Published online May 21, 2014. doi: 10.3748/wjg.v20.i19.5666
Revised: November 19, 2013
Accepted: January 6, 2014
Published online: May 21, 2014
Processing time: 231 Days and 18.9 Hours
Helicobacter pylori (H. pylori), a major pathogen colonizing the human stomach, shows great genetic variation. Comparative analysis of strains from different H. pylori populations revealed that the genome size of strains from East Asia decreased to 1.60 Mbp, which is significantly smaller than that from Europe or Africa. In parallel with the genome reduction, the number of protein coding genes was decreased, and the guanine-cytosine content was lowered to 38.9%. Elimination of non-essential genes by mutations is likely to be a major cause of the genome reduction. Bacteria with a small genome cost less energy. Thus, H. pylori strains from East Asia may have proliferation and growth advantages over those from Western countries. This could result in enhanced capacity of bacterial spreading. Therefore, the reduced genome size potentially contributes to the high prevalence of H. pylori in East Asia.
Core tip: Comparative analysis of strains from different Helicobacter pylori (H. pylori) populations revealed that the genome size of strains from East Asia was reduced. In parallel with this, the number of protein coding genes and the guanine-cytosine content were decreased. The reduced genome of H. pylori from East Asia potentially contributes to the high prevalence of H. pylori in East Asia.
-
Citation: Dong QJ, Wang LL, Tian ZB, Yu XJ, Jia SJ, Xuan SY. Reduced genome size of
Helicobacter pylori originating from East Asia. World J Gastroenterol 2014; 20(19): 5666-5671 - URL: https://www.wjgnet.com/1007-9327/full/v20/i19/5666.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i19.5666
Helicobacter pylori (H. pylori) is a major human pathogen causing chronic inflammation of gastric mucosa[1]. Infection by this bacterium is associated with an increased risk for the development of peptic ulcer and gastric cancer[2,3]. There is a dramatic geographical variation in the prevalence of the infection and H. pylori associated diseases. In Western countries, the infection rate is approximately 30%, while it is higher than 60% in Eastern countries[4]. The incidence of gastric cancer is much lower as well in Western countries[5].
It is believed that H. pylori has established its colonization in human stomachs 100000 years ago[6]. With the human migration out of the Africa, the bacterium carried by its hosts traced the route of migration reaching Asia through India, and then into South East Asia and Australia[7]. It arrived at Europe through Turkey[7]. The phylogenetic analysis of global strains of H. pylori reveals that the bacterium is subdivided into seven populations: hpAfrica1, hpAfrica2, hpEastAsia, hpEurope, hpNEAfrica, hpAsia2 and hpSahul[6-8]. These populations generally correspond to their geographical origins. Up to date, hundreds of strains have been sequenced or being sequenced[9-12]. The purpose of this review is to compare the genomic differences of H. pylori from different populations, and discuss their implications in the geographical variation of the prevalence of the infection.
To explore the genomic differences between H. pylori strains, we have determined the genome sequence of H. pylori strain D33 isolated from a Chinese patient with gastric cancer. Whole-genome sequencing was performed as described previously[9,10]. Briefly, the raw reads were trimmed and filtered, yielding a total of 2364383 reads with an average length of 260 nucleotides. This corresponded to about 297-fold genome coverage. A total of 74 contigs were assembled. The annotation of the genome was performed using the Glimmer program. This whole genome shotgun project has been deposited in DDBJ/EMBL/GenBank under the accession number ANIO00000000 (The version described here is the first version, ANIO01000000.) The genome of H. pylori D33 had a size of about 1555730 bp, with a guanine-cytosine (GC) content of 38.96%. It possessed 1570 protein coding genes. The D33 genome carries a complete cag pathogenicity island. It has been found that H. pylori from East Asia had a massive decay of molybdenum-related genes[13,14]. There are a total of 12 molybdenum-related genes including three genes encoding molybdenum transport proteins, eight genes involved in molybdenum cofactor synthesis, and only one molybdenum-containing enzyme[15]. Most of them have been fragmented[14]. Consistent with this, seven molybdenum-related genes of strain D33 were fragmented, while intact modB, moaE, moeB, mogA and mobA genes were present.
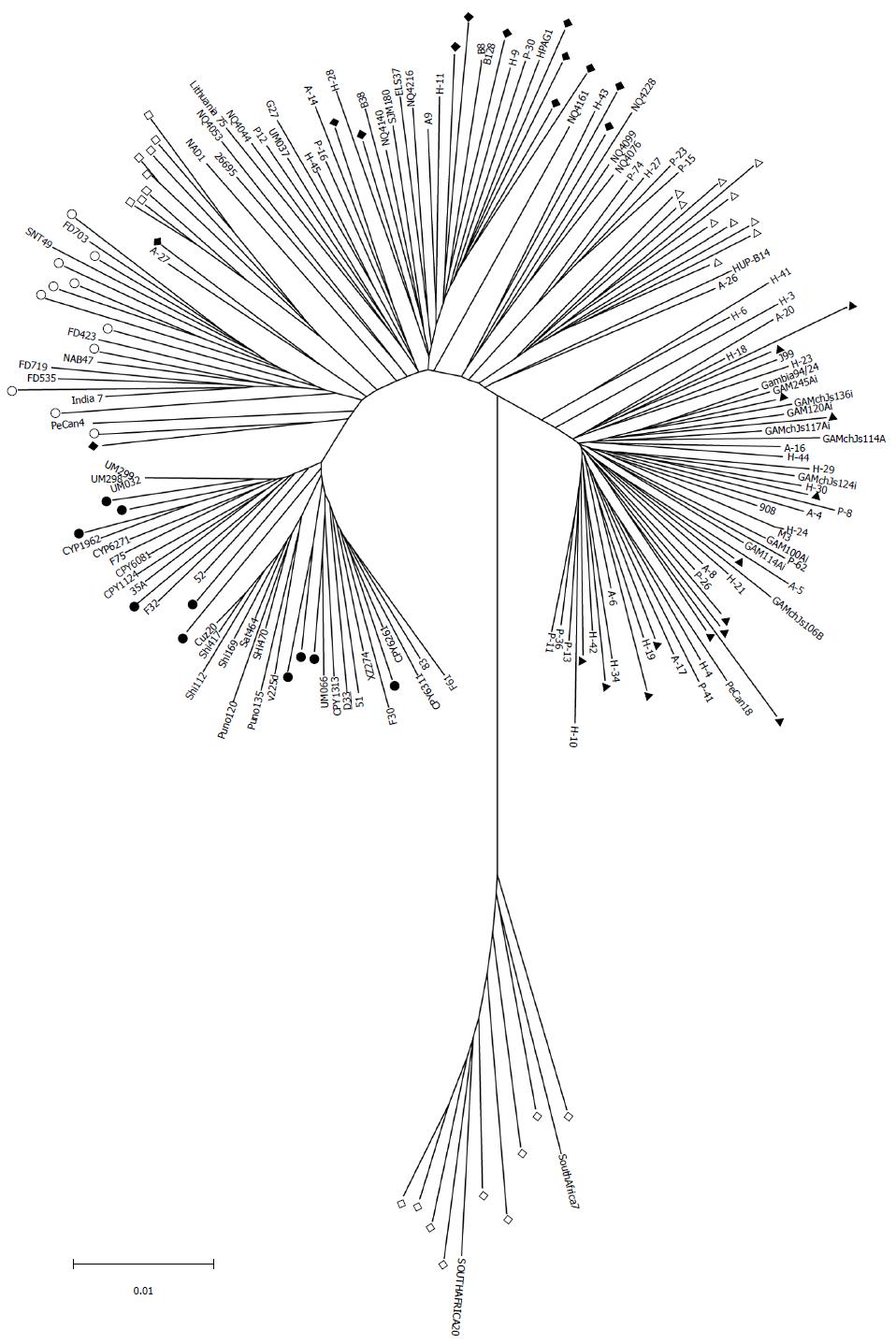
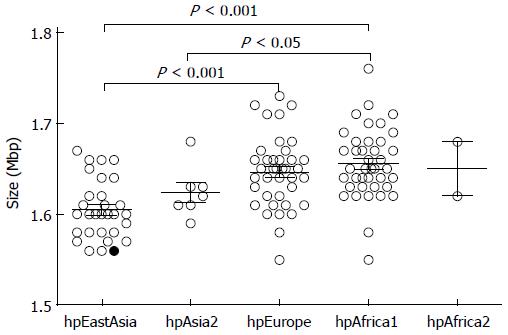
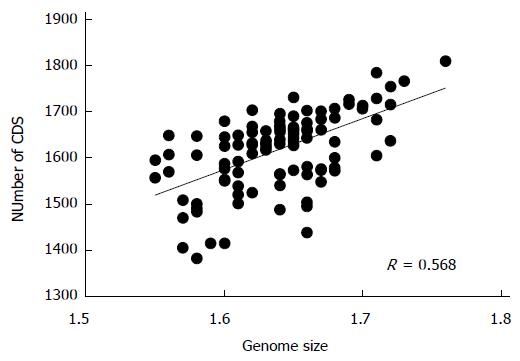
H. pylori D33 had a smaller genome compared to strains from Western countries. To identify the genomic size difference between different H. pylori populations, a total of 118 sequenced strains with required data available were assigned into different populations based on results of the phylogenetic analysis. Briefly, sequences of house-keeping genes atpA, efp, mutY, ppa, trpC, ureI and yphC were extracted from strain D33 sequenced in this study, 117 sequenced strains available from GenBank (http://www.ncbi.nlm.nih.gov), and 77 strains available from multilocus sequence typing (MLST) database (http://www.mlst.net). The sequences of these genes were concatenated and a neighbor-joining tree was constructed (Figure 1). The strains from the MLST database was used as a standard to assign those sequenced strains into different populations, since the population they belong to is known. These 118 sequenced strains could be assigned into five H. pylori populations, including hpEastAsia (30 strains), hpAsia2 (7 strains), hpEurope (39 strains), hpAfrica1 (40 strains) and hpAfrica2 (2 strains). No sequenced strains were assigned into the other two H. pylori populations, hpSahul and hpNEAfrica. The average genome size of H. pylori was about 1.64 Mbp. However, it is significantly different between the populations. Strains from hpEastAsia had a genome size of 1.60 Mbp on average (Table 1), which is significantly smaller than that from hpEurope (P < 0.001) or hpAfrica1 (P < 0.001) (Figure 2). The average genome size of strains from hpEastAsia was also smaller than that from hpAsia2 or hpAfrica2, although the difference is not statistically significant probably due to small sample size. These findings suggest that genome reduction occurs in strains of H. pylori from East Asia. As shown in Table 1, the genome of H. pylori decreased gradually with its spreading. In parallel to the genomic reduction, the number of protein coding genes was decreased (R = 0.568) (Figure 3), and the GC content of the genome declined gradually (Table 1).
| Population | n | Size (Mbp) | GC content | Protein coding genes |
| hpEastAsia | 30 | 1.6050 ± 0.0327 | 38.8167% ± 0.1206% | 1548.8000 ± 74.3762 |
| hpAsia2 | 71 | 1.6243 ± 0.0282 | 39.0286% ± 0.0756% | 1559.5000 ± 57.2757 |
| hpEurope | 391 | 1.6467 ± 0.0391 | 38.9231% ± 0.1287% | 1627.8684 ± 74.7411 |
| hpAfrica1 | 40 | 1.6558 ± 0.0384 | 39.1800% ± 0.1137% | 1660.0250 ± 58.9108 |
| hpAfrica2 | 2 | 1.6500 ± 0.0424 | 38.5000% ± 0.1414% | 1637.5000 ± 92.6310 |
| Total | 118 | 1.6379 ± 0.0417 | 38.9822% ± 0.1986% | 1637.5000 ± 71.3128 |
Bacterial genome size is mainly determined by gains or losses of genes[16]. Bacteria acquired new genes through duplication of genes or horizontal gene transfer[17,18]. Genes may be deleted through mutations or recombination events[19,20]. Genome reduction occurs when gene losses prevail gene gains. The life style of bacteria is a major factor contributing to the genome reduction. Bacterial species have a reduced genome when inhabiting in the host cells or in the extreme conditions[21,22]. It is, however, unusual that a particular bacterium living in similar environments has reduction of genome. Our comparative analyses of genome found that genome reduction occurs in H. pylori. This is unexpected as the bacterium only colonizes in the human stomach. It is likely that variation in host genetic backgrounds dramatically influences physiochemical properties, immune and inflammatory responses of the stomach, leading to genomic alterations of H. pylori.
Genome reduction may be caused by the deletion of intergenic regions[23]. This leads to an increased density of gene contents and thus a well compacted genome. It is most frequent, however, that genome reduces through removal of non-essential or redundant genes[24]. Our comparative analysis found that the number of protein coding genes of H. pylori was closely associated with the genome size. The number decreased with the reduced size of the genome. This demonstrates that the genome reduction in H. pylori is caused essentially by deletion of genes. Compared to strains from Western countries, two groups of genes are frequently removed from those from Eastern countries[14]. The first group consists of gene encoding outer membrane proteins, including oipA-2, hopN, babC, sabB, vacA2, homB and hopQ. All of these genes have their homologs and are variable in H. pylori[25,26]. Therefore, their functions are most likely redundant. The other group of genes deleted in Eastern strains are composed of those involved in central metabolism. These genes include molybdenum-related genes as found in our sequenced strain D33 and other Japanese strains[14], acoE encoding acetyl-CoA synthetase[27], tas encoding aldo-ketoreductase involved in carbonyl metabolism and[28] and jhp0585 encoding a putative hydroxyl-isobutyrate dehydrogenase which degrades valine[29]. Functions of these genes could be complemented by their paralogs in the genome. These findings suggest that H. pylori strains originating from Eastern countries could eliminate non-essential genes from their genome, resulting in the reduction of genome.
Bacterial genome size is well correlated with the GC content[30]. Events of G-C to adenosine-thymine (C-T) mutations are more common than those of C-T to A-G mutations[31-33]. Thus, GC contents significantly influence the genome size. Our comparative analysis found that the GC content drops with the gradual reduction in the genome size of strains from hpAfrica 1 and 2, to hpEurope and hpAsia2, and finally hpEastAsia. This indicates that mutation is a major factor contributing to the genome reduction in H. pylori. Mutations and recombination events frequently occur in this bacterium[34,35]. It has been estimated that the mutation rate is as high as 1.38 × 105 per site per year. The majority of genes deleted in Eastern strains are possibly caused by mutations. Recombination is also frequent in H. pylori[35]. However, recombination usually has no influence on the GC content. Thus, it is unlikely to be a major reason causing the genome reduction.
The prevalence of H. pylori shows considerable geographical variation[36]. In Eastern countries, the infection is much higher compared to Western countries. Family transmission is a major route for the bacterium to spread[37]. Spread of H. pylori is influenced by environmental conditions including socio-economic status, inhabiting conditions and hygiene levels[38,39]. These factors may lead to an increased exposure to the bacterium. Spread of the bacterium is also influenced by the individual susceptibility determined by host genetic backgrounds. The single nucleotide polymorphism of the Toll-like receptor gene is closely associated with increased susceptibility to the infection[40]. Bacterial factors related to the spread are, however, unclear.
Bacteria spend the majority of their energy for synthesis of proteins[41]. Genome reduction with reduced number of protein coding genes, thus, requires less cost of energy[42]. Furthermore, genome reduction would decrease the energy cost for the maintenance of DNA structure and replication[43]. These would promote bacterial growth and proliferation[44]. The lowered energy cost for bacteria means less nutrient needs. Therefore, bacteria with a reduced genome have an increased capacity of adapting to unfavorable environments[16]. Our comparative analysis found that strains form the hpEastAsia population have a smaller genome than those from hpEurope. Thus, H. pylori originating from East Asia could have an enhanced capacity of bacterial proliferation and growth, facilitating the spreading of the bacterium. It is conceivable that the reduced genome size contributes to the higher prevalence of H. pylori in East Asia.
H. pylori shows an unusual variation of the genome size. The reduced genome size of strains from East Asia potentially contributes to the high prevalence of the bacterium in the region. It is worth of further studies to investigate the influence of the genome reduction on the proliferation, growth and spread of H. pylori. This will benefit for the understanding of mechanism of the spreading and the prevention of the infection.
| 1. | Blaser MJ. Helicobacter pylori and gastric diseases. BMJ. 1998;316:1507-1510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 207] [Cited by in RCA: 199] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 2. | Blaser MJ, Atherton JC. Helicobacter pylori persistence: biology and disease. J Clin Invest. 2004;113:321-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 631] [Cited by in RCA: 622] [Article Influence: 28.3] [Reference Citation Analysis (1)] |
| 3. | Peek RM, Crabtree JE. Helicobacter infection and gastric neoplasia. J Pathol. 2006;208:233-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 368] [Cited by in RCA: 448] [Article Influence: 21.3] [Reference Citation Analysis (2)] |
| 4. | Lehours P, Yilmaz O. Epidemiology of Helicobacter pylori infection. Helicobacter. 2007;12 Suppl 1:1-3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 55] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 5. | Hu Y, Fang JY, Xiao SD. Can the incidence of gastric cancer be reduced in the new century? J Dig Dis. 2013;14:11-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 23] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 6. | Moodley Y, Linz B, Yamaoka Y, Windsor HM, Breurec S, Wu JY, Maady A, Bernhöft S, Thiberge JM, Phuanukoonnon S. The peopling of the Pacific from a bacterial perspective. Science. 2009;323:527-530. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 254] [Cited by in RCA: 215] [Article Influence: 12.6] [Reference Citation Analysis (1)] |
| 7. | Falush D, Wirth T, Linz B, Pritchard JK, Stephens M, Kidd M, Blaser MJ, Graham DY, Vacher S, Perez-Perez GI. Traces of human migrations in Helicobacter pylori populations. Science. 2003;299:1582-1585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 738] [Cited by in RCA: 716] [Article Influence: 31.1] [Reference Citation Analysis (1)] |
| 8. | Linz B, Balloux F, Moodley Y, Manica A, Liu H, Roumagnac P, Falush D, Stamer C, Prugnolle F, van der Merwe SW. An African origin for the intimate association between humans and Helicobacter pylori. Nature. 2007;445:915-918. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 720] [Cited by in RCA: 645] [Article Influence: 33.9] [Reference Citation Analysis (3)] |
| 9. | Sheh A, Piazuelo MB, Wilson KT, Correa P, Fox JG. Draft Genome Sequences of Helicobacter pylori Strains Isolated from Regions of Low and High Gastric Cancer Risk in Colombia. Genome Announc. 2013;1:pii: e00736-13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 10. | Armitano RI, Zerbetto De Palma G, Matteo MJ, Revale S, Romero S, Traglia GM, Catalano M. Draft Genome Sequences of Helicobacter pylori Strains HPARG63 and HPARG8G, Cultured from Patients with Chronic Gastritis and Gastric Ulcer Disease. Genome Announc. 2013;1:pii: e00700-13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 11. | Behrens W, Bönig T, Suerbaum S, Josenhans C. Genome sequence of Helicobacter pylori hpEurope strain N6. J Bacteriol. 2012;194:3725-3726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 23] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 12. | Guo Y, Wang H, Li Y, Song Y, Chen C, Liao Y, Ren L, Guo C, Tong W, Shen W. Genome of Helicobacter pylori strain XZ274, an isolate from a tibetan patient with gastric cancer in China. J Bacteriol. 2012;194:4146-4147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 13. | Dong QJ, Zhan SH, Wang LL, Xin YN, Jiang M, Xuan SY. Relatedness of Helicobacter pylori populations to gastric carcinogenesis. World J Gastroenterol. 2012;18:6571-6576. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 10] [Cited by in RCA: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 14. | Kawai M, Furuta Y, Yahara K, Tsuru T, Oshima K, Handa N, Takahashi N, Yoshida M, Azuma T, Hattori M. Evolution in an oncogenic bacterial species with extreme genome plasticity: Helicobacter pylori East Asian genomes. BMC Microbiol. 2011;11:104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 120] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 15. | Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539-547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2635] [Cited by in RCA: 2603] [Article Influence: 89.8] [Reference Citation Analysis (0)] |
| 16. | Moran NA. Microbial minimalism: genome reduction in bacterial pathogens. Cell. 2002;108:583-586. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 548] [Cited by in RCA: 529] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 17. | Gressmann H, Linz B, Ghai R, Pleissner KP, Schlapbach R, Yamaoka Y, Kraft C, Suerbaum S, Meyer TF, Achtman M. Gain and loss of multiple genes during the evolution of Helicobacter pylori. PLoS Genet. 2005;1:e43. [PubMed] |
| 18. | Furuta Y, Kawai M, Yahara K, Takahashi N, Handa N, Tsuru T, Oshima K, Yoshida M, Azuma T, Hattori M. Birth and death of genes linked to chromosomal inversion. Proc Natl Acad Sci USA. 2011;108:1501-1506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 54] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 19. | Björkholm B, Sjölund M, Falk PG, Berg OG, Engstrand L, Andersson DI. Mutation frequency and biological cost of antibiotic resistance in Helicobacter pylori. Proc Natl Acad Sci USA. 2001;98:14607-14612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 275] [Cited by in RCA: 268] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 20. | Aras RA, Kang J, Tschumi AI, Harasaki Y, Blaser MJ. Extensive repetitive DNA facilitates prokaryotic genome plasticity. Proc Natl Acad Sci USA. 2003;100:13579-13584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 100] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 21. | Kikuchi Y, Hosokawa T, Nikoh N, Meng XY, Kamagata Y, Fukatsu T. Host-symbiont co-speciation and reductive genome evolution in gut symbiotic bacteria of acanthosomatid stinkbugs. BMC Biol. 2009;7:2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 197] [Cited by in RCA: 177] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 22. | Stepkowski T, Legocki AB. Reduction of bacterial genome size and expansion resulting from obligate intracellular lifestyle and adaptation to soil habitat. Acta Biochim Pol. 2001;48:367-381. [PubMed] |
| 23. | Dufresne A, Garczarek L, Partensky F. Accelerated evolution associated with genome reduction in a free-living prokaryote. Genome Biol. 2005;6:R14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 259] [Cited by in RCA: 259] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 24. | Mendonça AG, Alves RJ, Pereira-Leal JB. Loss of genetic redundancy in reductive genome evolution. PLoS Comput Biol. 2011;7:e1001082. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 56] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 25. | Alm RA, Bina J, Andrews BM, Doig P, Hancock RE, Trust TJ. Comparative genomics of Helicobacter pylori: analysis of the outer membrane protein families. Infect Immun. 2000;68:4155-4168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 245] [Cited by in RCA: 248] [Article Influence: 9.5] [Reference Citation Analysis (1)] |
| 26. | Oleastro M, Cordeiro R, Ménard A, Yamaoka Y, Queiroz D, Mégraud F, Monteiro L. Allelic diversity and phylogeny of homB, a novel co-virulence marker of Helicobacter pylori. BMC Microbiol. 2009;9:248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 29] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 27. | Doig P, de Jonge BL, Alm RA, Brown ED, Uria-Nickelsen M, Noonan B, Mills SD, Tummino P, Carmel G, Guild BC. Helicobacter pylori physiology predicted from genomic comparison of two strains. Microbiol Mol Biol Rev. 1999;63:675-707. [PubMed] |
| 28. | Kratzer R, Wilson DK, Nidetzky B. Catalytic mechanism and substrate selectivity of aldo-keto reductases: insights from structure-function studies of Candida tenuis xylose reductase. IUBMB Life. 2006;58:499-507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 46] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 29. | Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, Smith DR, Noonan B, Guild BC, deJonge BL. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1387] [Cited by in RCA: 1363] [Article Influence: 50.5] [Reference Citation Analysis (1)] |
| 30. | Musto H, Naya H, Zavala A, Romero H, Alvarez-Valín F, Bernardi G. Genomic GC level, optimal growth temperature, and genome size in prokaryotes. Biochem Biophys Res Commun. 2006;347:1-3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 108] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 31. | Hildebrand F, Meyer A, Eyre-Walker A. Evidence of selection upon genomic GC-content in bacteria. PLoS Genet. 2010;6:e1001107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 289] [Cited by in RCA: 275] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 32. | Lind PA, Andersson DI. Whole-genome mutational biases in bacteria. Proc Natl Acad Sci USA. 2008;105:17878-17883. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 148] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 33. | Rocha EP, Feil EJ. Mutational patterns cannot explain genome composition: Are there any neutral sites in the genomes of bacteria? PLoS Genet. 2010;6:e1001104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 75] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 34. | Didelot X, Nell S, Yang I, Woltemate S, van der Merwe S, Suerbaum S. Genomic evolution and transmission of Helicobacter pylori in two South African families. Proc Natl Acad Sci USA. 2013;110:13880-13885. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 104] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 35. | Suerbaum S, Smith JM, Bapumia K, Morelli G, Smith NH, Kunstmann E, Dyrek I, Achtman M. Free recombination within Helicobacter pylori. Proc Natl Acad Sci USA. 1998;95:12619-12624. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 473] [Cited by in RCA: 448] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 36. | Goh KL, Chan WK, Shiota S, Yamaoka Y. Epidemiology of Helicobacter pylori infection and public health implications. Helicobacter. 2011;16 Suppl 1:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 207] [Cited by in RCA: 255] [Article Influence: 17.0] [Reference Citation Analysis (2)] |
| 37. | Kivi M, Tindberg Y. Helicobacter pylori occurrence and transmission: a family affair? Scand J Infect Dis. 2006;38:407-417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 64] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 38. | Tindberg Y, Bengtsson C, Granath F, Blennow M, Nyrén O, Granström M. Helicobacter pylori infection in Swedish school children: lack of evidence of child-to-child transmission outside the family. Gastroenterology. 2001;121:310-316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 78] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 39. | Brown LM. Helicobacter pylori: epidemiology and routes of transmission. Epidemiol Rev. 2000;22:283-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 620] [Cited by in RCA: 638] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 40. | Mayerle J, den Hoed CM, Schurmann C, Stolk L, Homuth G, Peters MJ, Capelle LG, Zimmermann K, Rivadeneira F, Gruska S. Identification of genetic loci associated with Helicobacter pylori serologic status. JAMA. 2013;309:1912-1920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 134] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 41. | Lane N, Martin W. The energetics of genome complexity. Nature. 2010;467:929-934. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 752] [Cited by in RCA: 824] [Article Influence: 51.5] [Reference Citation Analysis (0)] |
| 42. | Ranea JA. Genome evolution: micro(be)-economics. Heredity (Edinb). 2006;96:337-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 43. | Ranea JA, Grant A, Thornton JM, Orengo CA. Microeconomic principles explain an optimal genome size in bacteria. Trends Genet. 2005;21:21-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 43] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 44. | Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D, Bibbs L, Eads J, Richardson TH, Noordewier M. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309:1242-1245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 819] [Cited by in RCA: 766] [Article Influence: 36.5] [Reference Citation Analysis (0)] |
P- Reviewers: Franceschi F, Nakajima N S- Editor: Gou SX L- Editor: Wang TQ E- Editor: Liu XM