 32 | 30 | ||
| AFP level, ng/mL | ||||||
| ≤ 10 | 14 | 31 | 0.58 | 20 | 25 | 0.16 |
| > 10 | 42 | 71 | 65 | 48 | ||
| Unknown | 2 | 0 | 0 | 2 | ||
| Stage | ||||||
| I | 22 | 38 | 0.03 | 28 | 32 | 0.66 |
| II | 23 | 23 | 26 | 20 | ||
| III | 13 | 34 | 27 | 20 | ||
| IV | 0 | 7 | 4 | 3 | ||
| Invasion | ||||||
| No | 30 | 55 | 0.87 | 39 | 46 | 0.06 |
| Yes | 28 | 47 | 46 | 29 | ||
| Recurrence | ||||||
| No | 22 | 36 | 0.38 | 34 | 24 | 0.83 |
| Yes | 10 | 25 | 19 | 16 | ||
| Unknown | 26 | 41 | 32 | 35 | ||
| Survival | ||||||
| No | 25 | 46 | 0.47 | 38 | 33 | 0.26 |
| Yes | 7 | 20 | 18 | 9 | ||
| Unknown | 26 | 36 | 29 | 33 | ||
Recently, several high-resolution methods for genome-wide methylation analysis have been used in the study of HCC, such as methylated CpG island amplification microarray, bacterial artificial chromosome array-based methylated CpG island amplification, GlodenGate assay, and Infinium assay[8-17]. These results provide evidence that HCC tumors with specific DNA methylation patterns associated with risk factors or progression of HCC have important clinical applications. In our recent study, we also used the Infinium HumanMethylation27 BeadChip to analyze DNA methylation signatures of HCC and found 1968 genes that were hypermethylated in non-tumor tissue and/or tumor tissue with different viral etiologies. Among 34 genes selected for verification, we further identified that methylation of the HOXA9 gene could be a helpful biomarker to assist in HCC detection. In this study, we further identified that two novel genes, IRAK3 and GLOXD1, were frequently methylated in HCC. However, both of these two genes were undetectable in plasma. Moreover, IRAK3 methylation was statistically associated with tumor stage and poor 3-year disease-free survival of HCC patients.
IRAK3 encodes a member of the interleukin-1 receptor-associated kinase protein family that is an essential component of the Toll/IL-R immune signal transduction pathways. This gene is primarily expressed in monocytes and macrophages, and it is also detected in various adult human tissues including the liver[24]. It has been known that IRAK3 functions as a negative regulator in Toll-like receptor signaling and plays an important role in alcohol-induced liver injury[19,25]. In this study, we demonstrated that IRAK3 was mainly methylated in HCC, and its methylation was positively associated with tumor stage and poor 3-year disease-free survival of patients. Furthermore, the inverse correlation between IRAK3 expression and methylation status in HCC cell lines was also observed. Overall, our study indicates that IRAK3 methylation is associated with tumor stage and poor prognosis of patients and also implies that IRAK3 might play an important role in the development of HCC. Confirmation of this hypothesis requires further investigation.
GLOXD1 (the official gene symbol is HPDL) encodes a protein that may function like 4-hydroxyphenylpyruvate dioxygenase. Although the function of GLOXD1 is still unclear, 4-hydroxyphenylpyruvate dioxygenase is known as an important enzyme in the catabolic pathway of tyrosine in the liver, and defects in this gene will cause diseases such as tyrosinemia type 3[20]. Till now, there are no data regarding the GLOXD1 methylation in any cancer, even in HCC. We showed that GLOXD1 expression was down-regulated in HCC cell lines, which was inversely correlated with its methylation status, and GLOXD1 was frequently methylated in HCC tissues. All these results suggest that GLOXD1 expression might be down-regulated in HCC through the promoter methylation. However, the role of GLOXD1 in the development of HCC requires further investigation.
In this study, we used pyrosequencing to verify the actual methylation pattern of CpG sites within the promoter of the target genes, similar to previous studies. Then, we used the results of pyrosequencing to design a Q-MSP system for validation in a large clinical cohort. Therefore, we easily determined the methylation frequency of the target genes in 349 tissue samples, including 29 normal controls and 160 HCC tissues and their paired adjacent nontumor tissues. According to these results, our data indicate that this quantitative methylation analysis workflow is an efficient and economical approach to verify initially and validate further the data from high-throughput screening.
In summary, our data demonstrated that IRAK3 and GLOXD1 were frequently methylated in HCC tissues. Furthermore, IRAK3 methylation was statistically associated with tumor stage and a poor 3-year disease-free survival rate of HCC patients. This indicated that detection of IRAK3 methylation would be helpful in the prediction of patients’ survival as well as the follow-up of patients. Taken together, these findings reveal that methylation of IRAK3 and GLOXD1 has a potential clinical application.
Hepatocellular carcinoma (HCC) is a serious disease because it is difficult to detect and therefore leads to a very poor prognosis and high mortality rates. Studying the molecular mechanisms of HCC development can help us to design better strategies for disease detection or prognosis prediction.
Aberrant DNA methylation is associated with the development of HCC, suggesting that gene methylation could provide potential clinical applications in the diagnosis or prognosis of HCC. The authors’ previously identified that IRAK3 and GLOXD1 were frequently methylated in HCC using a methylation array. However, there are no quantitative data about the methylation level of two novel genes in HCC.
This study demonstrated frequent methylation of two novel genes [interleukin-1 receptor-associated kinase 3 (IRAK3) and GLOXD1] in HCC and further showed the potential value of IRAK3 methylation as a biomarker in the prognosis of HCC.
IRAK3 methylation would be helpful in prediction of patients’ survival as well as the follow-up of patients.
DNA methylation is a common epigenetic event that alters gene expression. Identification of DNA methylation pattern not only allows for a detailed understanding of the hepatocarcinogenesis but also provides potential clinical applications in the diagnosis or prognosis of HCC.
In this study, the authors demonstrated that IRAK3 and GLOXD1 gene expression was down-regulated in HCC cell lines and that it was partially restored after treatment with 5DAC. Importantly, they also found that IRAK3 methylation was statistically associated with tumor stage and with a trend of poor 3-year disease-free survival in HCC samples. Data are very interesting.
| 1. | Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893-2917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11128] [Cited by in RCA: 11886] [Article Influence: 792.4] [Reference Citation Analysis (6)] |
| 2. | Mínguez B, Lachenmayer A. Diagnostic and prognostic molecular markers in hepatocellular carcinoma. Dis Markers. 2011;31:181-190. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 41] [Reference Citation Analysis (1)] |
| 3. | Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683-692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3299] [Cited by in RCA: 3447] [Article Influence: 181.4] [Reference Citation Analysis (0)] |
| 4. | Herath NI, Leggett BA, MacDonald GA. Review of genetic and epigenetic alterations in hepatocarcinogenesis. J Gastroenterol Hepatol. 2006;21:15-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 118] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 5. | Pogribny IP, Rusyn I. Role of epigenetic aberrations in the development and progression of human hepatocellular carcinoma. Cancer Lett. 2014;342:223-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 147] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 6. | Anwar SL, Lehmann U. DNA methylation, microRNAs, and their crosstalk as potential biomarkers in hepatocellular carcinoma. World J Gastroenterol. 2014;20:7894-7913. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 65] [Cited by in RCA: 68] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 7. | Bibikova M, Le J, Barnes B, Saedinia-Melnyk S, Zhou L, Shen R, Gunderson KL. Genome-wide DNA methylation profiling using Infinium® assay. Epigenomics. 2009;1:177-200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 463] [Cited by in RCA: 465] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 8. | Gao W, Kondo Y, Shen L, Shimizu Y, Sano T, Yamao K, Natsume A, Goto Y, Ito M, Murakami H. Variable DNA methylation patterns associated with progression of disease in hepatocellular carcinomas. Carcinogenesis. 2008;29:1901-1910. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 99] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 9. | Arai E, Ushijima S, Gotoh M, Ojima H, Kosuge T, Hosoda F, Shibata T, Kondo T, Yokoi S, Imoto I. Genome-wide DNA methylation profiles in liver tissue at the precancerous stage and in hepatocellular carcinoma. Int J Cancer. 2009;125:2854-2862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 51] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 10. | Archer KJ, Mas VR, Maluf DG, Fisher RA. High-throughput assessment of CpG site methylation for distinguishing between HCV-cirrhosis and HCV-associated hepatocellular carcinoma. Mol Genet Genomics. 2010;283:341-349. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 47] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 11. | Hernandez-Vargas H, Lambert MP, Le Calvez-Kelm F, Gouysse G, McKay-Chopin S, Tavtigian SV, Scoazec JY, Herceg Z. Hepatocellular carcinoma displays distinct DNA methylation signatures with potential as clinical predictors. PLoS One. 2010;5:e9749. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 151] [Cited by in RCA: 156] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 12. | Shin SH, Kim BH, Jang JJ, Suh KS, Kang GH. Identification of novel methylation markers in hepatocellular carcinoma using a methylation array. J Korean Med Sci. 2010;25:1152-1159. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 40] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 13. | Tao R, Li J, Xin J, Wu J, Guo J, Zhang L, Jiang L, Zhang W, Yang Z, Li L. Methylation profile of single hepatocytes derived from hepatitis B virus-related hepatocellular carcinoma. PLoS One. 2011;6:e19862. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 52] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 14. | Ammerpohl O, Pratschke J, Schafmayer C, Haake A, Faber W, von Kampen O, Brosch M, Sipos B, von Schönfels W, Balschun K. Distinct DNA methylation patterns in cirrhotic liver and hepatocellular carcinoma. Int J Cancer. 2012;130:1319-1328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 77] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 15. | Shen J, Wang S, Zhang YJ, Kappil M, Wu HC, Kibriya MG, Wang Q, Jasmine F, Ahsan H, Lee PH. Genome-wide DNA methylation profiles in hepatocellular carcinoma. Hepatology. 2012;55:1799-1808. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 167] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 16. | Shen J, Wang S, Zhang YJ, Wu HC, Kibriya MG, Jasmine F, Ahsan H, Wu DP, Siegel AB, Remotti H. Exploring genome-wide DNA methylation profiles altered in hepatocellular carcinoma using Infinium HumanMethylation 450 BeadChips. Epigenetics. 2013;8:34-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 132] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 17. | Song MA, Tiirikainen M, Kwee S, Okimoto G, Yu H, Wong LL. Elucidating the landscape of aberrant DNA methylation in hepatocellular carcinoma. PLoS One. 2013;8:e55761. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 97] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 18. | Kuo CC, Lin CY, Shih YL, Hsieh CB, Lin PY, Guan SB, Hsieh MS, Lai HC, Chen CJ, Lin YW. Frequent methylation of HOXA9 gene in tumor tissues and plasma samples from human hepatocellular carcinomas. Clin Chem Lab Med. 2014;52:1235-1245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 19. | Wang Y, Hu Y, Chao C, Yuksel M, Colle I, Flavell RA, Ma Y, Yan H, Wen L. Role of IRAK-M in alcohol induced liver injury. PLoS One. 2013;8:e57085. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 18] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 20. | Rüetschi U, Cerone R, Pérez-Cerda C, Schiaffino MC, Standing S, Ugarte M, Holme E. Mutations in the 4-hydroxyphenylpyruvate dioxygenase gene (HPD) in patients with tyrosinemia type III. Hum Genet. 2000;106:654-662. [PubMed] |
| 21. | Shih YL, Shyu RY, Hsieh CB, Lai HC, Liu KY, Chu TY, Lin YW. Promoter methylation of the secreted frizzled-related protein 1 gene SFRP1 is frequent in hepatocellular carcinoma. Cancer. 2006;107:579-590. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 79] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 22. | Liao YP, Chen LY, Huang RL, Su PH, Chan MW, Chang CC, Yu MH, Wang PH, Yen MS, Nephew KP. Hypomethylation signature of tumor-initiating cells predicts poor prognosis of ovarian cancer patients. Hum Mol Genet. 2014;23:1894-1906. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 55] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 23. | Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, Danenberg PV, Laird PW. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. [PubMed] |
| 24. | Nishimura M, Naito S. Tissue-specific mRNA expression profiles of human toll-like receptors and related genes. Biol Pharm Bull. 2005;28:886-892. [PubMed] |
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
P- Reviewer: Alisi A, Lakatos PL, Sacco R S- Editor: Ma YJ L- Editor: Wang TQ E- Editor: Liu XM