Published online Dec 14, 2014. doi: 10.3748/wjg.v20.i46.17498
Revised: May 3, 2014
Accepted: July 24, 2014
Published online: December 14, 2014
Processing time: 263 Days and 22.7 Hours
AIM: To investigate the prognostic role of genomic stability and copy number alterations (CNAs) pancreatic neuroendocrine tumors (PanNETs).
METHODS: A high-resolution array-based comparative genomic hybridization approach was utilized in order to investigate and quantify chromosomal aberrations in a panel of 37 primary PanNET and 11 metastatic samples. DNA samples were extracted from formalin-fixed and paraffin-embedded tumor specimen. Genomic findings were correlated with histopathological and immunohistochemical data. Moreover, the dataset was subjected to employing an unsupervised hierarchical clustering analysis approach utilizing Euclidean distance and average linkage and associations between genomically defined tumor groups and recurrent CNAs or clinicopathological features of the study group were assessed.
RESULTS: Numerous chromosomal aberrations were recurrently detected in both, primary tumor samples and metastases. Copy number gains were most frequently observed at 06p22.2-p22.1 (27.1%), 17p13.1 (20.8%), 07p21.3-p21.2 (18.8%), 09q34.11 (18.8%). Genomic losses were significantly less frequent and the only recurrent aberration affected 08q24.3 (6.3%). Moreover, we detected a high degree of genomic heterogeneity between primary tumors and metastatic lesions. Unsupervised hierarchical clustering of loci affected by CNAs in more than 3 primary tumor samples revealed two genetically distinct tumor groups as well as two chromosomal clusters of genomic imbalances indicating a small subset of tumors with common molecular features (13.5%). Aberrations affecting 6p22.2-22.1, 8q24.3, 9q34.11 and 17p13.1 (P = 0.011; 0.003; 0.003; 0.001), were significantly associated with a poorer survival prognosis.
CONCLUSION: This study suggests that several frequent CNAs in numerous candidate regions are involved in the pathogenesis and metastatic progression of PanNET.
Core tip: In the current study, we characterized the genomic landscape of pancreatic neuroendocrine tumors (PanNETs). Analysis of recurrent genomic amplifications delineated two independent clusters of genomic aberrations as well as two patient groups characterized by significantly overlapping cytogenetic features. Copy number alterations affecting chromosomes 6, 8, 9 and 17 were shown to be associated with survival. A high degree of genomic heterogeneity between primary tumor samples and metastatic lesions demonstrates the need for a more focused molecular and cytogenetic characterization in the light of upcoming targeted therapy approaches. PanNETs appear to be genetically distinct from other types of neuroendocrine tumors.
- Citation: Gebauer N, Schmidt-Werthern C, Bernard V, Feller AC, Keck T, Begum N, Rades D, Lehnert H, Brabant G, Thorns C. Genomic landscape of pancreatic neuroendocrine tumors. World J Gastroenterol 2014; 20(46): 17498-17506
- URL: https://www.wjgnet.com/1007-9327/full/v20/i46/17498.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i46.17498
Pancreatic neuroendocrine tumors (PanNETs) are relatively rare. Their annual incidence, however, has increased substantially over the past two decades and these tumors now represent one of the most common entities of malignant primary pancreatic neoplasia, the second most common after ductal adenocarcinoma[1-3]. Moreover, PanNETs represent the second most common group of clinically relevant neuroendocrine tumors following small intestinal neuroendocrine tumors (midgut carcinoids) and accounting for approximately 7% of all neuroendocrine tumors[4]. PanNETs usually occur sporadically, but they have been described in the context of multiple endocrine neoplasia type 1 (MEN1) and less frequently in other hereditary syndromes, including von Hippel-Lindau and tuberous sclerosis[5]. Although the increasing incidence of PanNET has partially been attributed to both a rise in awareness among physicians and increased sensitivity of diagnostic methodology current numbers may still be an underestimation of the real occurrence. Several autopsy case studies, including extensive histopathological workup of the entire pancreas, revealed a prevalence ranging between approximately 1% and 10%[4,6]. The biological variability within the entity of PanNETs is best illustrated by the variety of clinical manifestations mostly determined by local stage and the specific hormone production of the tumors cell of origin[7]. Thus, tumors are primarily classified as functional or non-functional[8].
Patients with a PanNET frequently present with advanced stage disease including metastatic dissemination at the time of diagnosis. A complete resection, which is considered the therapeutic gold-standard, cannot be achieved in many of these patients[9]. Thus, the five-year survival rate for PanNET patients following surgical resection varies substantially depending on size, grading and proliferative activity of the primary tumor as well as the presence of metastatic disease.
The often low response rates to conventional chemotherapies as well as to novel targeted therapies further necessitate the improvement of our understanding of this heterogeneous entity at a genomic and molecular level[10]. While a number of comprehensive studies have been carried out, employing both array comparative genomic hybridization as well as single-nucleotide polymorphism approaches in order to elucidate cytogenetic aspects of small intestinal neuroendocrine tumors, no such attempts have been published for PanNETs[11-13].
In the present study, we aimed to determine the degree of genomic stability and copy number alterations (CNA) within the group of PanNETs, applying a high resolution array-based comparative genomic hybridization (a-CGH) approach to a panel of 37 primary PanNETs. The second major aim of our work was to assess chromosomal aberrations comparatively between pancreatic primary tumors and metastases (n = 11), in order to identify intraindividual genomic imbalances of potential therapeutic relevance.
Formalin-fixed and paraffin-embedded (FFPE) tissue specimen from 37 patients with PanNET and eleven corresponding metastases (six lymph node, three hepatic and three peritoneal metastases) from seven patients were retrieved from the registry of the Department of Pathology, University Hospital of Schleswig-Holstein, Campus Luebeck. All tissue samples were sent to the Department of Pathology as part of standard clinical care following resection in one of the local surgical departments. All patients underwent surgical resection, which was aimed to be complete. All studies were approved by the Ethics Commission at the University of Luebeck. All samples were revaluated and histopathological diagnosis was established in accordance with the current WHO classification of neuroendocrine tumors[14]. Twenty female and 17 male patients at a median age of 52 were included in the study group.
Immunohistochemical stains were performed on tissue micro arrays according to a standard three-step immunoperoxidase technique utilizing an automated TechMate system (DAKO, Glostrup, Denmark) and the BrightVision Kit (ImmunoLogic, Duiven, Netherlands).
Genomic DNA was obtained from FFPE specimen using the QiaAmp mini kit 250 (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s instructions. DNA concentration and purity was evaluated using a NanoDrop 1000 Spectrophotometer (Thermo Scientific, United States).
An a-CGH approach was applied on 37 PanNETs and 11 corresponding metastases from seven patients using 180K Oligo Arrays (Agilent Technologies Inc., Santa Clara, CA, United States). Male or female reference DNA (Agilent Technologies Inc.) was used in order to assess genomic imbalances (sex-matched). Array slides were analyzed on a Surescan high-resolution DNA micro-array scanner platform (Agilent Technologies Inc.). All procedures were performed according to the manufacturer’s instructions and protocol.
Genomic data were extracted from TIFF files using Feature Extraction and CytoGenomics V. 2.7.08 software (Agilent Technologies Inc.).
Definitions of genomic gain (+0.25), loss (-0.25), amplification (+1) and homozygous loss (-1) were established based on the log2 ratio thresholds and a minimum of three adjacent probes indicating the aberration. Moreover, the significance threshold P-value was set at a minimum of 5.0 × 10-6 as described[15]. Recurrent regions of CNAs were defined as those observed in four or more a-CGH profiles when reviewing both primary tumors and metastases combined[16].
Aberrations resembling known copy number variations in accordance with the UCSC Genome Bioinformatics database (http://genome.uscs.edu) were not reported and thus excluded from further analysis.
For cluster analysis of recurrently observed chromosomal imbalances, a binary matrix was constructed associating each primary tumor sample with each of the recurrently detectable aberrations. The status was coded as “0” for normal copy number or “1” for copy number variations (either gain or loss at a given candidate region) of each tumor sample individually. The matrix was subsequently analyzed employing an unsupervised hierarchical clustering analysis approach utilizing Euclidean distance and average linkage using Genesis 1.7.6[17].
Period of follow up was defined as the time interval between the date of primary diagnosis until time of death or the date of last clinical contact. In order to assess overall survival the log rank test was used and results were illustrated by Kaplan-Meier plots for both tumor groups as well as regions of recurrent genomic imbalance. Associations between genomically defined tumor groups and recurrent CNAs or clinicopathological features of the study group were assessed using the Fisher’s exact test and the Mann-Whitney U test respectively. All analyses were two-sided and the statistical significance level was set to 5% (P < 0.05). All statistical data analyses were performed using GraphPad Prism 5.
Primary tumors had a median size of 2.3 cm and median proliferative activity was < 1 mitosis per high-power field and 1.2% of Ki-67 positive staining cells. In accordance with the 2010 WHO classification of neuroendocrine tumors, 28 samples were diagnosed as NET G1, 8 tumors were NET G2 and one case was classified as NET G3. Clinical data were available for 29/37 patients with a median follow-up of 20 months. No significant difference in proliferative activity between primary tumors and metastatic lesions was observed (P = 0.21). A brief overview of all cases and corresponding metastatic lesions included in the current study is given in Tables 1 and 2.
| Case No. | Sex | Age | Diagnosis (WHO 2010) | Mitotic score | Ki67 | Tumor size (cm) | TNM (UICC) |
| P01 | F | 45 | NET G1 | < 1 | < 1% | 3.5 | pT2, pN0(0/10), pM1, L0, V0, pN0 |
| P02 | F | 77 | NET G2 | 2 | 3% | 14 | pT3, pN1, pMx, L1, V1, pN0 |
| P03 | M | 45 | NET G1 | < 1 | 1% | 2.3 | pT2, pNx, pMx, L0, V0, pN0 |
| P04 | M | 57 | NET G1 | 1 | 10% | 3.8 | pT3, pN0, pM1, L1, V0, pN0, G1 |
| P05 | M | 50 | NET G1 | < 1 | 0.4% | 1.1 | pT1, pNx, pMx, L0, V0, pN0, G1 |
| P06 | M | 61 | NET G1 | < 1 | 2.6% | 5 | pT3, pN0 (0/5), pMx, L0, V0, pN0, G1 |
| P07 | F | 77 | NET G3 | 25 | 35% | 10 | pT3, pN0, pMx, L1, V0, pN0 |
| P08 | M | 58 | NET G1 | < 1 | < 1% | 0.3 | pT1, pNx, pMx, L0, V0, pN0, G1 |
| P09 | M | 33 | NET G2 | 2 | 4.7% | 2.8 | pT2, pN1 (2/26), pMx, L1, V0, pN0, G2 |
| P10 | M | 50 | NET G2 | 5 | 12.4% | > 20 | pT4, pNx, pM1, L1, V1, pN1, G2 |
| P11 | F | 16 | NET G2 | 3 | 16.1% | 5 | pT3, pN1 (19/68), pM1, L1, V0, pN1, G2 |
| P12 | F | 33 | NET G2 | 2 | 1.2% | 2 | pT1, pN0, pMx, L0, V0, pN0, G2 |
| P13 | F | 38 | NET G2 | 2 | 7% | 5.5 | pT3, pN1 (2/15), pM1, L1, V0, pN1, G2 |
| P14 | M | 58 | NET G2 | 10 | 6.5% | 4.5 | pT3, pN1(8/23), pMx, L0, V1, pN1, G2 |
| P15 | M | 72 | NET G1 | < 1 | 1.5% | 1.5 | pT1, pNx, pM1, L0, V0, pN0, G1 |
| P16 | F | 60 | NET G1 | < 1 | 0.5% | 1.1 | pT1, pN0 (0/6), pMx, L0, V0, pN0, G1 |
| P17 | F | 77 | NET G1 | < 1 | 0.8% | 1 | pT1, pNx, pMx, L0, V0, pN0 ,G1 |
| P18 | F | 49 | NET G1 | < 1 | 1.4% | 2.7 | pT2, pN0 (0/5), pMx, L1, V0, pN0, G1 |
| P19 | F | 52 | NET G2 | 5 | 14.6% | 4 | pT3, pN0, pM1, L1, V1, pN1, G2 |
| P20 | F | 46 | NET G1 | 1 | 6% | 9.5 | pT2, pN0(0/9), pMx,L1,V1,pN0, G1 |
| P21 | M | 55 | NET G1 | < 1 | 2.9% | 3 | pT3, pN1 /1/15), pM1, V0, L1, pN0, G1 |
| P22 | M | 31 | NET G1 | 1 | 2.2% | 0.9 | pT1, pN0 (0/5), pMx, L0, V0, pN0, G1 |
| P23 | M | 56 | NET G1 | < 1 | 1% | 0.4 | pT1, pN0, pM0, L0, V0, pN0, G1 |
| P24 | M | 42 | NET G1 | 0 | 0.3% | 0.2 | pT1(m), pN0 (0/3), pMx, L0,V0, pN0, G1 |
| P25 | M | 68 | NET G1 | < 1 | 1.2% | 2.5 | pT2, pNx, pMx, L0, V0, pN0, G1 |
| P26 | F | 33 | NET G1 | < 1 | 1.3% | 2 | pT1, pNx, pMx, L0, V0, pN0, G1 |
| P27 | F | 50 | NET G1 | < 1 | 2% | 2.7 | pT2, pNx, pMx, L0, V0, pN0, G1 |
| P28 | M | 59 | NET G1 | 1 | 1.2% | 3.5 | pT2, pN1(1/20), pMx, L1, V1, pN0, G1 |
| P29 | F | 57 | NET G1 | < 1 | 1.5% | 0.8 | pT1, pN0, pMx, L0, V0, pN1, G1 |
| P30 | F | 59 | NET G1 | < 1 | 1% | 2.1 | pT2, pN0 (0/1), pMx L0, V0, pN0, G1 |
| P31 | M | 86 | NET G1 | < 1 | 1% | 0.7 | pT1(m), pNx, pMx, L0, V0, pN0, G1 |
| P32 | F | 81 | NET G1 | < 1 | 0% | 0.8 | pT1, pN0 (0/14), pMx, L0, V0, pN0, G1 |
| P33 | F | 51 | NET G1 | 1 | 0.5% | 9 | pT2, pN1 (7/26), pMx, L1, V0, pN0, G1 |
| P34 | F | 45 | NET G1 | 1 | 0.9% | 2 | pT1(m), pN0, pM1, L0, V0, pN0, G1 |
| P35 | F | 41 | NET G1 | < 1 | < 1% | 2 | pT1, pNx, pMx, L0, V0, pN0, G1 |
| P36 | F | 40 | NET G1 | < 1 | 0.8% | 0.6 | pT1, pN0 (0/21), pM0, L0, V0, pN0, G1 |
| P37 | M | 70 | NET G1 | < 1 | 1% | 0.5 | pT1, pN0 (0/12), pM0, L0, V0, pN0 |
| Case No. | Primary tumor | Localization | Mitotic score | Ki67 | Number of CNAs in primary tumor | Number of CNAs in metastases |
| M_P8.1 | P08 | LN | < 1 | 1% | 0 | 7 |
| M_P8.2 | P08 | LN | 2 | 4% | 0 | 4 |
| M_P10.1 | P10 | LN | 2 | 7% | 22 | 5 |
| M_P10.2 | P10 | Peritoneal | 3 | 21% | 19 | 7 |
| M_P12.1 | P12 | LN | 2 | 4% | 28 | 4 |
| M_P12.2 | P12 | Liver | < 1 | < 1% | 28 | 0 |
| M_P13.1 | P13 | Liver | 5 | 15% | 1 | 37 |
| M_P32.1 | P32 | LN | < 1 | 1% | 56 | 70 |
| M_P32.2 | P32 | Peritoneal | < 1 | 1% | 59 | 14 |
| M_P33.1 | P33 | Peritoneal | < 1 | 1% | 6 | 2 |
| M_P36.1 | P36 | Liver | 8 | 20% | 11 | 3 |
We performed genome-wide high-resolution screening for CNAs in 37 PanNET primary tumor samples and eleven corresponding metastases from seven patients employing an array-based comparative genomic hybridization approach.
CNAs were detected in all samples at an average of 30.35 CNAs per sample in primary tumors and 43.09 in metastases. Yet, this difference in CNA frequency (P = 0.16) failed to reach statistical significance, potentially due to the size of the study group. The highest number of independent CNAs was found in P6 (n = 152), whereas P24 revealed only one CNA.
Most genomic losses and gains displayed extensive aberrations recurrently affecting total or near total chromosomes and involving a mean of 1212 probes per CNA in primary tumors and 1288 in metastases. Gains were more frequent than losses in both primary tumors (2.66:1) and metastases (6.68:1).
Recurrent copy number gains were observed on chromosome 6 at p22.2-p22.1 (all tumors: 13/48, 27.1%; primary tumors: 9/37, 24.3%; metastases 4/11, 36.4%), chromosome 17 at p13.1 (all tumors: 10/48, 20.8%; primary tumors: 7/37, 18.9%; metastases 3/11, 27.3%), chromosome 7 at p21.3-p21.2 (all tumors 9/48, 18.8%; primary tumors: 6/37, 16.2%; metastases 3/11, 27.3%) and chromosome 9 at q34.11 (all tumors 9/48, 18.8%; primary tumors: 6/37, 16.2%; metastases 3/11, 27.3%). Several genes with integral functions in oncogenesis and biological processes including regulation of development, differentiation, growth and apoptosis are located at these loci. Affected genes of specific interest in the context of PanNET include regulatory genes controlling transcription, intracellular signaling and epigenetic regulation e.g., EFNA1, MUC1, ABT1, ZNF322A, DGKB, DMTF1, CBFB, CLDN7 and HIST-family genes. Several genes implicated in cell-cell adhesion and migration activity such as ADAM 15, THBS3, CD36, HGF, DNM1 and ELMO3 exhibited significant amplification. Moreover, multiple genes involved in cell cycle regulation, growth and proliferation were affected by copy number gains, including CKS1B, FGF11, ETV1 and TNFSF12.
Copy number gains were detectable at loci known to include HFE and TP53.
Genomic losses were far less common among both primary PanNET tumor samples as well as metastases and the only regions affected in more than one sample were 08q24.3 (3/48, 8.1%), 11q13.1-q13.2 (2/37, 5.4%) and 16p11.2 (2/37, 5.4%). Of note, none of these aberrations were detectable in metastatic lesions.
Genes affected by these aberrations with potential implications in PanNET include genes with functions in intracellular signaling such as ARL2, BANF1, epigenetic regulators of gene expression including BRMS1 and genes implicated in inflammation and tumorgenesis as well as cytosceletal properties, e.g., CFL1. Notably, four patients presented with copy number alterations (two losses and two gains) affecting the MEN1 gene and CD248 (potentially implicated in tumorgenesis), one of which exhibited an additional MEN1 germline mutation. Recurrent aberrations are summarized in Table 3.
| Recurrent region | Primary tumor | Metastases | All tumors |
| n | 37 | 11 | 48 |
| Gains | |||
| 01q21.3-q22 | 4 (10.8) | 1 (9.1) | 5 (10.4) |
| 03q24 | 4 (10.8) | 0 (0) | 4 (8.3) |
| 06p22.2-p22.1 | 9 (24.3) | 4 (36.4) | 13 (27.1) |
| 07p21.3-p21.2 | 6 (16.2) | 3 (27.3) | 9 (18.8) |
| 07q21.11-q21.12 | 2 (5.4) | 3 (27.3) | 5 (10.4) |
| 08q24.3 | 4 (10.8) | 1 (9.1) | 5 (10.4) |
| 09q34.11 | 6 (16.2) | 3 (27.3) | 9 (18.8) |
| 11q13.1-q13.2 | 2 (5.4) | 2 (18.2) | 4 (8.3) |
| 16p11.2 | 2 (5.4) | 3 (27.3) | 5 (10.4) |
| 16q22.1 | 5 (13.5) | 1 (9.1) | 6 (12.5) |
| 17p13.1 | 7 (18.9) | 3 (27.3) | 10 (20.8) |
| 18q12.1 | 5 (13.5) | 0 (0) | 5 (10.4) |
| 19p13.3 | 4 (10.8) | 0 (0) | 4 (8.3) |
| 22q13.33 | 1 (2.7) | 2 (18.2) | 3 (6.3) |
| Losses | |||
| 08q24.3 | 3 (8.1) | 0 (0) | 3 (6.3) |
| 11q13.1-q13.2 | 2 (5.4) | 0 (0) | 2 (4.2) |
| 16p11.2 | 2 (5.4) | 0 (0) | 2 (4.2) |
| 16q22.1 | 1 (2.7) | 0 (0) | 1 (2.1) |
| 17p13.1 | 1 (2.7) | 0 (0) | 1 (2.1) |
| 22q13.33 | 0 (0) | 1 (9.1) | 1 (2.1) |
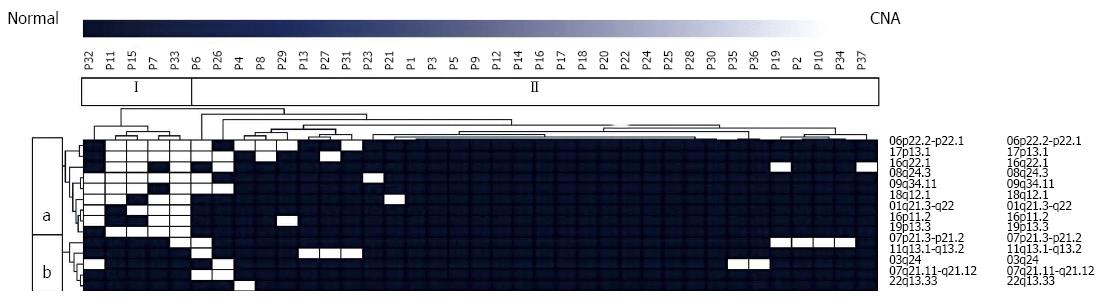
Genomic data of primary PanNETs obtained by a-CGH were subjected to unsupervised hierarchical cluster analysis in order to detect sub-groups of tumors exhibiting a significant overlap with regard to recurrently observed CNAs. Two genomically distinct types of PanNET were identified. Tumor group I included 5 tumors and group II consisted of 32 tumor samples.
Moreover, we detected two distinct chromosomal clusters (a, b). Cluster a consisted of 9 loci recurrently harboring aberrations whereas cluster b included 5 regions with frequent genomic imbalances (Figure 1).
Tumor group I was significantly enriched for all aberrations included in chromosomal cluster a. 6p22.2, p22.1 (P = 0.0084), 17p13.1 (P = 0.0025), 16q22.1 (P = 0.0224), 8q24.3 (P < 0.0001), 9q34.11 (P = 0.0011), 18q12.1 (P = 0.0004), 1q21.2, 22 (P < 0.0001), 16p11.2 (P = 0.0049), 19p13.3 (P < 0.0001). With the exception of CNAs at chromosomes 1 and 19 all of the above were also detected, albeit at a much lower frequency in isolated cases of tumor group II.
There was a significant overlap between CNAs and their frequencies in primary tumor samples and metastases. With the exception of aberrations on chromosomes 3, 18 and 19, which were observed in a minor subset of primary tumor samples (10.8%, 13.5% and 10.8%, respectively), all CNAs were found in both groups and no significant differences with regard to their frequency were identified.
Subjecting couples of primary tumors and metastases to comparative analysis, however, we detected a significant degree of heterogeneity with an average of only 9.33% of shared aberrations. Analyzing the four cases with two available metastases, a high degree of intermetastatic genomic variability became apparent as well. Moreover, no genomic losses were reproducibly detected in samples of both primary tumors and metastases.
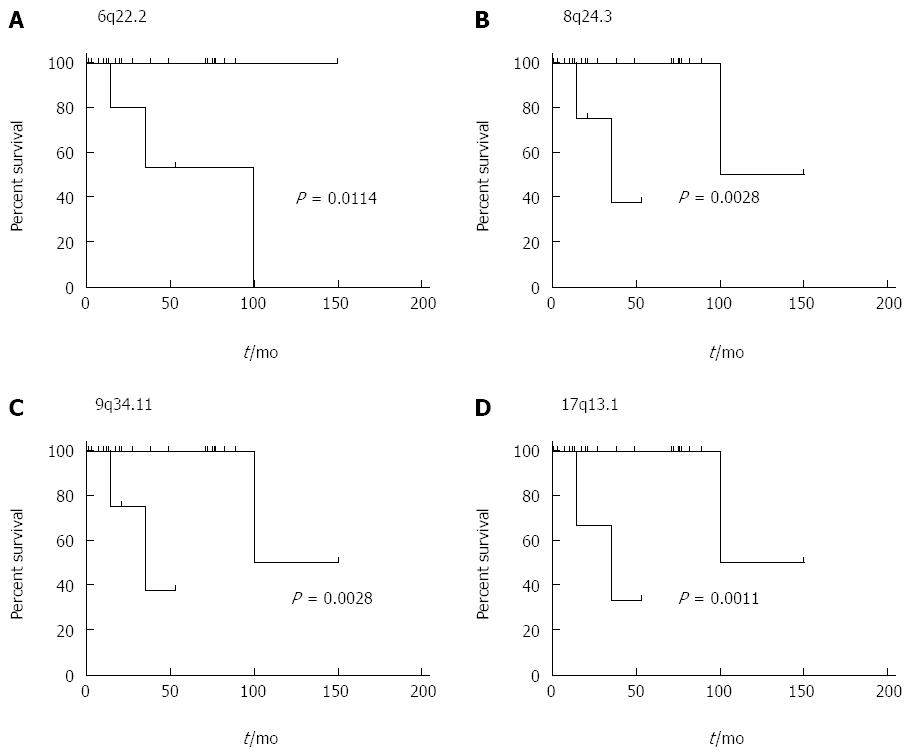
We further investigated potential associations between recurrent chromosomal aberrations and patient survival employing the Mantel-Cox test. CNAs affecting 6p22.2-22.1, 8q24.3, 9q34.11 and 17p13.1 were shown to be associated with a significantly worse survival (P = 0.0114; 0.0028; 0.0028; 0.0011). Moreover, patients harboring CNAs at 6p22.2-22.1 were significantly older and patients showing CNAs at 6p22.2-22.1 and 17p13.1 displayed a trend towards accelerated mitotic activity assessed by both count of mitotic figures per 10 high-power fields as well as percentage of MIB1 positive staining cells. These trends however failed to reach statistical significance. The impact of CNA affecting 6p22.2-22.1, 8q24.3, 9q34.11 and 17p13.1 on overall survival are visualized in Figure 2.
In recent years, several studies have demonstrated the need for a paradigm shift in the definition of pathological entities, changing from primarily histopathological to genomic and molecular based criteria. Through the application of increasingly refined cytogenetic and molecular approaches, significant intraindividual and more recently, intratumor heterogeneity on a genomic and molecular level have become apparent in samples previously indistinguishable by means of microscopic evaluation[18]. These developments demonstrate the need for a novel cytogenetic and molecular definition of a patient’s malignancy, facilitating not only individually but intraindividually customized targeted therapy approaches in order to devise therapeutic concepts for targeting multiple sub clones in a patient.
In the current study we report, for the first time, on genetic findings in an extensive cohort of PanNET patients with special consideration of differences and similarities between primary tumor samples and metastases.
CNAs were detected in all samples with a slightly increased frequency in metastases compared to primary tumors. This trend, however, failed to reach statistical significance, potentially due to the relatively small size of the study group. These observations appear to be consistent with the mild increase in proliferative activity between the two groups.
Copy number gains were most frequently observed on chromosomes 6, 7, 9 and 17. Genomic losses were significantly less frequent and the only recurrent aberration affected 08q24.3 (3/48, 6.3%).
Gains affecting the 6p22.2-22.1 region were present in 24% of the primary tumor samples and 36% of metastases included in the study group and thus constitute the most prominent feature of PanNETs in our data set. Genes affected by this recurrent aberration include ABT1 and ZNF322A as well as the histone gene cluster. Similar aberrations have been implicated in the recurrence of menigiomas[19]. The significance and clinical impact of these findings is further emphasized by the apparent effect of this chromosomal lesion on overall survival as well as proliferative activity of the tumor.
Amplifications on the short arm of chromosome 7 have previously been implicated in a variety of malignant tumors including several types of sarcomas and carcinomas[20,21]. Candidate genes in this region with a possible role in PanNET pathogenesis and progression include CD36 as a prominent adhesion molecule and hepatocyte growth factor, a regulatory protein implicated in cell growth, proliferation, motility and matrix invasion rendering it a central element in tumorogenesis and tumor dissemination[22,23].
The 8q24.3 locus was altered in approximately 20% of both primary tumor samples and metastases. A more refined investigation of the detectable aberrations, however, revealed no complete overlap between these lesions and both losses and gains were found. Thus, the pathophysiological implications of genomic gains within this region remain widely obscure and need to be clarified in future studies on more extensive cohorts.
Genomic imbalances located on the long arm of chromosome 9 were previously reported in atypical lung carcinoids and post-irradiation thyroid carcinomas[24,25].
Although deletions and/or mutations affecting the 17p13.1 locus and TP53 as its most prominent gene are by far the most recurrent features in malignancies, genomic amplifications affecting this region have been described for a variety of tumors including bladder cancer as well[26,27].
In addition, we identified 11q13.1-q13.2 as a region of elevated genomic instability in PanNETs with both copy number losses and gains being frequently detectable in the present study. A more refined investigation revealed it to contain several candidate genes with potential implications in PanNET tumor development including MEN1 and endosialin (CD248)[28]. In addition to its role in hereditary endocrine disorders, MEN1 was recently characterized as a somatic mutational hot-spot and a recurrent site of germline and/or somatic deletions in PanNETs and other malignancies[29,30]. Copy number gains encompassing this region have been implicated in tumor development as well, albeit far less frequently[31].
The elevated frequency of aberrations affecting chromosomes 6, 7, 9, 11, 16, 17 and 22 when comparing primary tumor samples and metastatic lesions is suggestive of potential implications of these aberrations with disease progression and predisposes them as regions of interest for future drug design approaches.
Our findings of CNAs affecting chromosomes 3, 18 and 19 in a subset of primary tumors but not in metastases suggest a role of these CNAs in tumor initiation but not in further tumor growth and spread.
Interestingly, we detected several recurrent genomic imbalances; none of these however was detectable in more than one fourth of all primary samples included in this study. From this observation we deduce the possibility that the aberrations we identified constitute recurrent secondary genetic defects associated with distinct aspects of biological behavior.
Unsupervised hierarchical cluster analysis revealed two independent and genomically distinct sub-groups of tumors exhibiting a significant overlap with regard to recurrently observed CNAs. Moreover, two distinct chromosomal clusters (a, b) were detected. These clusters of recurrent CNAs are suggestive of distinct subsets of secondary genomic imbalances associated with different types of biological behavior within the heterogeneous group of PanNETs. Future, and if possible functional studies will be needed to clarify this complex issue, especially in the light of therapeutic applicability.
Interestingly, one of the tumor groups was significantly enriched for all aberrations included in chromosomal cluster a. It is tempting to speculate, that these findings are suggestive of a small proportion of PanNETs sharing a distinct subset of chromosomal aberrations.
With regard to the association of CNAs affecting 6p22.2-22.1, 8q24.3, 9q34.11 and 17p13.1 with reduced overall survival, the relatively small size of the study cohort and the generally favorable prognosis of the entity have to be taken into account. As the accuracy of Kaplan-Meier calculations is highly dependent on a sufficiently large number of cases, our findings need to be confirmed in further studies including larger cohorts of patients and a longer period of follow-up in order to better determine the prognostic potential and implications of these cytogenetic aberrations in the context of PanNETs. A trend associating said tumor group with worse overall survival failed to reach statistical significance in the current study. Yet again we believe that the investigation of a more extensive cohort is likely to clarify this issue and contribute to further cytogenetic classification of panNET, as well as future prognostic strategy development. Another interesting aspect would be the prognostic impact of CNAs detected in primary tumor samples and their potential association with later metastatic progression. Such a concept could not be derived from our current data set; most likely due to the abovementioned limitations of the study.
Mutations in death domain-associated protein gene (DAXX) or ATR-X gene (ATRX) (which both encode proteins involved in chromatin remodeling) have been detected in 40% of PanNETs, in association with activation of alternative lengthening of telomeres. Moreover, Marinoni et al[32], recently proposed a concept associating loss of DAXX and/or ATRX with tumor stage and metastasis, reduced time of relapse-free survival, and decreased time of tumor-associated survival. We were unable to reproduce these observations in our current study, however, the smaller size of the study group as well as the overall lower grading of cases included in our investigations is to be considered in this context.
In comparison to recently published data on CNAs in small intestinal neuroendocrine tumors (SI-Nets), our results reveal a low degree of accordance between these two entities as none of the prominent features of SI-NETs (including genomic losses affecting chromosomes 18 and 16 as well as gains on chromosomes 4, 5, 7 and 14) were detectable in a significant subset of PanNET samples[33,34]. Thus, PanNETs and SI-NETs seem to be very different from a genomic point of view. Moreover, we believe that these results add further evidence to the previously postulated necessity of a more refined molecular based diagnostic algorithm for malignant neoplasia.
In summary, we identified multiple recurrent genomic amplifications delineating two independent clusters of genomic aberrations as well as two groups of primary PanNETs characterized by significantly overlapping cytogenetic features. Several of the genomic regions affected by CNAs were shown to encompass potent tumor suppressors and oncogenes, hinting at a significant translational potential of our findings for the development of more refined therapy approaches.
CNAs affecting chromosomes 6, 8, 9 and 17 in primary tumor samples were shown to be associated with significant impairment of overall survival. The high degree of genomic heterogeneity between primary tumor samples and multiple metastatic lesions demonstrate the need for a more refined molecular characterization in the light of upcoming targeted therapy approaches. PanNETS appear to be genetically distinct from other types of neuroendocrine tumors.
We thank Annette Aufseß and Tanja Oeltermann for their skilled and dedicated technical assistance.
Pancreatic neuroendocrine tumors (PanNETs) are still considered a rare entity. The annual incidence, however, has increased substantially over the past two decades.
The biological characterization of PanNETs as well as the prognostic role of genomic stability and copy number alterations (CNAs), detectable by array-based comparative genomic hybridization (a-CGH), remains widely rudimental.
In comparison to recently published data on CNAs in small intestinal neuroendocrine tumors which have greatly advanced the understanding of this entity, the results reveal a low degree of accordance between these two types of neuroendocrine neoplasia.
In summary, authors identified multiple recurrent genomic amplifications delineating two independent clusters of genomic aberrations as well as two groups of primary PanNETs characterized by significantly overlapping cytogenetic features. Several of the CNAs detected in this study are shown to encompass potent tumor suppressors and oncogenes, rendering them potentially vital targets for future therapeutic approaches.
The authors employed high resolution a-CGH to reveal aberrant chromosomal copy number in primary pancreatic neuroendocrine tumors and corresponding metastases. This is a very interesting article illustrating the use of genomic analysis to help predict clinical outcome. This study provides essential and previously unreported information of genomic alteration present in PanNETs and identifies critical genomic region for further investigation of their role in PanNET.
| 1. | Metz DC, Jensen RT. Gastrointestinal neuroendocrine tumors: pancreatic endocrine tumors. Gastroenterology. 2008;135:1469-1492. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 637] [Cited by in RCA: 552] [Article Influence: 30.7] [Reference Citation Analysis (2)] |
| 2. | Oberg K. Pancreatic endocrine tumors. Semin Oncol. 2010;37:594-618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 122] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 3. | Yao JC, Hassan M, Phan A, Dagohoy C, Leary C, Mares JE, Abdalla EK, Fleming JB, Vauthey JN, Rashid A. One hundred years after “carcinoid”: epidemiology of and prognostic factors for neuroendocrine tumors in 35,825 cases in the United States. J Clin Oncol. 2008;26:3063-3072. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3022] [Cited by in RCA: 3320] [Article Influence: 184.4] [Reference Citation Analysis (0)] |
| 4. | Sadaria MR, Hruban RH, Edil BH. Advancements in pancreatic neuroendocrine tumors. Expert Rev Gastroenterol Hepatol. 2013;7:477-490. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 5. | Francalanci P, Diomedi-Camassei F, Purificato C, Santorelli FM, Giannotti A, Dominici C, Inserra A, Boldrini R. Malignant pancreatic endocrine tumor in a child with tuberous sclerosis. Am J Surg Pathol. 2003;27:1386-1389. [PubMed] |
| 6. | Kimura W, Kuroda A, Morioka Y. Clinical pathology of endocrine tumors of the pancreas. Analysis of autopsy cases. Dig Dis Sci. 1991;36:933-942. [PubMed] |
| 7. | Reid MD, Balci S, Saka B, Adsay NV. Neuroendocrine tumors of the pancreas: current concepts and controversies. Endocr Pathol. 2014;25:65-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 87] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 8. | Wang SE, Su CH, Kuo YJ, Shyr YM, Li AF, Chen TH, Wu CW, Lee CH. Comparison of functional and nonfunctional neuroendocrine tumors in the pancreas and peripancreatic region. Pancreas. 2011;40:253-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 31] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 9. | Burns WR, Edil BH. Neuroendocrine pancreatic tumors: guidelines for management and update. Curr Treat Options Oncol. 2012;13:24-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 70] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 10. | Kouvaraki MA, Ajani JA, Hoff P, Wolff R, Evans DB, Lozano R, Yao JC. Fluorouracil, doxorubicin, and streptozocin in the treatment of patients with locally advanced and metastatic pancreatic endocrine carcinomas. J Clin Oncol. 2004;22:4762-4771. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 433] [Cited by in RCA: 411] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 11. | Andersson E, Swärd C, Stenman G, Ahlman H, Nilsson O. High-resolution genomic profiling reveals gain of chromosome 14 as a predictor of poor outcome in ileal carcinoids. Endocr Relat Cancer. 2009;16:953-966. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 62] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 12. | Kytölä S, Höög A, Nord B, Cedermark B, Frisk T, Larsson C, Kjellman M. Comparative genomic hybridization identifies loss of 18q22-qter as an early and specific event in tumorigenesis of midgut carcinoids. Am J Pathol. 2001;158:1803-1808. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 98] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 13. | Zhao J, de Krijger RR, Meier D, Speel EJ, Saremaslani P, Muletta-Feurer S, Matter C, Roth J, Heitz PU, Komminoth P. Genomic alterations in well-differentiated gastrointestinal and bronchial neuroendocrine tumors (carcinoids): marked differences indicating diversity in molecular pathogenesis. Am J Pathol. 2000;157:1431-1438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 66] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 14. | Bosman FT, Carneiro F, Hruban RH, Theise ND. WHO Classification of Tumours of the Digestive System. 4th Ed ed. Lyon: IARC Press 2010; . |
| 15. | Salawu A, Ul-Hassan A, Hammond D, Fernando M, Reed M, Sisley K. High quality genomic copy number data from archival formalin-fixed paraffin-embedded leiomyosarcoma: optimisation of universal linkage system labelling. PLoS One. 2012;7:e50415. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 17] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 16. | Rouveirol C, Stransky N, Hupé P, Rosa PL, Viara E, Barillot E, Radvanyi F. Computation of recurrent minimal genomic alterations from array-CGH data. Bioinformatics. 2006;22:849-856. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 61] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 17. | Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207-208. [PubMed] |
| 18. | Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883-892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6102] [Cited by in RCA: 6087] [Article Influence: 434.8] [Reference Citation Analysis (0)] |
| 19. | Pérez-Magán E, Rodríguez de Lope A, Ribalta T, Ruano Y, Campos-Martín Y, Pérez-Bautista G, García JF, García-Claver A, Fiaño C, Hernández-Moneo JL. Differential expression profiling analyses identifies downregulation of 1p, 6q, and 14q genes and overexpression of 6p histone cluster 1 genes as markers of recurrence in meningiomas. Neuro Oncol. 2010;12:1278-1290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 45] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 20. | Aragane H, Sakakura C, Nakanishi M, Yasuoka R, Fujita Y, Taniguchi H, Hagiwara A, Yamaguchi T, Abe T, Inazawa J. Chromosomal aberrations in colorectal cancers and liver metastases analyzed by comparative genomic hybridization. Int J Cancer. 2001;94:623-629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 59] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 21. | Schmidt H, Taubert H, Würl P, Kappler M, Lange H, Bartel F, Bache M, Holzhausen HJ, Hinze R. Gains of 12q are the most frequent genomic imbalances in adult fibrosarcoma and are correlated with a poor outcome. Genes Chromosomes Cancer. 2002;34:69-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 22. | Hale JS, Li M, Sinyuk M, Jahnen-Dechent W, Lathia JD, Silverstein RL. Context dependent role of the CD36--thrombospondin--histidine-rich glycoprotein axis in tumor angiogenesis and growth. PLoS One. 2012;7:e40033. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 23. | Wu X, Chen X, Zhou Q, Li P, Yu B, Li J, Qu Y, Yan J, Yu Y, Yan M. Hepatocyte growth factor activates tumor stromal fibroblasts to promote tumorigenesis in gastric cancer. Cancer Lett. 2013;335:128-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 79] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 24. | Kimmel RR, Zhao LP, Nguyen D, Lee S, Aronszajn M, Cheng C, Troshin VP, Abrosimov A, Delrow J, Tuttle RM. Microarray comparative genomic hybridization reveals genome-wide patterns of DNA gains and losses in post-Chernobyl thyroid cancer. Radiat Res. 2006;166:519-531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 21] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 25. | Swarts DR, Claessen SM, Jonkers YM, van Suylen RJ, Dingemans AM, de Herder WW, de Krijger RR, Smit EF, Thunnissen FB, Seldenrijk CA. Deletions of 11q22.3-q25 are associated with atypical lung carcinoids and poor clinical outcome. Am J Pathol. 2011;179:1129-1137. [PubMed] |
| 26. | Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413-431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2178] [Cited by in RCA: 2507] [Article Influence: 147.5] [Reference Citation Analysis (0)] |
| 27. | Gallucci M, Guadagni F, Marzano R, Leonardo C, Merola R, Sentinelli S, Ruggeri EM, Cantiani R, Sperduti I, Lopez Fde L. Status of the p53, p16, RB1, and HER-2 genes and chromosomes 3, 7, 9, and 17 in advanced bladder cancer: correlation with adjacent mucosa and pathological parameters. J Clin Pathol. 2005;58:367-371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 28. | Valdez Y, Maia M, Conway EM. CD248: reviewing its role in health and disease. Curr Drug Targets. 2012;13:432-439. [PubMed] |
| 29. | Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, Schulick RD, Tang LH, Wolfgang CL, Choti MA. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199-1203. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1432] [Cited by in RCA: 1379] [Article Influence: 91.9] [Reference Citation Analysis (0)] |
| 30. | Raef H, Zou M, Baitei EY, Al-Rijjal RA, Kaya N, Al-Hamed M, Monies D, Abu-Dheim NN, Al-Hindi H, Al-Ghamdi MH. A novel deletion of the MEN1 gene in a large family of multiple endocrine neoplasia type 1 (MEN1) with aggressive phenotype. Clin Endocrinol (Oxf). 2011;75:791-800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 31. | Paris PL, Sridharan S, Hittelman AB, Kobayashi Y, Perner S, Huang G, Simko J, Carroll P, Rubin MA, Collins C. An oncogenic role for the multiple endocrine neoplasia type 1 gene in prostate cancer. Prostate Cancer Prostatic Dis. 2009;12:184-191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 17] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 32. | Marinoni I, Kurrer AS, Vassella E, Dettmer M, Rudolph T, Banz V, Hunger F, Pasquinelli S, Speel EJ, Perren A. Loss of DAXX and ATRX are associated with chromosome instability and reduced survival of patients with pancreatic neuroendocrine tumors. Gastroenterology. 2014;146:453-460.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 347] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 33. | Hashemi J, Fotouhi O, Sulaiman L, Kjellman M, Höög A, Zedenius J, Larsson C. Copy number alterations in small intestinal neuroendocrine tumors determined by array comparative genomic hybridization. BMC Cancer. 2013;13:505. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 34. | Löllgen RM, Hessman O, Szabo E, Westin G, Akerström G. Chromosome 18 deletions are common events in classical midgut carcinoid tumors. Int J Cancer. 2001;92:812-815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 91] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
P- Reviewer: Abbott DE, Chen YC S- Editor: Gou SX L- Editor: A E- Editor: Liu XM