INTRODUCTION
Gastric cancer is the fourth most common malignancy in the world, with 934000 new cases occurring each year; unfortunately, the overall five-year survival rate remains below 20%[1]. The poor prognosis is in part due to tumor invasion and metastasis, with contributions from abnormalities in tumor intracellular signaling pathways[2]. Signaling molecules, such as matrix metalloproteinase (MMP)-2, MMP-9 and vascular endothelial growth factor, have been shown to affect tumor metastasis[3]. It has been hypothesized that the dysregulation of these signaling pathways results from mutations in genes coding for these molecules and their interactions with environmental factors[1]. However, recent studies indicate that abnormal expression of microRNAs (miRNAs) is also an important factor[4].
miRNAs are a class of small non-coding RNA molecules approximately 22 nucleotides in length that are widely expressed in many organisms[5]. miRNAs bind to partially complementary sites on target mRNAs to promote their degradation or inhibit translation[6]. More than half of the human miRNA genes are located at tumor-related chromosomal loci[4], and recent studies have identified altered regulation of many miRNAs in gastric cancer. For example, miR-20[7], miR-34b[8], miR-128[9] and miR-21[10] are abnormally expressed during gastric carcinoma induction, while miR-15, miR-16[11], miR-372[12] and miR-143[13] can affect the apoptosis and drug resistance of gastric cancer cells, and miR-218[14], miR-141[15], mir-375[16], miR-101[17] and miR-25[18] contribute to tumor differentiation, invasion and metastasis. Thus, further investigation of aberrant miRNA expression could aid in the development of novel strategies for gastric cancer prevention, diagnosis and treatment. In the current study, miRNA expression in gastric cancer cell lines was profiled in order to reveal new miRNAs for further clinical applications. Furthermore, the role of miR-374b-5p (a newly identified aberrantly expressed miRNA) in cancer cell invasion and metastasis was investigated.
MATERIALS AND METHODS
Cell lines and culture
The human gastric carcinoma cell line MGC-803, provided by the Cell Bank of Chinese Academy of Sciences (Shanghai, China), was cultured in RPMI-1640 medium. The human gastric adenocarcinoma cell line SGC-7901 and the normal human gastric epithelial cell line GES-1 were provided by the Cell Bank of Xiangya Central Laboratory at Central South University (Changsha, Hunan, China) and cultured in Dulbecco’s modified Eagle’s medium (DMEM; Hyclone of Thermo Fisher Scientific Inc., Waltham, MA, United States). All cell lines were cultured at 37 °C with 5% CO2 in media containing 100 U/mL penicillin, 100 μg/mL streptomycin (Sigma-Aldrich, St Louis, MO, United States) and 10% fetal bovine serum (FBS; Invitrogen of Thermo Fisher Scientific Inc.).
RNA isolation and miRNA microarray profiling
Total cellular RNA was isolated from cultured cells using Trizol reagent (Invitrogen) and purified with an RNeasy Mini kit (Qiagen, Hilden, Germany). The RNA samples were labeled with a MiRCURY Array Power Labeling kit (Exiqon, Vedbaek, Denmark) and used as probes for hybridization in a Hybridization Chamber II (Ambion of Thermo Fisher Scientific Inc.). Hybridized miRNA microarrays were scanned on a GenePix 4000B microarray scanner and analyzed with GenePix Pro software V6.0 (Molecular Devices, Sunnyvale, CA, United States). MeV software (V4.6, TIGR) was used to analyze the differential miRNA expression (fold change > 1.50 or ratio < 0.667) and differential expression was confirmed by quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR).
qRT-PCR
Total cellular RNA was reverse transcribed using a Hairpin-it miRNA qPCR Quantitation kit (GenePharma, Shanghai, China). Amplification conditions were 95 °C for 3 min followed by 40 cycles of 95 °C for 30 s and 62 °C for 40 s using primers obtained from GenePharma: miR-374b-5p (80 bp), 5’-TCAGCGGATATAATACAACCTGC-3’ and 5’-TATCGTTGTTCTCCACTCCTTCAC-3’; reversion-inducing cysteine-rich protein with Kazal motifs (RECK) (90 bp), 5’-TCTGATGGCTGGGTTGGCTTAG-3’ and 5’-ATCATTCTTTGAAGATGCCTGCTTG-3’; U6 (70 bp, used as an internal control), 5’-ATTGGAACGATACAGAGAAGATT-3’ and 5’-GGAACGCTTCACGAATTTG-3’; and β-actin (70 bp, used as an internal control), 5’-CCCTGGCACCCAGCAC-3’ and 5’-AGTCCTTCCACGATACCAAAGT-3’.
Antisense oligonucleotide transfection
MGC-803 cells were seeded in six-well plates and cultured with Lipofectamine 2000 transfection reagent (Invitrogen) and vectors (20 nmol/L) containing negative control (5’-CAGUACUUUUGUGUAGUACAA-3’) and anti-miR-374b-5p (5’-CACUUAGCAGGUUGUAUUAUAU-3’) sequences, according to the manufacturer’s protocol. The control conditions included untreated (phosphate-buffered saline only), transfection reagent only, and transfection with the negative control vector. The transfection rate after 48 h was confirmed by qRT-PCR.
Cell counting kit -8 cell viability assay
Cells from each group (untreated, transfection reagent only, control vector, and anti-miR-374b-5p) were added to 10 μL of cell counting kit (CCK)-8 solution (Yiyuan Biotech, Guangzhou, China) and incubated at 37 °C in 5% CO2 for 1 h according to the manufacturer’s protocol. An Elx-800 enzyme-linked immunoassay analyzer (Bio-Tek, Winooski, VT, United States) was used to measure the optical densities at 450 nm. The average and standard deviation of absorbances in each group were presented as a histogram.
Transwell tumor cell invasion assay
Cells from each group (untreated, transfection reagent only, control vector, and anti-miR-374b-5p) were plated on transwell chambers (PengBo Biotech, Changsha, China) coated with Matrigel Basement Membrane Matrix (BD Biocoat; Becton, Dickenson and Co., Franklin Lakes, NJ, United States) according to the manufacturer’s protocol. The upper chamber contained the plated cells in 100 μL of media, whereas the lower chamber contained 500 μL of DMEM with 20% FBS. After 48 h, the cells remaining on the upper side of the membrane were cleared, and the invaded cells on the lower side of the membrane were fixed and stained with Coomassie Brilliant Blue. The number of cells from five random microscopic fields was quantified for each group.
Wound scratch assay
Cells from each group (untreated, transfection reagent only, control vector, and anti-miR-374b-5p) that were seeded on a six-well plate were scraped with a 100 μL pipette tip to create two linear regions devoid of cells. Cells were stained with Coomassie Brilliant Blue and the scratch distance between the two linear regions was imaged and measured at different time points using ImagePro 6.0 (Media Cybernetic, Rockville, MD, United States). Five random non-overlapping images in each well were selected and quantitated for statistical analysis.
Dual luciferase reporter assay
MGC-803 cells were co-transfected with RECK 3’UTR-pmirGLO plasmid and a miR-374b-5p mimic or hsa-miR-NC vector using Lipofectamine 2000 according to the manufacturer’s instructions. After 48 h, luciferase activity was detected on a Glomax 20/20 luminometer using a Dual-Luciferase Reporter Assay System (Promega, Madison, WI, United States). For this assay, relative activity was defined by the ratio of firefly to renilla luciferase activity. Statistical analyses were then performed using: Δ luciferase activity = relative activity with miR-374b-5p/relative activity with hsa-miR-NC.
Protein extraction and Western blot
Total cellular protein was extracted using RIPA buffer (Beyotime Biotech, Shanghai, China) and samples were separated via 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto a polyvinylidene difluoride membrane (Beyotime Biotech). After blocking with 3% bovine serum albumin, the membranes were blotted overnight with a primary antibody against RECK (1:1000; Proteintech Group, Chicago, IL, United States) or GAPDH (1:3000; Sigma-Aldrich), and incubated with a secondary antibody (1:5000; Jackson ImmunoResearch, West Grove, PA, United States). Protein bands were visualized with enhanced chemiluminescence (Thermo Fisher Scientific Inc.) and imaged and analyzed with Alphalmager 2200 image software (UVP, Upland, CA, United States).
Statistical analysis
All statistical analyses were performed using SPSS 16.0 statistical software (SPSS, Chicago, IL, United States). One-way ANOVA and least significant difference tests were used to investigate the difference between groups, with P < 0.05 indicating statistical significance. Data are presented as mean ± SD.
RESULTS
Expression of miR-374b-5p in gastric cancer cells
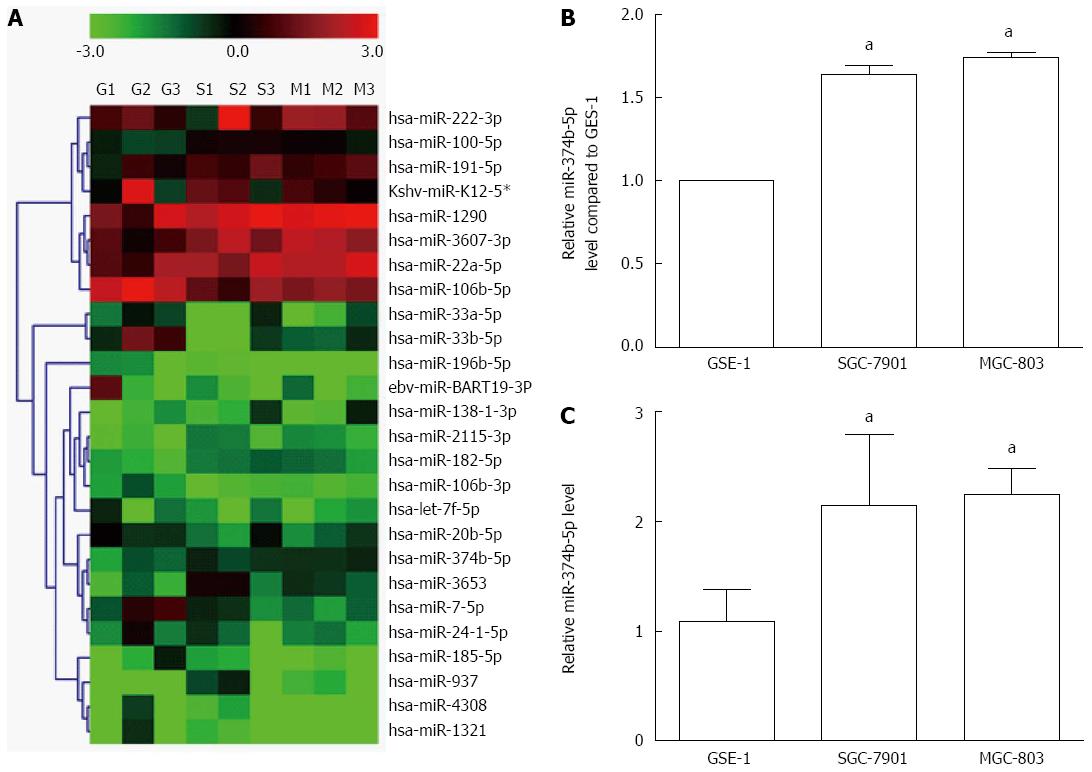
Fourteen miRNAs were significantly reduced in gastric cancer cells (MGC-803 and SGC-7901) compared to normal gastric epithelial cells (GES-1), including hsa-miR-20b-5p, ebv-miR-BART19-3p, hsa-miR-33a-5p, hsa-miR-33b-5p, hsa-miR-196b-5p, hsa-miR-4308, hsa-miR-106b-3p, kshv-miR-K12-5*, hsa-miR-106b-5p, hsa-let-7f-5p, hsa-miR-7-5p, hsa-miR-24-1-5p, hsa-miR-185-5p and hsa-miR-1321 (ratio < 0.667, P < 0.05) (Figure 1A). In contrast, 12 miRNAs were increased in MGC-803 and SGC-7901 cell lines, including hsa-miR-1290, hsa-miR-2115-3p, hsa-miR-3607-3p, hsa-miR-182-5p, hsa-miR-138-1-3p, hsa-miR-222-3p, hsa-miR-937, hsa-miR-100-5p, hsa-miR-20a-5p, hsa-miR-3653 and hsa-miR-191-5p (ratio > 1.50, P < 0.05). The expression of hsa-miR-374b-5p was increased by fold changes of 1.75 and 1.64 in MGC-803 and SGC-7901 cells, respectively (Figure 1B), which was further confirmed via qRT-PCR (Figure 1C).
Figure 1 Differential expression of microRNAs in gastric cancer cells.
A: microRNA (miRNA) expression in gastric cancer cell lines SGC-7901 and MGC-803 was compared with the normal GES-1 cell line using cluster analysis. All cell samples were detected in triplicate, such that the nine lanes include GES-1 (G1-G3), SGC-7901 (S1-S3) and MGC-803 (M1-M3). The color scale denotes the extent of relative upregulation (red) and downregulation (green); B: Histogram of miR-374b-5p expression detected by cluster analysis; C: Confirmation of miR-374b-5p expression via quantitative real-time reverse transcription-polymerase chain reaction (n = 3); aP < 0.05 vs GSE-1.
miR-374b-5p inhibits gastric cancer cell metastasis and invasion
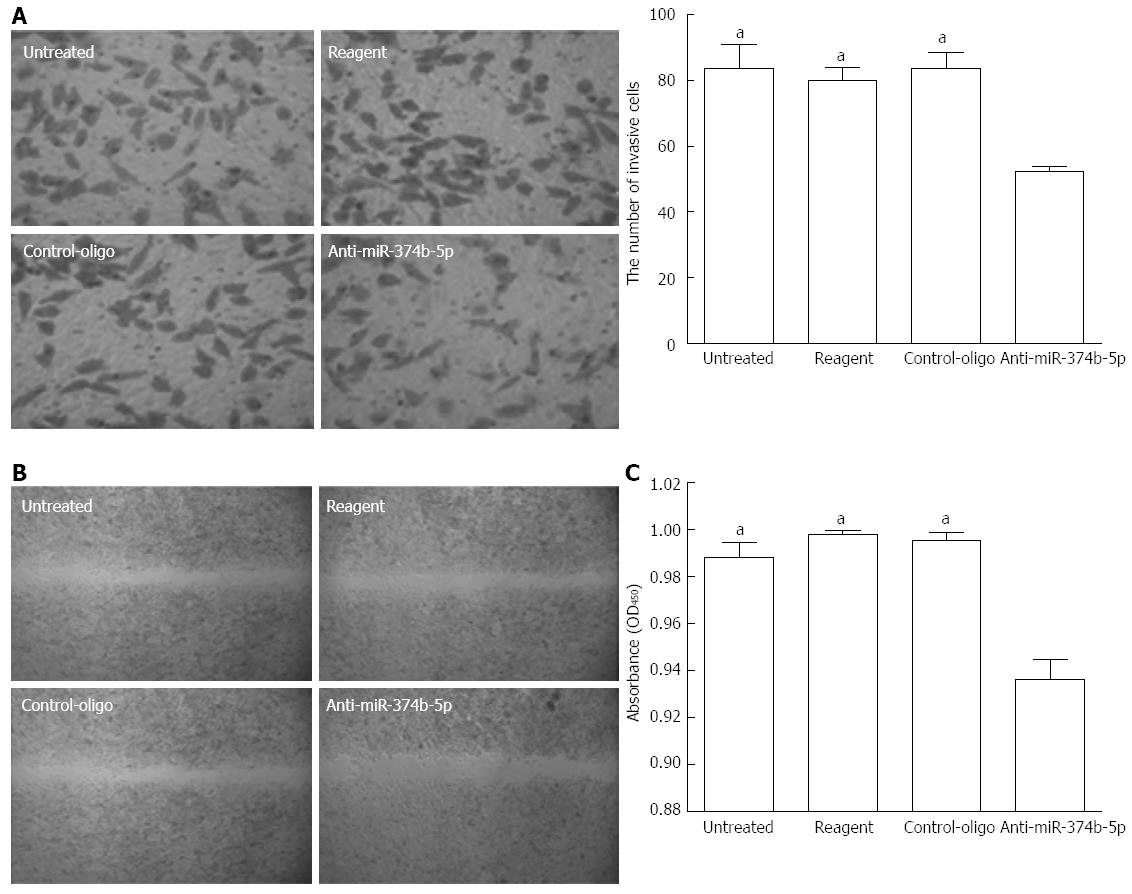
To examine the function of upregulated miR-374b-5p in cancer cells, its expression was suppressed in MGC-803 cells with antisense oligonucleotides (confirmed by qRT-PCR; data not shown). Compared to the control groups, the anti-miR-374b-5p group showed a significant reduction in transwell invasion after 48 h (Figure 2A). A scratch assay to evaluate tumor cell metastasis ability revealed that suppression of miR-374b-5p expression inhibited cell migration into the barren substratum (Figure 2B). Furthermore, the proliferation of MGC-803 cells as assessed by the CCK-8 assay was suppressed by miR-374b-5p antisense, which was statistically significant 96 h after transfection (Figure 2C).
Figure 2 Effects of miR-374b-5p on MGC-803 cell metastasis and invasion.
MGC-803 cells were transfected with a miR-374b-5p antisense oligonucleotide or negative control-oligo vector, or treated with phosphate-buffered saline (untreated), or transfection reagent only. A: Cells were subjected to a transwell invasion assay and detected at 48 h after seeding (left: Coomassie Brilliant Blue staining, × 200; right, quantification); B: Wound margin distance was measured after 48 h in a scratch assay (Coomassie Brilliant Blue staining, × 40); C: Absorbances in a cell counting kit-8 assay 96 h after transfection; (n = 3), aP < 0.05 vs Anti-miR-374b-5p.
Regulation of RECK expression by miR-374b-5p
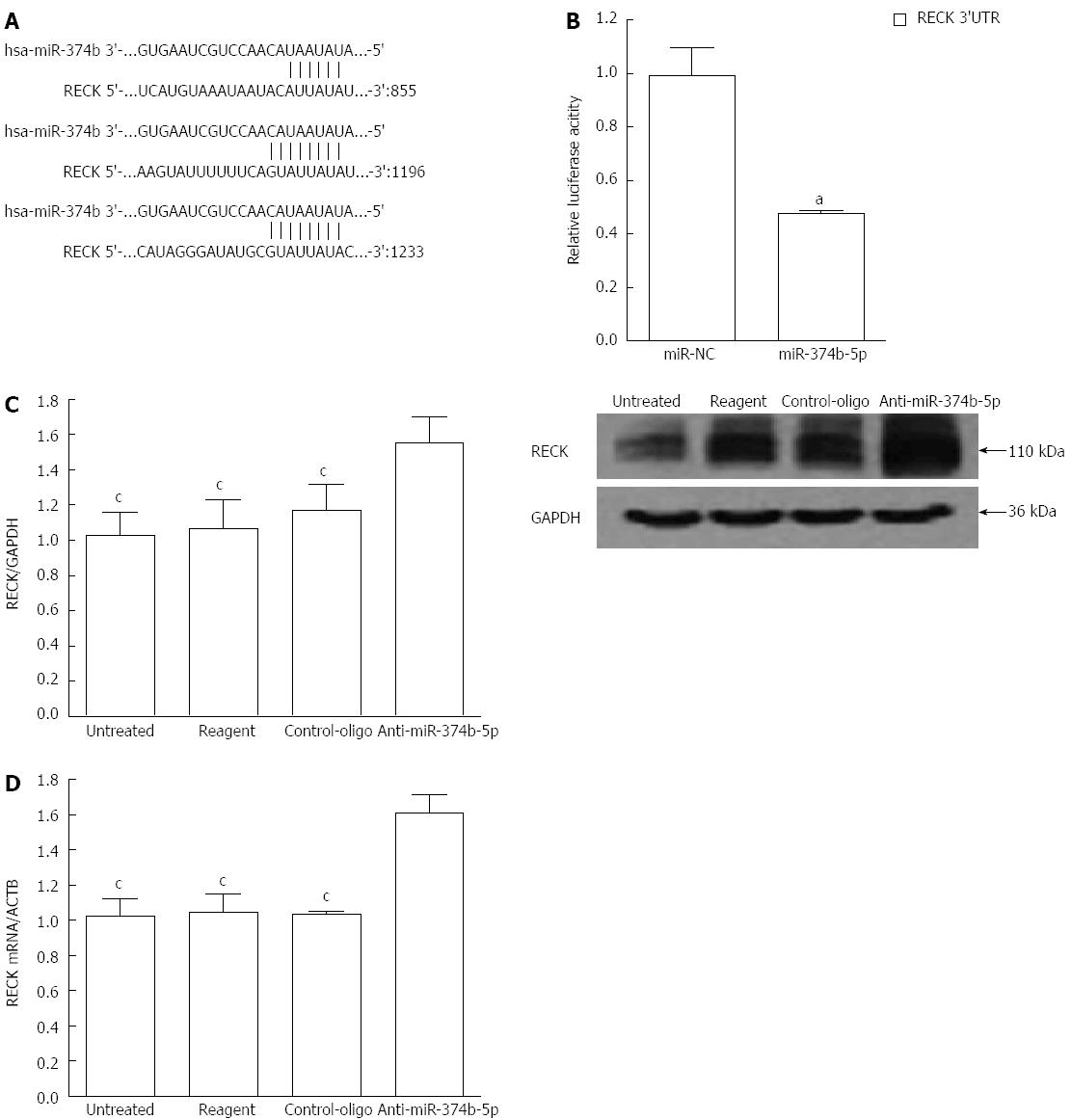
To identify the effect of miR-374b-5p on gene regulation in gastric cancer cells, a bioinformatic analysis was performed to predict direct tumor development targets. miRanda (http://www.microrna.org/microrna/home.do), PicTar (http://pictar.mdc-berlin.de) and TargetScan (http://www.targetscan.org) software revealed that RECK, a gene involved in tumor metastasis and angiogenesis, contains three target sequences for miR-374b-5p in the 3’ untranslated region (UTR) (Figure 3A). This was confirmed by a dual luciferase reporter assay showing that co-transfection with miR-374b-5p significantly reduced RECK-driven luciferase activity (Figure 3B). Moreover, RECK protein and mRNA levels were increased in cells transfected with miR-374b-5p antisense oligonucleotides (P < 0.05) (Figure 3C, D).
Figure 3 Regulation of RECK expression in MGC-803 cells by miR-374b-5p.
A: The miR-374b-5p binding site within the 3′ UTR of human (hsa) RECK cDNA was predicted with PicTar, TargetScan and miRanda software; B: Cells were co-transfected with RECK 3’UTR-pmirGLO and a plasmid containing either miR-374b-5p or miR-NC plasmid, aP < 0.05 vs miR-NC (n = 3); C: Western blot of proteins; D: qRT-PCR from cells transfected for 48 h with anti-miR-374b-5p or a negative control vector, or treated with phosphate-buffered saline (untreated), or transfection reagent only; (n = 3), cP < 0.05 vs Anti-miR-374b-5p.
DISCUSSION
Many miRNAs have expression patterns that are unique within certain cancer tissues[19]. miR-222-3p is upregulated in endometrial carcinoma, promoting proliferation and invasion, but downregulated in prostate cancer[20,21]. Some researchers suggest that these miRNA expression characteristics are potential biomarkers and powerful diagnostic tools for tumor classification and diagnosis[19]. Therefore, we profiled miRNA expression in gastric cancer cells, comparing two cancer cell lines with normal mucosal cells. From this analysis, we identified upregulation of miR-374b-5p and determined that it is involved in tumor cell metastasis and invasion. Furthermore, we confirmed that a target of miR-374b-5p is the 3’ UTR of RECK, which has been implicated in tumor metastasis and angiogenesis[22], and that suppression of miR-374b-5p activity increases the expression of RECK mRNA and protein. These results suggest a role of aberrant miR-374b-5p upregulation and an underlying molecular mechanism in gastric cancer progression.
Aberrant expression of various miRNAs is associated with carcinogenesis and cancer progression, and can therefore serve as efficient diagnostic biomarkers for analyses of histologic type, drug resistance and clinical prognosis[23,24]. Evaluation of the expression of 200 miRNAs is sufficient to molecularly classify human cancers and may help in the assessment of clinical diagnosis and treatment[23,25]. However, the miRNA expression profile of gastric cancer is not well defined. The analyses presented here identified upregulation of 12 and downregulation of 14 miRNAs in two gastric cancer cell lines. Many of these findings were consistent with previous reports concerning the upregulation of miR-1290[26], miR-182-5p[27], miR-138-1-3p[28], miR-20a-5p[29], miR-100-5p[30] and miR-191-5p[31] and the downregulation of miR-33a-5p[32], miR-33b-5p[32], miR-4308[33], kshv-miR-K12-5*[34], miR-106b-5p[35], let-7f-5p[31] and miR-7-5p[36]. However, our results differ from reports on miR-222-3p[30], miR-937[37], miR-185-5p[38], miR-3607-3p[39], miR-3653[40], miR-20b-5p[38], miR-106b-3p[41], ebv-miR-BART19-3p[42], miR-196b-5p[43] and miR-1321[39], possibly due to differences in cell lines, cancer cell differentiation, or the screening criteria for miRNA. There have been no previous reports regarding gastric cancer and expression of miR-24-1-5p, miR-2115-3p, or miR-374b-5p, which was selected for further study.
We previously reported that downregulation of miR-7 in gastric cancer affects tumor metastasis, though antisense treatment did not completely inhibit the metastasis of cancer cells[44], suggesting that other miRNAs may also be involved. This study shows that miR-374b-5p expression was significantly upregulated in SGC-7901 cells, and to a greater extent in MGC-803 cells. Furthermore, the adherence, migration, and invasion of MGC-803 cells were decreased significantly after suppression of miR-374b-5p expression. Abnormal miR-374b-5p expression in luminal-HER2-positive breast cancer cells can be used for classifying clinicopathologic subtypes of breast cancer[35], and its upregulation in blood may be an underlying diagnostic molecular marker for invasive urinary bladder cancer[45]. Taken together, these data indicate that miR-374b-5p plays an important role in tumorigenesis within different cancers.
After demonstrating the effects of miR-374b-5p on gastric cancer cell growth and migration, we identified a functional target through bio-informatic analysis and dual luciferase reporter assays. The results suggest that miR-374b-5p can bind to the 3’ UTR of RECK and consequently downregulate its expression. Previous studies have shown that RECK is a membrane-anchored glycoprotein that binds and negatively regulates a number of MMPs, included MMP-2 and MMP-9[22,46]. MMPs proteolytically degrade extracellular matrix proteins, which is critical for tumor metastasis and invasion[22]. Clinical research indicates that RECK expression negatively correlates with invasion-related clinicopathologic factors in hepatocellular carcinoma[47] and breast cancer[35], and tumor tissues with higher RECK mRNA expression are less invasive and associated with better patient survival[48]. Some miRNAs, such as miRNA-222[49] and miRNA-221[50], have been shown to regulate RECK expression and affect tumor invasion and metastasis. The results of the present study indicate that miR-374b-5p also can post-transcriptionally suppress RECK expression and thus promote the invasion and metastasis of gastric cancer cells. Future studies will be performed to verify this regulation in animal models of gastric cancer and investigate the relative mechanism.
In conclusion, the results of this study demonstrate that human gastric cancer cells have significant alterations of miRNA expression, including upregulation of miR-374b-5p. Furthermore, miR-374b-5p can facilitate the invasion and metastasis of gastric cancer cells, likely through inhibition of RECK expression.
COMMENTS
Background
MircroRNAs (miRNAs) play an important role in the post-transcriptional regulation of gene expression and human carcinogenesis. Although there have been significant advances, the function of miRNAs in gastric cancer still needs further research.
Research frontiers
Abnormal expression of miR-374b-5p has been reported in luminal-HER2-positive breast cancer cells and invasive urinary bladder cancer. However, little is known about the expression and mechanism of miR-374b-5p as a metastasis-promoting factor in gastric cancer.
Innovations and breakthroughs
This study profiles the expression of, and identifies significantly altered, miRNAs in two human gastric cancer cell lines. The results suggest that upregulation of miR-374b-5p facilitates the invasion and metastasis of gastric cancer cells, and identify inhibition of reversion-inducing cysteine-rich protein with Kazal motifs (RECK) as a possible molecular mechanism.
Applications
miR-374b-5p may be a potential biomarker and powerful diagnostic tool for gastric cancer classification and diagnosis.
Terminology
RECK is a membrane-anchored glycoprotein that binds and negatively regulates a number of matrix metalloproteinases that break down the extracellular matrix.
Peer review
This study investigated the alterations of miRNA expression in gastric cancer cells and found that upregulation of miR-374b-5p regulates the invasion and metastasis of cancer cells. Moreover, RECK was identified as a target of miR-374b-5p, which indicates a mechanism for the tumorigenic effects. These results are interesting and suggest that miR-374b-5p may be a potential biomarker and diagnostic tool in gastric cancer.