Published online May 21, 2014. doi: 10.3748/wjg.v20.i19.5610
Revised: December 4, 2013
Accepted: January 19, 2014
Published online: May 21, 2014
Processing time: 204 Days and 13.7 Hours
Helicobacter pylori (H. pylori) colonizes the stomach of humans and causes chronic infection. The majority of bacteria live in the mucus layer overlying the gastric epithelial cells and only a small proportion of bacteria are found interacting with the epithelial cells. The bacteria living in the gastric mucus may act as a reservoir of infection for the underlying cells which is essential for the development of disease. Colonization of gastric mucus is likely to be key to the establishment of chronic infection. How H. pylori manages to colonise and survive in the hostile environment of the human stomach and avoid removal by mucus flow and killing by gastric acid is the subject of this review. We also discuss how bacterial and host factors may together go some way to explaining the susceptibility to colonization and the outcome of infection in different individuals. H. pylori infection of the gastric mucosa has become a paradigm for chronic infection. Understanding of why H. pylori is such a successful pathogen may help us understand how other bacterial species colonise mucosal surfaces and cause disease.
Core tip: Colonization of gastric mucus by Helicobacter pylori (H. pylori) key to the establishment of chronic infection. How H. pylori manages to colonise and survive in the hostile environment of the human stomach and avoid removal by “mucus flow” and killing by gastric acid is the subject of this review. We also discuss how bacterial and host factors may together go some way to explaining the susceptibility to colonization and the outcome of infection in different individuals. Understanding of how H. pylori causes chronic infection will likely serve as a valuable reference system for how other bacteria colonise mucosal surfaces.
-
Citation: Dunne C, Dolan B, Clyne M. Factors that mediate colonization of the human stomach by
Helicobacter pylori . World J Gastroenterol 2014; 20(19): 5610-5624 - URL: https://www.wjgnet.com/1007-9327/full/v20/i19/5610.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i19.5610
Helicobacter pylori (H. pylori) is a Gram negative, microaerophillic, spiral shaped bacterium, which colonizes the human stomach. H. pylori is one of the most common infections in the world, persistently colonizing the gastric mucosa of over 50% of the global population. Colonization with H. pylori induces a chronic gastritis in all infected individuals[1]. The majority of infections are asymptomatic, however long-term infection increases the risk of developing site-specific disease. 10%-15% of infections result in the development of peptic ulcer disease and H. pylori is associated with 95% of duodenal ulcers and 80% of gastric ulcers[2]. Infection with H. pylori is a significant risk factor for the development of gastric cancer and 1%-3% of infected individuals develop the disease[3,4]. As a result, H. pylori is classified as a group 1 carcinogen by the World Health Organisation[5]. Infection is associated in particular with intestinal-type (approximately 90% of patients) rather than diffuse-type gastric cancers (approximately 32% of patients)[6]. The risk of developing gastric cancer is reduced in patients with duodenal ulcers[7]. The gastric mucosa does not normally contain any mucosa-associated lymphoid tissue (MALT), however the pan-gastric inflammation induced by H. pylori infection results in the development of MALT. In < 0.1% of infected individuals this develops into B cell MALT lymphoma[3], however at early stages the lymphoma can be cured by eradication of H. pylori[8,9].
H. pylori exhibit a very strict tissue tropism. It colonizes the gastric mucosa of humans and is only found colonizing other sites in the body where gastric metaplasia occurs[10]. Within the gastric mucosa the majority of organisms are found living in gastric mucus[11] and we suggest that these organisms act as a reservoir of infection for the underlying epithelial cells. Only a small percentage of the organisms colonizing are found in association with epithelial cells but the interaction of the bacteria with the cells is essential for the development of disease. How H. pylori causes chronic infection, which can persist for the lifetime of the host, in the hostile acidic environment of the stomach while avoiding removal by both mucus, which is constantly “turning over”, and the immune response is not completely understood. Evasion of the host innate immune response by H. pylori has recently been covered in two excellent reviews[12,13]. This review will focus on what we know about other specific bacterial and host factors that promote H. pylori survival in the stomach and colonization of the gastric mucosa, causing disease in some individuals but asymptomatic infection in others.
H. pylori is not an acidophile and key to its ability to overcome the acidic conditions of the gastric lumen is the production of a very potent urease enzyme. The expression of urease[14] and its activation[15] is essential for colonization of the gastric mucosa. H. pylori survives at a pH range between 4.0 and 8.0 in the absence of urea[16]. However, in the presence of urea the organism can survive at a pH as low as 2.5. The urease enzyme of H. pylori hydrolyses urea to NH3 and CO2 and has a Km value for urea of 0.8 mmol/L[17], meaning that it displays a much higher affinity for its substrate than that of ureases produced by other bacterial species. This allows for the utilization of the limited amounts of urea (5 mmol/L) present in the human stomach. The generation of NH3 provides both acid-neutralising and acid buffering capabilities, enabling H. pylori to raise the pH in its microenvironment and periplasm thus maintaining the proton motive force. The biosynthesis of urease is controlled by a seven gene cluster. The urease enzyme, estimated to be approximately 600 kDa in size[18], consists of a complex of 12 UreA and UreB subunits[19], which individually are 30 and 62 kDa in size[18]. The protein is originally produced as an immature apoenzyme, and activation takes place when four chaperone proteins, UreE, UreF, UreG and UreH assemble the catalytic site of the protein[20]. The insertion of 24 nickel ions into the enzyme is essential for complete activation of the protein, as well as GTP hydrolysis[19]. The genes encoding UreAB are located in one operon[21], while the ureEFGH genes are found on another[22]. Recent studies have shown that the ureAB operon can yield a 2.7 kB transcript that produces a functional enzyme and also a 1.4 kB transcript, the product of which exhibits much lower urease activity and is generated by cleavage of the 3’ureB region. The expression of this smaller transcript was shown to be influenced by pH, the presence of the histidine kinase ArsS and the phosphorylation state of the response regulator ArsR, illustrating the influence that pH has on the expression of an active urease[23]. Urease is found in both the cytoplasm of H. pylori and on the surface of the bacteria due to the lysis of some organisms[24-26]. Intracellular urease, regulated by external pH, acts to increase the pH of the periplasm and increase membrane potential thereby allowing protein synthesis at low pH[27,28]. A proton-gated channel, UreI, which regulates the uptake of urea[29], is only active at acidic pH and therefore does not allow for the transport of urea into the bacterial cell at neutral pH, thus preventing lethal alkalinisation of the cytoplasm[30]. Buffering of the periplasm also occurs through the conversion of CO2 to HCO3- by the periplasmic α-carbonic anhydrase[31]. HCO3- acts in conjunction with NH3 to buffer the periplasmic and cytoplasmic pH, generating neutral conditions. This acid acclimation feature of the bacteria, maintaining an intracellular neutral pH while the pH of the environment is acidic, is unique to H. pylori, and is critical to survival of the organism in the stomach. The data above is strong evidence that the urease enzyme of H. pylori is a factor absolutely essential for survival of the organism in the human stomach. Indeed it may be a factor that explains why H. pylori is not found readily at other sites in the body, as production of ammonia by the urease enzyme at sites that are not acidic could increase the local pH above pH 7.0 and it has been shown that H. pylori is sensitive to alkaline conditions[30].
Another key colonization factor shown to be absolutely essential for colonization is the possession of polar flagella which confer motility on H. pylori[32]. Non-motile mutants lacking flagella are unable to establish persistent infection in animal models[33,34]. Studies focusing on the motor protein MotB, have shown that it is the motility conferred on the organism by the flagella that contributes to colonization and the presence of flagella alone is not sufficient[35]. H. pylori possesses two to six sheathed unipolar flagella. The structures extend 3-5 μm from the bacterial surface, with bulb-like structures often seen at the tip of the filaments[32]. The sheath consists of both proteins and lipopolysaccharide, and is thought to be an extension of the bacterial outer membrane that protects the flagellar filaments from acid in the stomach[36]. Expression of the two major flagellar proteins, FlaA and FlaB, are required for full motility of the bacteria. FlaA mutants exhibit a greater decrease in motility than that of FlaB mutants[37]. Other components of the flagellar structure which have also been shown to be essential for motility and colonization include the hook protein FlgE[38] and FliD, which functions as a hook-associated protein[39].
In keeping with flagella being an essential colonization factor for H. pylori flagellar biogenesis is a very well regulated process, dysfunction of which can have a strong impact on motility and infection. For example, deletion of the regulator FlhA, which controls expression of flaA, flaB and flgE, leads to the generation of non-motile mutants[40]. Of interest FlhA also regulates urease expression, and deletion of this membrane protein leads to a decrease in colonization rates[41]. Thus flagellar biosynthesis and urease activity, two key essential colonization factors of H. pylori, are linked. The response regulator FlgR, which is part of the two-component FlgRS system, controls expression of RpoN-regulated genes, including flgE and flaB, and deletion of this regulator abolishes expression of its target genes[42]. H. pylori utilises the histidine kinase FlgS to detect changes in its environment which in turn activates FlgR to modify transcription of flagellar genes. Interestingly, it does so independently of pH, so there must be other environmental triggers that activate transcription of flagellar genes[43]. More recently, production of auto-inducer 2 (AI-2) protein, a product of the luxS gene, has been shown to regulate expression of flagellar genes. Auto-inducers play a role in quorum sensing, and so it is possible that H. pylori regulates expression of its flagellar genes in response to bacterial numbers and it’s environment through this system[44,45]. This is supported by evidence that mutants deficient in luxS exhibit reduced motility and infectivity rates compared to the wild-type[46]. Thus efficient and controlled expression and synthesis of the flagellar components is essential for successful motility and colonization.
Modifications to the flagella that impact on motility and colonization can also occur at a post-transcriptional level. The glycosylation state of the structure has been shown to impact on both, with H. pylori mutants deficient in a deglycosylase HP0518, exhibiting increased levels of the pseudaminic acid on FlaA and displaying a hypermotile phenotype. Infection studies showed that the mutants are able to associate more closely with epithelial cells and induce rapid activation of NF-κB, and also exhibit increased in vivo colonization rates. Thus increased levels of flagellin glycosylation in turn seem to increase the ability of H. pylori to colonize[47]. Other genes, including HP0326B and HP0178, involved in the biosynthesis of the precursors to pseudaminic acid, have been shown to have a similar role to HP0518. Inactivation of these also affects motility and colonization[48]. As well as modifying sugars found directly on flagellin proteins, changes in bacterial peptidoglycan by transglycosylases affects the functioning of the flagella by disturbing the localisation of MotB, thus preventing the flagellum from moving[49]. Thus, like the urease enzyme, motility is an essential colonization factor for H. pylori. The structure of the flagella with an outer sheath to protect against the effect of acid, stringent regulation of flagellar biogenesis and post translational modifications to flagella which result in enhanced motility and ability to colonise all underline the pivotal role of motility and flagella in H. pylori colonization.
The mucus layer that overlies the epithelial cells in the gastrointestinal tract is a physical barrier which acts to prevent pathogens from colonizing and interacting with the underlying epithelium. Pathogens which infect mucosal surfaces share two main goals: (1) to overcome the mucus barrier; and (2) to interact with the underlying epithelial cells which results in disease. The majority (approximately 80%) of infecting H. pylori are found living in gastric mucus rather than in contact with the underlying epithelium[11].
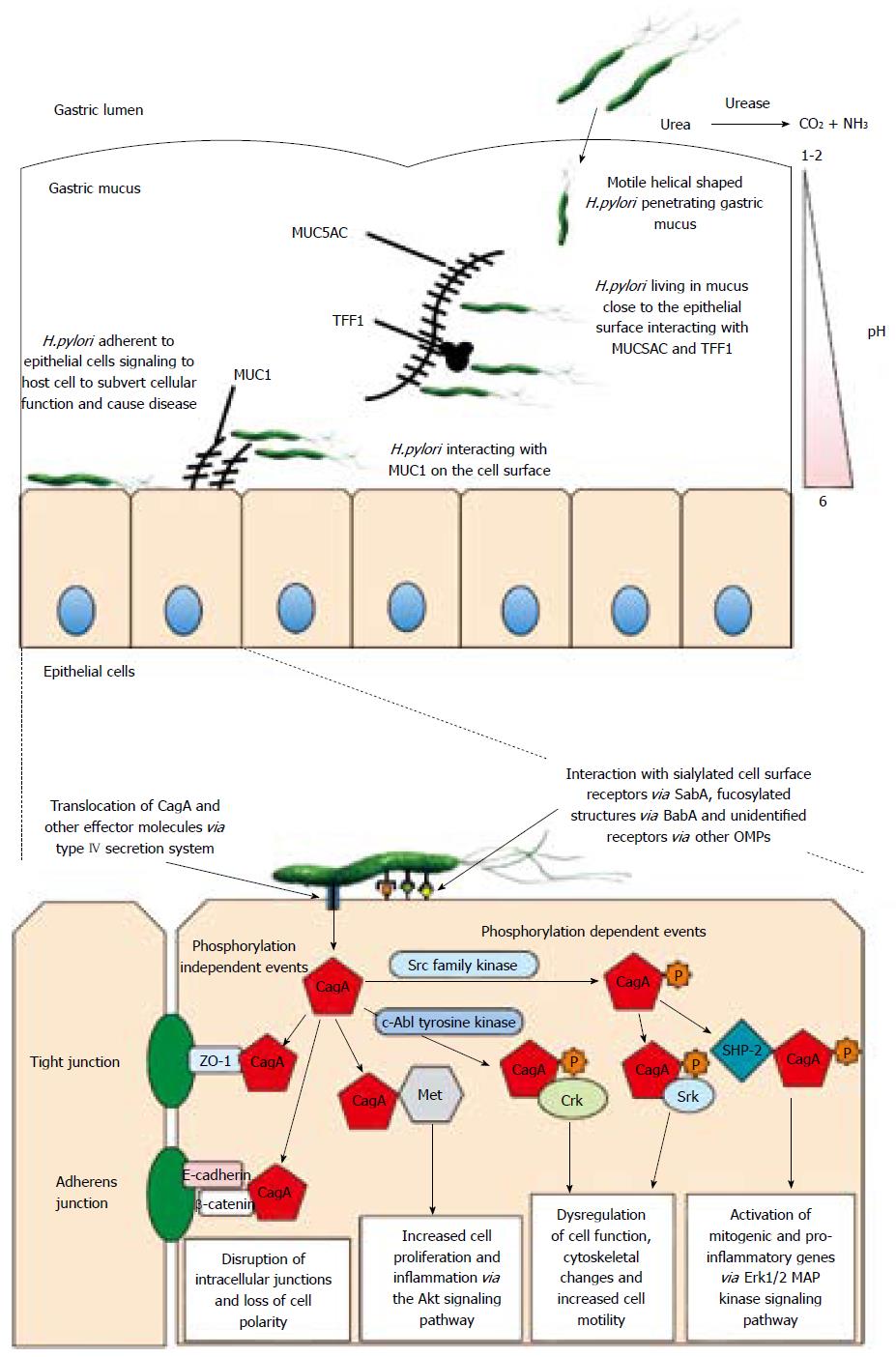
The entire epithelial surface of the gastrointestinal tract is covered in a thick layer of secreted mucus[50]. The mucus layer in the stomach is approximately 300 μm thick[50], and H. pylori has to penetrate it in order to colonise and gain access to the underlying epithelium. A pH gradient exists across the gastric mucus layer with the pH being approximately neutral at the epithelium but very acidic (pH 1-2) close to the lumen. H. pylori can lose motility rapidly when it encounters acidic conditions[51], therefore it is imperative that it penetrates the gastric mucus quickly and establish persistent colonization in an area close to the epithelium[52].
H. pylori is able to alter mucus structure which may aid movement through the viscoelastic mucus gel layer. A thioredoxin system that specifically reduces interchain disulphide bonds of mucins has been identified[53]. This reduces the gel-forming capabilities of mucins and therefore the viscoelastic properties of mucus aiding travel of the bacterium through mucus. In addition the rheology of gastric mucin exhibits a reversible pH-dependent transition. At a strongly acidic pH the viscoelastic properties of mucus increase whereas the mucus becomes less gel-like as the pH increases above approximately 4.0[54]. The ability of H. pylori to utilise urea to raise the pH in its microenvironment modifies mucus so that it is less gel-like, enabling the bacterium to move quickly through it[55]. H. pylori is usually found close to the gastric epithelium[56]. When the pH gradient that exists across the gastric mucus layer was disrupted in Mongolian gerbils H. pylori were no longer found close to the epithelium but were scattered throughout the mucus layer, suggesting that pH plays an important role in maintaining the particular localization of H. pylori[56]. The characteristic helical shape of H. pylori is also thought to play a role in mediating penetration of gastric mucus. Alterations in the cross linking of peptidoglycan in the outer membrane has been shown to modify the shape of H. pylori. Some cell shape mutants are unable to colonise as efficiently as helical shaped bacteria despite displaying similar motility to wildtype bacteria in vitro[57]. However mutants which exhibited the most dramatic changes in cell shape had reduced motility compared to that of the wild-type and other mutants, and they exhibited further reduced ability to colonise mice compared to mutants that retained wild type motility. This led to the hypothesis that the characteristic helical shape of H. pylori allows the bacteria to penetrate gastric mucus in a corkscrew like motion[58].
H. pylori has been shown to form microcolonies within the mucus secreted by the surface epithelium of the gastric mucosa[59]. MUC5AC is the predominant secreted gel forming mucin expressed by gastric surface epithelial cells and H. pylori has been shown to co-localise with MUC5AC in vivo[60]. Lower mucus neck cells found in the antral glands produce MUC6. α1,4-linked N-acetylglucosamine capped O-linked glycans expressed on the surface of MUC6 have anti-microbial activity towards H. pylori, inhibiting the synthesis of cholesteryl-α-D-glucopyranoside a vital cell wall component[61]. Aberrant expression of MUC2 occurs occasionally in the stomach in areas of intestinal metaplasia, however H. pylori is not found in association with areas of complete intestinal metaplasia, although it is sometimes found in areas of incomplete metaplasia[62,63]. This suggests that MUC5AC or a molecule co-expressed with it may explain the preference of H. pylori for gastric mucus.
H. pylori has been shown to interact with the Lewisb blood group antigen structure found on the surface of MUC5AC in gastric mucus and this interaction, which has been extensively studied, is mediated through the bacterial outer membrane protein BabA[64]. H. pylori can also interact with sialylated structures found on mucins via the sialic-acid binding adhesin (SabA). This 66 kDa outer membrane protein is a major adhesin of H. pylori and mediates binding to sialyl-Lewisx and sialyl-Lewisa glycoconjugates[65]. During chronic infection by H. pylori there is an increase in the proportion of sialylated structures present in the gastric mucosa, which is attributed to mucosal inflammation and transformation[66], and so SabA is thought to play a role in promoting chronic infection.
H. pylori can also interact with the membrane bound mucin MUC1 and this has been shown to be mediated via both BabA and SabA outer membrane adhesins. A ΔbabA strain of H. pylori displayed significantly reduced adherence to MUC1 from two independent sources[67]. In addition, mutants lacking SabA, also displayed some reduction in binding suggesting that binding to sialylated structures play a role in the interaction of H. pylori with MUC1 in vivo[67]. MUC1 has been shown to be important in limiting infection by Helicobacter in a mouse model of infection[68]. Over expression of MUC1 was shown to counter regulate H. pylori induced gastric inflammation[69]. The interaction of H. pylori with MUC1 blocks H. pylori stimulated β-catenin nuclear translocation and attenuates IL-8 production and neutrophil induced gastric inflammation[70]. Speculation that augmented expression of MUC1 could be used for treatment of H. pylori gastritis highlights the need for model systems that express adherent mucus layers and appropriate mucins in order to test such hypotheses. Using an in vitro infection model we recently showed a clear interaction between H. pylori and the membrane bound mucin MUC1 present on the apical surface of the mucus secreting cell line HT29-MTX-E12[71] suggesting that this might be a useful model system to study the interaction of H. pylori with mucus and mucins.
Infection of mice with H. pylori demonstrated a reduction in the rate of mucin turnover and decreased levels of MUC1 in infected animals, thus creating a more stable niche for the bacteria to colonise and increasing the ability of the organisms to interact with the epithelial cells[72]. The bacterial factors which signal to the cells to modulate mucin synthesis and turnover have not yet been elucidated.
Gastric mucins have been shown to promote the proliferation of H. pylori and to alter gene expression[73]. We looked at the interaction of H. pylori with purified native mucins from different animals that were printed on a microarray slide. Three strains of H. pylori were examined, strains J99, G27 and 26695. Strain 26695 is known not to express active BabA or SabA proteins and is therefore unable to bind to Lewisb or Sialyl Lewisx. Surprisingly all three strains bound to porcine gastric mucins equally well. This finding suggests that in addition to BabA and SabA other bacterial adhesins must exist which can mediate the binding of H. pylori strains such as strain 26695 to mucin.
While the interaction of H. pylori with gastric mucins has been well characterized the interaction of the organism with other non-mucin components of mucus has not received as much attention. In addition to mucin H. pylori has recently been shown to interact with mucus bound glycolipids[74] and we have previously shown that H. pylori interacts directly with TFF1, a member of the trefoil peptide family of proteins which is co-expressed with MUC5AC in the stomach, and this interaction is mediated by the core oligosaccharide portion of H. pylori LPS[75,76]. The optimum pH for H. pylori binding to TFF1 was 5.0-6.0[76]. The pH dependence of this interaction indicates that binding of H. pylori to TFF1 in the stomach could promote colonization of the mucus layer adjacent to the gastric epithelial surface. The interaction of H. pylori with TFF1 may explain the distinct tropism that the organism exhibits for gastric tissue and specifically for the gastric mucin MUC5AC. A role for TFF1 in colonization of mucosal surfaces by H. pylori was confirmed in a study which showed that H. pylori mutants expressing a truncated core oligosaccharide unable to interact with TFF1 have a reduced ability to colonise the mucus layer produced by the mucus secreting HT29-MTX-E12 cell line compared to wildtype strains[71]. In summary H. pylori interacts with both secreted and membrane bound mucins in the intestinal tract. These interactions are mediated via the well described outer membrane adhesins BabA and SabA interacting with fucosylated and sialylated glycans found on mucins and possibly other as yet undescribed adhesins. H. pylori has also been shown to interact with TFF1 a small protein found co-expressed with MUC5AC in the stomach. This suggests that the interaction of bacteria with non mucin components of mucus warrants further investigation.
While the majority of H. pylori organisms live in gastric mucus and only a small percentage of infecting organisms interact with gastric epithelial cells, it is widely accepted that the organisms in contact with the epithelial cells cause disease. A number of bacterial outer membrane proteins have been identified that can act as adhesins. Following binding of H. pylori to epithelial cells the organism signals to the cells to subvert host cell function and this results in the development of pathology.
BabA together with SabA, both already mentioned above, are the two best characterized adhesins of H. pylori. Binding of H. pylori to fucosylated structures, including the H-1 type and Lewisb blood group antigen[77] is mediated via BabA, a 78 kDa outer membrane protein[64]. Two genes encode for a BabA protein, babA1 and babA2. Only the protein encoded by babA2 is functionally active, due to the presence of a 10 bp insert in the gene that encodes for a translational initiation codon[64].
A high level of heterogeneity is found in the BabA protein amongst strains, with various polymorphisms being identified, and different levels of Lewisb binding observed[78]. There is also a high level of allelic variation in bab genes. H. pylori possesses a closely related gene to babA, babB, and both proteins are highly similar in their N- and C-terminus regions, but vary quite significantly in their central region. The BabB protein does not bind Lewisb, indicating that the central region of the two proteins confers unique functions[79].
Geographic location can also influence the binding specificities of BabA, with strains from specific regions often exhibiting a specialist phenotype in which they only bind to certain blood group antigens, whereas strains from other regions are more generalised in their BabA-mediated binding and interact with numerous different blood-group antigens. This differential binding is thought to be a result of a selective pressure that led to evolution of the BabA protein, with the predominant blood group of a specific region influencing the binding specificity of the BabA adhesin[80]. Furthermore, strains have been identified which possess an active babA2 gene, but do not produce a functional protein[81].
BabA expression is extremely dynamic in vivo. Experimental infection of Rhesus macaques showed a loss of babA expression post-infection, caused by either a change in the number of CT dinucleotide repeats in the 5’ region of babA, or replacement of babA by the uncharacterised babB. Both events yielded a non-functional BabA protein that exhibited no Lewisb binding[82]. Another study showed loss of BabA expression six months after infection of Mongolian gerbils, and this was attributed to nucleotide changes that introduced a stop codon into the sequence, which in turn yielded a truncated BabA protein[83]. When these results were elaborated on, experiments showed that modification in six amino acids eliminated binding of H. pylori BabA to Lewisb following infection of Rhesus macaques. The babA2 gene underwent gene conversion in which a portion of the gene was replaced by a portion of the non-functioning babA1 gene. While the strains still expressed BabA, these six amino acid changes were enough to eliminate BabA binding[84]. A recent study reported inter-micro-niche variation in H. pylori infected patients, with isolates of the same strain exhibiting differences in babA and babB copy number and gene location, as well as BabA/B chimeras[85]. BabA is thus a highly variable protein that is easily susceptible to change at both a genetic and protein level.
H. pylori binds to sialylated structures present on gastric mucin and on epithelial cells via the sialic-acid binding adhesin, SabA. SabA also binds to sialylated receptors on neutrophils, which leads to nonopsonic activation of the neutrophils, phagocytosis of the bacteria and induction of the oxidative burst response[86]. Furthermore, the adhesin exhibits haemagglutinating activity, binding to gangliosides on erythrocytes in mucosal blood vessels. Differences between strains in their ability to bind to sialylated carbohydrates has been seen, and such differences may allow for the pathogen to adapt to changing glycosylation patterns in the host during infection[87]. Similar to BabA there is a high degree of genetic diversity in SabA. The gene has a number of poly-T tracts in its promoter region and a stretch of CT dinucleotide repeats in its coding region[88]. Phase variation can occur at these sites, and leads to allelic variation in the SabA locus.
Differences in the length of the CT dinucleotide repeats found in the coding regions are often seen, giving rise to various alleles. The number of repeats differs among strains, and was originally thought to determine the functionality of the sabA gene, with seven repeats yielding a functional gene, while six or eight repeats leads to the gene being turned off[89]. However, later findings speculated that other sequences in the gene rather than the number of repeats were playing a role in the functionality of SabA. The sequence following the CT repeat region has been shown to play a role in expression, with strains possessing seven CT repeats having both in frame and out of frame sabA genes depending on the nucleotide sequence following the repeat region[88].
The poly-T tracts which are found upstream of the sabA gene also undergo phase variation. Recently it was shown that the length of these tracts can vary between strains, and that the length can influence the promoter activity of the sabA gene. This variation also arises through slip-strand mispairing, similar to the CT repeats[90]. This variation could be very beneficial to H. pylori, for example when it is exposed to acidic conditions. SabA is regulated by the acid-responsive two-component ArsRS system[91]. Expression of the gene correlates inversely with the acid secretion in the stomach[92]. Therefore, when acidic conditions prevail, there is low SabA expression. The ArsRS system itself appears to repress expression of SabA, with a major increase in adherence dependent on SabA seen in mutants lacking the histidine kinase ArsS. However, this repressive effect on SabA expression only occurs in strains that have an in-frame SabA allele[93]. This highlights the role that phase variation plays in infection, as it may allow H. pylori to alter its adhesin expression in response to a change in environmental conditions.
Another mechanism that H. pylori utilises to modify SabA expression is gene duplication. It was recently shown that an increase in the copy number of sabA leads to greater production of the protein, which in turn leads to greater adherence by the bacteria in vitro. It is thought this gene conversion event occurs due to natural uptake of the DNA by competent bacteria[94]. The sabA locus is therefore a highly heterogenic one, enabling H. pylori to alter its adhesin profile depending on the microenvironment and what host defences it comes in contact with.
AlpA and AlpB are two closely related proteins carried on the same operon in the H. pylori genome[95]. Loss of these proteins was found to influence the ability of H. pylori to colonise the guinea pig stomach[96] and to bind to gastric tissue, indicating a role in adhesion of the bacteria[97]. Virtually all strains express AlpA and AlpB, indicating they have an essential function[98]. Recently, these two proteins were shown to contribute to the ability of H. pylori to bind host laminin[99]. While their role in colonization is established, the effect AlpA/B has on infection remains unclear.
Helicobacter expresses up to 33 outer membrane proteins (OMPs) or Hops (Helicobacter outer membrane porins). HopZ was identified as an OMP of H. pylori that plays a role in colonization[100]. Two allelic variants of the hopZ gene were identified, with a 20 amino acid region present in only one allele. The sequence of hopZ consists of a number of dinucleotide repeats in the signal peptide. These have also been found in other outer membrane proteins of H. pylori, and it is thought that they allow for slip-strand mispairing, which in turn generates phenotypic variation between strains. The number of repeats varies among strains, and determines functionality of the protein, with 7 or 10 dinucleotides allowing for expression of an intact protein[100]. The role of HopZ in infection has been largely unexplored. There is strong selection in vivo for HopZ expression as hopZ ON variants were recovered from volunteers challenged with a hopZ OFF strain, BCS 100[101]. Transmission of H. pylori within families has also been associated with a status change of hopZ. In contrast, hopZ sequences obtained from 26 sets of sequential isolates from chronically infected individuals showed no changes of status, suggesting that the hopZ status selected during early infection is subsequently stable[101].
Bacterial factors that increase the virulence of certain strains are responsible in part for the outcome of infection with H. pylori. The best characterized virulence factor is the product of the cytotoxin-associated gene A, CagA[102]. Strains expressing CagA are associated with more severe forms of disease and expression is closely related to that of the vacuolating cytotoxin, VacA[103]. Following binding of H. pylori to epithelial cells strains that express the CagA protein can inject it into the cells via a Type IV secretion system (T4SS) encoded by genes present on a pathogenicity island termed the cagPAI. Upon translocation into host cells CagA can be phosphorylated by host cell kinases and act to subvert host cell signaling mechanisms[104].
The cagPAI contains approximately 30 genes, many of which are involved in synthesis of the T4SS[105]. CagL is a highly conserved protein amongst H. pylori strains that forms the tip of the pilus of the T4SS, allowing for CagA translocation into host cells. It possesses an arginine-glycine-aspartate (RGD) motif, which allows the bacteria to bind to the α5β1 integrin on the surface of target cells[106]. A closely related protein, CagI, has also been shown to be an essential part of the T4SS, and the expression of both proteins is influenced by the expression of other cagPAI products, indicating that their expression requires partial assembly of the T4SS[107]. CagI however is not required for transport of CagA from the cytoplasm to the bacterial membrane[108]. The recently annotated transmembrane spanning CagM protein also appears to contribute to CagA translocation, with mutants deficient in the protein exhibiting little to no CagA translocation upon contact with host cells[109]. Non-CagPAI proteins have also been shown to be involved in the translocation process. For example the outer membrane protein HopQ is required for CagA injection as well as the intracellular responses induced by CagA[110]. While not essential, the iron-regulator Fur was also shown to influence CagA expression and cellular phenotypic changes induced upon CagA injection using adherence assays with AGS cells[111]. CagA positive strains of H. pylori which possess a functional BabA along with VacA are thought to be more virulent in the disease type they induce[112]. BabA-mediated adherence contributes to the pathogenesis of disease by inducing proinflammatory cytokines and precancerous related factors (such as CDX2), a phenotype which is dependent upon expression of the T4SS[113]. While BabA appears to promote CagA mediated pathogenesis, in contrast, binding of H. pylori via the AlpAB outer membrane proteins may signal to restrict the amount of CagA protein injected into the host cell[114]. AlpAB also seems to influence signalling cascades and immune responses upon infection, with differences seen between strains of different origins[115].
EPIYA motifs present on CagA are phosphorylated and the 145 kDa phosphorylated protein induces a strong immune response[116]. The number of motifs varies amongst strains, and strains with a higher number of motifs are more biologically active and hence more virulent, potentially playing a role in the development of gastric carcinoma[117]. Each EPIYA motif can be classified into a certain type, namely EPIYA-A,-B,-C and -D motifs[118]. Studies have shown that host kinases can be specific in the EPIYA motif that they target, with c-Src kinases exhibiting preference for EPIYA-C and EPIYA-D motifs, while c-Abl is more general and phosphorylates all four motifs[119].
CagA also exhibits phosphorylation-independent effects, many of which remain elusive. A conserved-motif in the C-terminus of the non-phosphorylated protein has recently been identified, and was shown to interact with the host hepatocyte growth factor receptor Met, which contributes to cellular proliferation and inflammation via the Akt signalling pathway, which activates NF-κB and β-catenin[120]. CagA also disrupts Par1b kinase activation, a protein involved in maintaining cell polarity and adherens junctions, independently of tyrosine phosphorylation[121]. The complex formed between E-cadherin and β-catenin in the adherens junctions of cells is also targeted by H. pylori in a CagA phosphorylation-independent manner. A physical interaction between CagA and E-cadherin disrupts the complex, and leads to a cytoplasmic accumulation of β-catenin, a protein which, when deregulated, has been shown to contribute to carcinogenesis[122]. Taken together the above suggests that binding of H. pylori to epithelial cells is a dynamic process and bacteria can alter the expression of adhesins depending on the availability of receptors. Close association of the bacteria with epithelial cells enables the translocation of the bacterial CagA protein across the cell membrane where it can be phosphorylated and lead to subversion of cell signaling. CagA can also cause disruption of epithelial cell junctions in a phosphorylation independent manner. CagA is thus a key virulence factor of H. pylori. Figure 1 illustrates H. pylori colonization of gastric mucus, the interaction of bacteria with host cells and the events triggered by that interaction.
The ability of H. pylori to cause disease depends not just on bacterial factors but also on environmental and host factors. A number of host gene polymorphisms have been identified which are thought to increase H. pylori colonization and increase susceptibility to disease. Some polymorphisms have been identified as risk factors for H. pylori associated gastric cancer. The IL-1 gene cluster located on chromosome 2q is composed of three genes which encode the pro-inflammatory cytokines IL-1α and IL-1β as well as their endogenous receptor antagonist IL-1ra. IL-1β is a potent inhibitor of gastric acid secretion in vivo[123,124]. H. pylori infection results in an upregulation of IL-1β, which plays an important role in initiating and amplifying the immune response[125,126]. Three diallelic polymorphisms in the IL-1β gene have been identified all of which are C-T substitutions at positions -511, -31 and +3954 from the transcriptional start site. Polymorphisms at position -511 and -31 are associated with increased IL-1β secretion and subsequent hypochlorhydria in the presence of H. pylori[127], symptoms which have been implicated in the development of gastric cancer. A recent study in Venezuela has reported that infection with the more virulent cagA+ H. pylori strains is associated with individuals harbouring the +3954 polymorphism[128].
Polymorphisms in IFN-γ were associated with infection by cag positive strains and polymorphism in TNF-α were associated with development of peptic ulcer disease while polymorphisms in IL-10 favoured the development of intestinal metaplasia and non-cardia gastric cancer[129]. Polymorphisms which effect cytokine function may explain the highly variable outcomes of H. pylori infection, highlighting the importance of genetic background of the host in disease progression and susceptibility to infection by specific H. pylori strains.
Polymorphisms in genes involved in the innate and adaptive immune response play a role in host susceptibility to infection and disease progression. IL-2 plays an important role in mediating the T lymphocyte response including the Th1 phenotype which is known to dominate the immune response to H. pylori infection. An increase in IL-2 production as a result of a gene polymorphism, T330G, has been shown to be negatively associated with H. pylori infection in adults[130]. Autophagy plays a critical role in the modulation of host immunity/inflammatory responses and is thought to serve as an innate defence mechanism against infection[131,132]. A single nucleotide polymorphism in the autophagy gene, ATg16L1 (T300A), has been identified as a causal risk variant for Crohn’s disease. The functional relevance of this polymorphism is not known but it is thought to result in production of an unstable ATg16L1 protein leading to impaired cytokine responses and antimicrobial autophagy. Induction of autophagy in response to the H. pylori VacA cytotoxin is significantly reduced in cells harbouring the T300A polymorphism and a survey of Scottish and German individuals showed that the T300A polymorphism is associated with a higher odds ratio of H. pylori infection[133], suggesting that the T300A polymorphism in ATg16L1 confers a modest but significant host genetic risk of H. pylori infection.
There is an association between polymorphisms in the gene encoding the membrane-associated mucin MUC1 which result in shorter MUC1 alleles and H. pylori related gastritis[134]. Short MUC1 alleles have also been linked to the development of gastric adenocarcinoma[135] and intestinal metaplasia in patients with chronic gastritis[136]. In support of the hypothesis that a link exists between short MUC1 alleles and H. pylori related pathologies it has been shown that Muc1-/- mice display increased colonization by H. pylori, compared to wildtype controls, and also display increased severity of H. pylori induced gastritis. Shorter MUC1 alleles encode proteins with a much smaller extracellular domain, thereby reducing the ability of MUC1 on the surface of cells to maintain a distance between the microbe and the cell surface and thus protect the cell from microbial adhesion and the subsequent development of disease[67].
H. pylori is rarely found in the mucus covering the gastric gland[59] where mucins are capped with a terminal α-1,4-linked N-acetylglucosamine[137]. Glycans possessing this residue suppress H. pylori growth in vitro[61]. The transfer of α-1,4-linked N-acetylglucosamine is mediated by a transferase encoded by the gene a4GnT[137]. Polymorphisms in this gene may lead to altered expression or function of the encoded transferase. An analysis of single nucleotide polymorphisms in a4GnT found that changes in H. pylori seropositivity were associated with changes in a4GnT and may be related to an increased risk of H. pylori infection[138].
The secretor status of an individual is also thought to influence their susceptibility to infection. A Fucα1,2-glycan is common to all three ABH blood group antigens. These carbohydrate structures are expressed along the gastro-intestinal tract of individuals with a positive secretor status[139,140]. Secretor individuals express the secretor-(fucosyl)transferase, an enzyme involved in the synthesis of Fucα1,2-glycan. Non-secretors lack expression of this fucosyl-transferase[141] while weak secretors express a mutated secretor-(fucosyl)transferase with reduced enzymatic activity. Studies involving infected secretor and non-secretor mice have revealed that the non-secretor mice display reduced adhesion of H. pylori to gastric tissue[142]. In addition experimental infection of Rhesus monkeys has revealed that the weak secretor phenotype displays reduced H. pylori density and reduced inflammation[143]. Non-secretor phenotypes have been shown to express higher inflammatory reactivity and expression of sialylated antigens in response to H. pylori infection, and this may explain the higher incidence of peptic ulcer disease previously reported in these individuals[144]. Bacterial factors alone cannot explain why some individuals develop disease upon H. pylori infection whereas others remain asymptomatic. The discovery of host factors such as the polymorphisms described above go some way to explaining the different outcomes of infection in different individuals.
H. pylori has evolved with humans over thousands of years and is a paradigm of chronic infection. Both bacterial and host factors play a role in mediating bacterial virulence and susceptibility to infection and can explain somewhat the outcomes of infection in different individuals. However, we suggest that H. pylori colonization of gastric mucus is key to the ability of the organism to cause chronic infection in humans. The establishment of a reservoir of bacteria in gastric mucus close to the epithelial surface would allow H. pylori to avoid removal by mucus flow and killing by gastric acid. The interaction of H. pylori with gastric mucus could limit the number of bacteria that can interact with gastric epithelial cells and thus also limit the inflammatory response and promote chronic infection. Modulation of the interaction of H. pylori with gastric mucus may be a viable alternative or even adjuvant therapy to antimicrobials for prevention of colonization and the eradication of the organism. Given that the majority of infections in humans and animals occur through mucosal surfaces combined with the results of recent studies which show that alterations in the composition of the gut microflora, sometimes referred to as dysbiosis, are associated with a number of chronic diseases including obesity[145], diabetes[146] and inflammatory disease[147] there is now intense interest in how bacteria living in mucus cause disease. Studies on H. pylori are likely to serve as a valuable reference system for how other organisms colonise and infect mucosal surfaces.
| 1. | Buck GE, Gourley WK, Lee WK, Subramanyam K, Latimer JM, DiNuzzo AR. Relation of Campylobacter pyloridis to gastritis and peptic ulcer. J Infect Dis. 1986;153:664-669. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 164] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 2. | Kuipers EJ, Thijs JC, Festen HP. The prevalence of Helicobacter pylori in peptic ulcer disease. Aliment Pharmacol Ther. 1995;9 Suppl 2:59-69. [PubMed] |
| 3. | Peek RM, Crabtree JE. Helicobacter infection and gastric neoplasia. J Pathol. 2006;208:233-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 368] [Cited by in RCA: 448] [Article Influence: 21.3] [Reference Citation Analysis (2)] |
| 4. | Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, Yamakido M, Taniyama K, Sasaki N, Schlemper RJ. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345:784-789. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3126] [Cited by in RCA: 3247] [Article Influence: 129.9] [Reference Citation Analysis (1)] |
| 5. | Schistosomes , liver flukes and Helicobacter pylori. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Lyon, 7-14 June 1994. IARC Monogr Eval Carcinog Risks Hum. 1994;61:1-241. [PubMed] |
| 6. | Parsonnet J, Vandersteen D, Goates J, Sibley RK, Pritikin J, Chang Y. Helicobacter pylori infection in intestinal- and diffuse-type gastric adenocarcinomas. J Natl Cancer Inst. 1991;83:640-643. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 248] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 7. | Hansson LE, Nyrén O, Hsing AW, Bergström R, Josefsson S, Chow WH, Fraumeni JF, Adami HO. The risk of stomach cancer in patients with gastric or duodenal ulcer disease. N Engl J Med. 1996;335:242-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 432] [Cited by in RCA: 431] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 8. | Wotherspoon AC, Doglioni C, Diss TC, Pan L, Moschini A, de Boni M, Isaacson PG. Regression of primary low-grade B-cell gastric lymphoma of mucosa-associated lymphoid tissue type after eradication of Helicobacter pylori. Lancet. 1993;342:575-577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1564] [Cited by in RCA: 1390] [Article Influence: 42.1] [Reference Citation Analysis (0)] |
| 9. | Stolte M, Bayerdörffer E, Morgner A, Alpen B, Wündisch T, Thiede C, Neubauer A. Helicobacter and gastric MALT lymphoma. Gut. 2002;50 Suppl 3:III19-III24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 131] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 10. | Wyatt JI, Rathbone BJ, Sobala GM, Shallcross T, Heatley RV, Axon AT, Dixon MF. Gastric epithelium in the duodenum: its association with Helicobacter pylori and inflammation. J Clin Pathol. 1990;43:981-986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 126] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 11. | Hessey SJ, Spencer J, Wyatt JI, Sobala G, Rathbone BJ, Axon AT, Dixon MF. Bacterial adhesion and disease activity in Helicobacter associated chronic gastritis. Gut. 1990;31:134-138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 222] [Cited by in RCA: 207] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 12. | Müller A, Oertli M, Arnold IC. H. pylori exploits and manipulates innate and adaptive immune cell signaling pathways to establish persistent infection. Cell Commun Signal. 2011;9:25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 63] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 13. | Salama NR, Hartung ML, Müller A. Life in the human stomach: persistence strategies of the bacterial pathogen Helicobacter pylori. Nat Rev Microbiol. 2013;11:385-399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 405] [Cited by in RCA: 502] [Article Influence: 38.6] [Reference Citation Analysis (2)] |
| 14. | Eaton KA, Brooks CL, Morgan DR, Krakowka S. Essential role of urease in pathogenesis of gastritis induced by Helicobacter pylori in gnotobiotic piglets. Infect Immun. 1991;59:2470-2475. [PubMed] |
| 15. | Eaton KA, Krakowka S. Effect of gastric pH on urease-dependent colonization of gnotobiotic piglets by Helicobacter pylori. Infect Immun. 1994;62:3604-3607. [PubMed] |
| 16. | Meyer-Rosberg K, Scott DR, Rex D, Melchers K, Sachs G. The effect of environmental pH on the proton motive force of Helicobacter pylori. Gastroenterology. 1996;111:886-900. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 146] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 17. | Dunn BE, Campbell GP, Perez-Perez GI, Blaser MJ. Purification and characterization of urease from Helicobacter pylori. J Biol Chem. 1990;265:9464-9469. [PubMed] |
| 18. | Turbett GR, Høj PB, Horne R, Mee BJ. Purification and characterization of the urease enzymes of Helicobacter species from humans and animals. Infect Immun. 1992;60:5259-5266. [PubMed] |
| 19. | Ha NC, Oh ST, Sung JY, Cha KA, Lee MH, Oh BH. Supramolecular assembly and acid resistance of Helicobacter pylori urease. Nat Struct Biol. 2001;8:505-509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 337] [Cited by in RCA: 367] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 20. | Mobley HL, Island MD, Hausinger RP. Molecular biology of microbial ureases. Microbiol Rev. 1995;59:451-480. [PubMed] |
| 21. | Labigne A, Cussac V, Courcoux P. Shuttle cloning and nucleotide sequences of Helicobacter pylori genes responsible for urease activity. J Bacteriol. 1991;173:1920-1931. [PubMed] |
| 22. | Cussac V, Ferrero RL, Labigne A. Expression of Helicobacter pylori urease genes in Escherichia coli grown under nitrogen-limiting conditions. J Bacteriol. 1992;174:2466-2473. [PubMed] |
| 23. | Wen Y, Feng J, Scott DR, Marcus EA, Sachs G. A cis-encoded antisense small RNA regulated by the HP0165-HP0166 two-component system controls expression of ureB in Helicobacter pylori. J Bacteriol. 2011;193:40-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 31] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 24. | Marcus EA, Scott DR. Cell lysis is responsible for the appearance of extracellular urease in Helicobacter pylori. Helicobacter. 2001;6:93-99. [PubMed] |
| 25. | Phadnis SH, Parlow MH, Levy M, Ilver D, Caulkins CM, Connors JB, Dunn BE. Surface localization of Helicobacter pylori urease and a heat shock protein homolog requires bacterial autolysis. Infect Immun. 1996;64:905-912. [PubMed] |
| 26. | Bode G, Malfertheiner P, Nilius M, Lehnhardt G, Ditschuneit H. Ultrastructural localisation of urease in outer membrane and periplasm of Campylobacter pylori. J Clin Pathol. 1989;42:778-779. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 21] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 27. | Scott DR, Weeks D, Hong C, Postius S, Melchers K, Sachs G. The role of internal urease in acid resistance of Helicobacter pylori. Gastroenterology. 1998;114:58-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 200] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 28. | Stingl K, Altendorf K, Bakker EP. Acid survival of Helicobacter pylori: how does urease activity trigger cytoplasmic pH homeostasis? Trends Microbiol. 2002;10:70-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 137] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 29. | Weeks DL, Eskandari S, Scott DR, Sachs G. A H+-gated urea channel: the link between Helicobacter pylori urease and gastric colonization. Science. 2000;287:482-485. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 364] [Cited by in RCA: 340] [Article Influence: 13.1] [Reference Citation Analysis (1)] |
| 30. | Clyne M, Labigne A, Drumm B. Helicobacter pylori requires an acidic environment to survive in the presence of urea. Infect Immun. 1995;63:1669-1673. [PubMed] |
| 31. | Marcus EA, Moshfegh AP, Sachs G, Scott DR. The periplasmic alpha-carbonic anhydrase activity of Helicobacter pylori is essential for acid acclimation. J Bacteriol. 2005;187:729-738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 145] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 32. | Geis G, Leying H, Suerbaum S, Mai U, Opferkuch W. Ultrastructure and chemical analysis of Campylobacter pylori flagella. J Clin Microbiol. 1989;27:436-441. [PubMed] |
| 33. | Eaton KA, Morgan DR, Krakowka S. Motility as a factor in the colonisation of gnotobiotic piglets by Helicobacter pylori. J Med Microbiol. 1992;37:123-127. [PubMed] |
| 34. | Eaton KA, Suerbaum S, Josenhans C, Krakowka S. Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect Immun. 1996;64:2445-2448. [PubMed] |
| 35. | Ottemann KM, Lowenthal AC. Helicobacter pylori uses motility for initial colonization and to attain robust infection. Infect Immun. 2002;70:1984-1990. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 218] [Cited by in RCA: 244] [Article Influence: 10.2] [Reference Citation Analysis (3)] |
| 36. | Geis G, Suerbaum S, Forsthoff B, Leying H, Opferkuch W. Ultrastructure and biochemical studies of the flagellar sheath of Helicobacter pylori. J Med Microbiol. 1993;38:371-377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 79] [Article Influence: 2.4] [Reference Citation Analysis (1)] |
| 37. | Josenhans C, Labigne A, Suerbaum S. Comparative ultrastructural and functional studies of Helicobacter pylori and Helicobacter mustelae flagellin mutants: both flagellin subunits, FlaA and FlaB, are necessary for full motility in Helicobacter species. J Bacteriol. 1995;177:3010-3020. [PubMed] |
| 38. | O'Toole PW, Kostrzynska M, Trust TJ. Non-motile mutants of Helicobacter pylori and Helicobacter mustelae defective in flagellar hook production. Mol Microbiol. 1994;14:691-703. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 78] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 39. | Kim JS, Chang JH, Chung SI, Yum JS. Molecular cloning and characterization of the Helicobacter pylori fliD gene, an essential factor in flagellar structure and motility. J Bacteriol. 1999;181:6969-6976. [PubMed] |
| 40. | Schmitz A, Josenhans C, Suerbaum S. Cloning and characterization of the Helicobacter pylori flbA gene, which codes for a membrane protein involved in coordinated expression of flagellar genes. J Bacteriol. 1997;179:987-997. [PubMed] |
| 41. | McGee DJ, Coker C, Testerman TL, Harro JM, Gibson SV, Mobley HL. The Helicobacter pylori flbA flagellar biosynthesis and regulatory gene is required for motility and virulence and modulates urease of H. pylori and Proteus mirabilis. J Med Microbiol. 2002;51:958-970. [PubMed] |
| 42. | Spohn G, Scarlato V. Motility of Helicobacter pylori is coordinately regulated by the transcriptional activator FlgR, an NtrC homolog. J Bacteriol. 1999;181:593-599. [PubMed] |
| 43. | Wen Y, Feng J, Scott DR, Marcus EA, Sachs G. The pH-responsive regulon of HP0244 (FlgS), the cytoplasmic histidine kinase of Helicobacter pylori. J Bacteriol. 2009;191:449-460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 41] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 44. | Rader BA, Campagna SR, Semmelhack MF, Bassler BL, Guillemin K. The quorum-sensing molecule autoinducer 2 regulates motility and flagellar morphogenesis in Helicobacter pylori. J Bacteriol. 2007;189:6109-6117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 81] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 45. | Shen F, Hobley L, Doherty N, Loh JT, Cover TL, Sockett RE, Hardie KR, Atherton JC. In Helicobacter pylori auto-inducer-2, but not LuxS/MccAB catalysed reverse transsulphuration, regulates motility through modulation of flagellar gene transcription. BMC Microbiol. 2010;10:210. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 37] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 46. | Osaki T, Hanawa T, Manzoku T, Fukuda M, Kawakami H, Suzuki H, Yamaguchi H, Yan X, Taguchi H, Kurata S. Mutation of luxS affects motility and infectivity of Helicobacter pylori in gastric mucosa of a Mongolian gerbil model. J Med Microbiol. 2006;55:1477-1485. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 52] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 47. | Asakura H, Churin Y, Bauer B, Boettcher JP, Bartfeld S, Hashii N, Kawasaki N, Mollenkopf HJ, Jungblut PR, Brinkmann V. Helicobacter pylori HP0518 affects flagellin glycosylation to alter bacterial motility. Mol Microbiol. 2010;78:1130-1144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 51] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 48. | Schirm M, Soo EC, Aubry AJ, Austin J, Thibault P, Logan SM. Structural, genetic and functional characterization of the flagellin glycosylation process in Helicobacter pylori. Mol Microbiol. 2003;48:1579-1592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 220] [Cited by in RCA: 241] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 49. | Roure S, Bonis M, Chaput C, Ecobichon C, Mattox A, Barrière C, Geldmacher N, Guadagnini S, Schmitt C, Prévost MC. Peptidoglycan maturation enzymes affect flagellar functionality in bacteria. Mol Microbiol. 2012;86:845-856. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 44] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 50. | Atuma C, Strugala V, Allen A, Holm L. The adherent gastrointestinal mucus gel layer: thickness and physical state in vivo. Am J Physiol Gastrointest Liver Physiol. 2001;280:G922-G929. [PubMed] |
| 51. | Schreiber S, Bücker R, Groll C, Azevedo-Vethacke M, Garten D, Scheid P, Friedrich S, Gatermann S, Josenhans C, Suerbaum S. Rapid loss of motility of Helicobacter pylori in the gastric lumen in vivo. Infect Immun. 2005;73:1584-1589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 58] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 52. | Thomsen LL, Gavin JB, Tasman-Jones C. Relation of Helicobacter pylori to the human gastric mucosa in chronic gastritis of the antrum. Gut. 1990;31:1230-1236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 60] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 53. | Windle HJ, Fox A, Ní Eidhin D, Kelleher D. The thioredoxin system of Helicobacter pylori. J Biol Chem. 2000;275:5081-5089. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 62] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 54. | Celli JP, Turner BS, Afdhal NH, Ewoldt RH, McKinley GH, Bansil R, Erramilli S. Rheology of gastric mucin exhibits a pH-dependent sol-gel transition. Biomacromolecules. 2007;8:1580-1586. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 233] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 55. | Celli JP, Turner BS, Afdhal NH, Keates S, Ghiran I, Kelly CP, Ewoldt RH, McKinley GH, So P, Erramilli S. Helicobacter pylori moves through mucus by reducing mucin viscoelasticity. Proc Natl Acad Sci USA. 2009;106:14321-14326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 261] [Cited by in RCA: 296] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 56. | Schreiber S, Konradt M, Groll C, Scheid P, Hanauer G, Werling HO, Josenhans C, Suerbaum S. The spatial orientation of Helicobacter pylori in the gastric mucus. Proc Natl Acad Sci USA. 2004;101:5024-5029. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 249] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 57. | Sycuro LK, Pincus Z, Gutierrez KD, Biboy J, Stern CA, Vollmer W, Salama NR. Peptidoglycan crosslinking relaxation promotes Helicobacter pylori’s helical shape and stomach colonization. Cell. 2010;141:822-833. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 234] [Cited by in RCA: 217] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 58. | Sycuro LK, Wyckoff TJ, Biboy J, Born P, Pincus Z, Vollmer W, Salama NR. Multiple peptidoglycan modification networks modulate Helicobacter pylori’s cell shape, motility, and colonization potential. PLoS Pathog. 2012;8:e1002603. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 122] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 59. | Hidaka E, Ota H, Hidaka H, Hayama M, Matsuzawa K, Akamatsu T, Nakayama J, Katsuyama T. Helicobacter pylori and two ultrastructurally distinct layers of gastric mucous cell mucins in the surface mucous gel layer. Gut. 2001;49:474-480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 71] [Article Influence: 2.8] [Reference Citation Analysis (1)] |
| 60. | Van den Brink GR, Tytgat KM, Van der Hulst RW, Van der Loos CM, Einerhand AW, Büller HA, Dekker J. H pylori colocalises with MUC5AC in the human stomach. Gut. 2000;46:601-607. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 112] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 61. | Kawakubo M, Ito Y, Okimura Y, Kobayashi M, Sakura K, Kasama S, Fukuda MN, Fukuda M, Katsuyama T, Nakayama J. Natural antibiotic function of a human gastric mucin against Helicobacter pylori infection. Science. 2004;305:1003-1006. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 253] [Cited by in RCA: 256] [Article Influence: 11.6] [Reference Citation Analysis (1)] |
| 62. | Genta RM, Gürer IE, Graham DY, Krishnan B, Segura AM, Gutierrez O, Kim JG, Burchette JL. Adherence of Helicobacter pylori to areas of incomplete intestinal metaplasia in the gastric mucosa. Gastroenterology. 1996;111:1206-1211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 65] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 63. | Teixeira A, David L, Reis CA, Costa J, Sobrinho-Simões M. Expression of mucins (MUC1, MUC2, MUC5AC, and MUC6) and type 1 Lewis antigens in cases with and without Helicobacter pylori colonization in metaplastic glands of the human stomach. J Pathol. 2002;197:37-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 41] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 64. | Ilver D, Arnqvist A, Ogren J, Frick IM, Kersulyte D, Incecik ET, Berg DE, Covacci A, Engstrand L, Borén T. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science. 1998;279:373-377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 872] [Cited by in RCA: 850] [Article Influence: 30.4] [Reference Citation Analysis (1)] |
| 65. | Mahdavi J, Sondén B, Hurtig M, Olfat FO, Forsberg L, Roche N, Angstrom J, Larsson T, Teneberg S, Karlsson KA. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science. 2002;297:573-578. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 639] [Cited by in RCA: 674] [Article Influence: 28.1] [Reference Citation Analysis (0)] |
| 66. | Ota H, Nakayama J, Momose M, Hayama M, Akamatsu T, Katsuyama T, Graham DY, Genta RM. Helicobacter pylori infection produces reversible glycosylation changes to gastric mucins. Virchows Arch. 1998;433:419-426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 67] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 67. | Lindén SK, Sheng YH, Every AL, Miles KM, Skoog EC, Florin TH, Sutton P, McGuckin MA. MUC1 limits Helicobacter pylori infection both by steric hindrance and by acting as a releasable decoy. PLoS Pathog. 2009;5:e1000617. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 184] [Cited by in RCA: 215] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 68. | McGuckin MA, Every AL, Skene CD, Linden SK, Chionh YT, Swierczak A, McAuley J, Harbour S, Kaparakis M, Ferrero R. Muc1 mucin limits both Helicobacter pylori colonization of the murine gastric mucosa and associated gastritis. Gastroenterology. 2007;133:1210-1218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 157] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 69. | Guang W, Ding H, Czinn SJ, Kim KC, Blanchard TG, Lillehoj EP. Muc1 cell surface mucin attenuates epithelial inflammation in response to a common mucosal pathogen. J Biol Chem. 2010;285:20547-20557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 80] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 70. | Guang W, Twaddell WS, Lillehoj EP. Molecular Interactions between MUC1 Epithelial Mucin, β-Catenin, and CagA Proteins. Front Immunol. 2012;3:105. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 71. | Dolan B, Naughton J, Tegtmeyer N, May FE, Clyne M. The interaction of Helicobacter pylori with the adherent mucus gel layer secreted by polarized HT29-MTX-E12 cells. PLoS One. 2012;7:e47300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 37] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 72. | Navabi N, Johansson ME, Raghavan S, Lindén SK. Helicobacter pylori infection impairs the mucin production rate and turnover in the murine gastric mucosa. Infect Immun. 2013;81:829-837. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 69] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 73. | Skoog EC, Sjöling Å, Navabi N, Holgersson J, Lundin SB, Lindén SK. Human gastric mucins differently regulate Helicobacter pylori proliferation, gene expression and interactions with host cells. PLoS One. 2012;7:e36378. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 73] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 74. | Naughton JA, Mariño K, Dolan B, Reid C, Gough R, Gallagher ME, Kilcoyne M, Gerlach JQ, Joshi L, Rudd P. Divergent mechanisms of interaction of Helicobacter pylori and Campylobacter jejuni with mucus and mucins. Infect Immun. 2013;81:2838-2850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 50] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 75. | Clyne M, Dillon P, Daly S, O’Kennedy R, May FE, Westley BR, Drumm B. Helicobacter pylori interacts with the human single-domain trefoil protein TFF1. Proc Natl Acad Sci USA. 2004;101:7409-7414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 69] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 76. | Reeves EP, Ali T, Leonard P, Hearty S, O’Kennedy R, May FE, Westley BR, Josenhans C, Rust M, Suerbaum S. Helicobacter pylori lipopolysaccharide interacts with TFF1 in a pH-dependent manner. Gastroenterology. 2008;135:2043-2054, 2054.e1-2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 73] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 77. | Borén T, Falk P, Roth KA, Larson G, Normark S. Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science. 1993;262:1892-1895. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 840] [Cited by in RCA: 812] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 78. | Hennig EE, Mernaugh R, Edl J, Cao P, Cover TL. Heterogeneity among Helicobacter pylori strains in expression of the outer membrane protein BabA. Infect Immun. 2004;72:3429-3435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 58] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 79. | Pride DT, Meinersmann RJ, Blaser MJ. Allelic Variation within Helicobacter pylori babA and babB. Infect Immun. 2001;69:1160-1171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 99] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 80. | Aspholm-Hurtig M, Dailide G, Lahmann M, Kalia A, Ilver D, Roche N, Vikström S, Sjöström R, Lindén S, Bäckström A. Functional adaptation of BabA, the H. pylori ABO blood group antigen binding adhesin. Science. 2004;305:519-522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 290] [Cited by in RCA: 304] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 81. | Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539-547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2635] [Cited by in RCA: 2603] [Article Influence: 89.8] [Reference Citation Analysis (0)] |
| 82. | Solnick JV, Hansen LM, Salama NR, Boonjakuakul JK, Syvanen M. Modification of Helicobacter pylori outer membrane protein expression during experimental infection of rhesus macaques. Proc Natl Acad Sci USA. 2004;101:2106-2111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 186] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 83. | Ohno T, Vallström A, Rugge M, Ota H, Graham DY, Arnqvist A, Yamaoka Y. Effects of blood group antigen-binding adhesin expression during Helicobacter pylori infection of Mongolian gerbils. J Infect Dis. 2011;203:726-735. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 41] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 84. | Styer CM, Hansen LM, Cooke CL, Gundersen AM, Choi SS, Berg DE, Benghezal M, Marshall BJ, Peek RM, Borén T. Expression of the BabA adhesin during experimental infection with Helicobacter pylori. Infect Immun. 2010;78:1593-1600. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 81] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 85. | Matteo MJ, Armitano RI, Romeo M, Wonaga A, Olmos M, Catalano M. Helicobacter pylori bab genes during chronic colonization. Int J Mol Epidemiol Genet. 2011;2:286-291. [PubMed] |
| 86. | Unemo M, Aspholm-Hurtig M, Ilver D, Bergström J, Borén T, Danielsson D, Teneberg S. The sialic acid binding SabA adhesin of Helicobacter pylori is essential for nonopsonic activation of human neutrophils. J Biol Chem. 2005;280:15390-15397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 89] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 87. | Aspholm M, Olfat FO, Nordén J, Sondén B, Lundberg C, Sjöström R, Altraja S, Odenbreit S, Haas R, Wadström T. SabA is the H. pylori hemagglutinin and is polymorphic in binding to sialylated glycans. PLoS Pathog. 2006;2:e110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 129] [Cited by in RCA: 143] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 88. | Lehours P, Ménard A, Dupouy S, Bergey B, Richy F, Zerbib F, Ruskoné-Fourmestraux A, Delchier JC, Mégraud F. Evaluation of the association of nine Helicobacter pylori virulence factors with strains involved in low-grade gastric mucosa-associated lymphoid tissue lymphoma. Infect Immun. 2004;72:880-888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 99] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 89. | de Jonge R, Pot RG, Loffeld RJ, van Vliet AH, Kuipers EJ, Kusters JG. The functional status of the Helicobacter pylori sabB adhesin gene as a putative marker for disease outcome. Helicobacter. 2004;9:158-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 41] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 90. | Kao CY, Sheu SM, Sheu BS, Wu JJ. Length of thymidine homopolymeric repeats modulates promoter activity of sabA in Helicobacter pylori. Helicobacter. 2012;17:203-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 91. | Forsyth MH, Cao P, Garcia PP, Hall JD, Cover TL. Genome-wide transcriptional profiling in a histidine kinase mutant of Helicobacter pylori identifies members of a regulon. J Bacteriol. 2002;184:4630-4635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 41] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 92. | Yamaoka Y, Ojo O, Fujimoto S, Odenbreit S, Haas R, Gutierrez O, El-Zimaity HM, Reddy R, Arnqvist A, Graham DY. Helicobacter pylori outer membrane proteins and gastroduodenal disease. Gut. 2006;55:775-781. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 191] [Cited by in RCA: 176] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 93. | Goodwin AC, Weinberger DM, Ford CB, Nelson JC, Snider JD, Hall JD, Paules CI, Peek RM, Forsyth MH. Expression of the Helicobacter pylori adhesin SabA is controlled via phase variation and the ArsRS signal transduction system. Microbiology. 2008;154:2231-2240. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 67] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 94. | Talarico S, Whitefield SE, Fero J, Haas R, Salama NR. Regulation of Helicobacter pylori adherence by gene conversion. Mol Microbiol. 2012;84:1050-1061. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 47] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 95. | Odenbreit S, Till M, Hofreuter D, Faller G, Haas R. Genetic and functional characterization of the alpAB gene locus essential for the adhesion of Helicobacter pylori to human gastric tissue. Mol Microbiol. 1999;31:1537-1548. [PubMed] |
| 96. | de Jonge R, Durrani Z, Rijpkema SG, Kuipers EJ, van Vliet AH, Kusters JG. Role of the Helicobacter pylori outer-membrane proteins AlpA and AlpB in colonization of the guinea pig stomach. J Med Microbiol. 2004;53:375-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 66] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 97. | Odenbreit S, Faller G, Haas R. Role of the alpAB proteins and lipopolysaccharide in adhesion of Helicobacter pylori to human gastric tissue. Int J Med Microbiol. 2002;292:247-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 67] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 98. | Odenbreit S, Swoboda K, Barwig I, Ruhl S, Borén T, Koletzko S, Haas R. Outer membrane protein expression profile in Helicobacter pylori clinical isolates. Infect Immun. 2009;77:3782-3790. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 86] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 99. | Senkovich OA, Yin J, Ekshyyan V, Conant C, Traylor J, Adegboyega P, McGee DJ, Rhoads RE, Slepenkov S, Testerman TL. Helicobacter pylori AlpA and AlpB bind host laminin and influence gastric inflammation in gerbils. Infect Immun. 2011;79:3106-3116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 89] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 100. | Peck B, Ortkamp M, Diehl KD, Hundt E, Knapp B. Conservation, localization and expression of HopZ, a protein involved in adhesion of Helicobacter pylori. Nucleic Acids Res. 1999;27:3325-3333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 133] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 101. | Kennemann L, Brenneke B, Andres S, Engstrand L, Meyer TF, Aebischer T, Josenhans C, Suerbaum S. In vivo sequence variation in HopZ, a phase-variable outer membrane protein of Helicobacter pylori. Infect Immun. 2012;80:4364-4373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 40] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 102. | Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G, Massone A, Papini E, Xiang Z, Figura N. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci USA. 1993;90:5791-5795. [PubMed] |
| 103. | Xiang Z, Censini S, Bayeli PF, Telford JL, Figura N, Rappuoli R, Covacci A. Analysis of expression of CagA and VacA virulence factors in 43 strains of Helicobacter pylori reveals that clinical isolates can be divided into two major types and that CagA is not necessary for expression of the vacuolating cytotoxin. Infect Immun. 1995;63:94-98. [PubMed] |
| 104. | Odenbreit S, Püls J, Sedlmaier B, Gerland E, Fischer W, Haas R. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science. 2000;287:1497-1500. [PubMed] |
| 105. | Censini S, Lange C, Xiang Z, Crabtree JE, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA. 1996;93:14648-14653. [PubMed] |
| 106. | Kwok T, Zabler D, Urman S, Rohde M, Hartig R, Wessler S, Misselwitz R, Berger J, Sewald N, König W. Helicobacter exploits integrin for type IV secretion and kinase activation. Nature. 2007;449:862-866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 473] [Cited by in RCA: 515] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 107. | Pham KT, Weiss E, Jiménez Soto LF, Breithaupt U, Haas R, Fischer W. CagI is an essential component of the Helicobacter pylori Cag type IV secretion system and forms a complex with CagL. PLoS One. 2012;7:e35341. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 48] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 108. | Kumar N, Shariq M, Kumari R, Tyagi RK, Mukhopadhyay G. Cag type IV secretion system: CagI independent bacterial surface localization of CagA. PLoS One. 2013;8:e74620. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 24] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 109. | Ling F, Wang X, Dai D, Yu M, Chen C, Qian J, Liu C, Zhang Y, Ding J, Guan XW. The Helicobacter pylori protein CagM is located in the transmembrane channel that is required for CagA translocation. Curr Microbiol. 2013;67:531-536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 110. | Belogolova E, Bauer B, Pompaiah M, Asakura H, Brinkman V, Ertl C, Bartfeld S, Nechitaylo TY, Haas R, Machuy N. Helicobacter pylori outer membrane protein HopQ identified as a novel T4SS-associated virulence factor. Cell Microbiol. 2013;15:1896-1912. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 54] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 111. | Raghwan R. Host cell contact induces fur-dependent expression of virulence factors CagA and VacA in Helicobacter pylori. Helicobacter. 2014;19:17-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 112. | Gerhard M, Lehn N, Neumayer N, Borén T, Rad R, Schepp W, Miehlke S, Classen M, Prinz C. Clinical relevance of the Helicobacter pylori gene for blood-group antigen-binding adhesin. Proc Natl Acad Sci USA. 1999;96:12778-12783. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 431] [Cited by in RCA: 436] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 113. | Ishijima N, Suzuki M, Ashida H, Ichikawa Y, Kanegae Y, Saito I, Borén T, Haas R, Sasakawa C, Mimuro H. BabA-mediated adherence is a potentiator of the Helicobacter pylori type IV secretion system activity. J Biol Chem. 2011;286:25256-25264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 155] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 114. | Jiménez-Soto LF, Clausen S, Sprenger A, Ertl C, Haas R. Dynamics of the Cag-type IV secretion system of Helicobacter pylori as studied by bacterial co-infections. Cell Microbiol. 2013;15:1924-1937. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 115. | Lu H, Wu JY, Beswick EJ, Ohno T, Odenbreit S, Haas R, Reyes VE, Kita M, Graham DY, Yamaoka Y. Functional and intracellular signaling differences associated with the Helicobacter pylori AlpAB adhesin from Western and East Asian strains. J Biol Chem. 2007;282:6242-6254. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 69] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 116. | Asahi M, Azuma T, Ito S, Ito Y, Suto H, Nagai Y, Tsubokawa M, Tohyama Y, Maeda S, Omata M. Helicobacter pylori CagA protein can be tyrosine phosphorylated in gastric epithelial cells. J Exp Med. 2000;191:593-602. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 359] [Cited by in RCA: 376] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 117. | Higashi H, Tsutsumi R, Fujita A, Yamazaki S, Asaka M, Azuma T, Hatakeyama M. Biological activity of the Helicobacter pylori virulence factor CagA is determined by variation in the tyrosine phosphorylation sites. Proc Natl Acad Sci USA. 2002;99:14428-14433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 434] [Cited by in RCA: 462] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 118. | Xia Y, Yamaoka Y, Zhu Q, Matha I, Gao X. A comprehensive sequence and disease correlation analyses for the C-terminal region of CagA protein of Helicobacter pylori. PLoS One. 2009;4:e7736. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 78] [Article Influence: 4.6] [Reference Citation Analysis (1)] |
| 119. | Mueller D, Tegtmeyer N, Brandt S, Yamaoka Y, De Poire E, Sgouras D, Wessler S, Torres J, Smolka A, Backert S. c-Src and c-Abl kinases control hierarchic phosphorylation and function of the CagA effector protein in Western and East Asian Helicobacter pylori strains. J Clin Invest. 2012;122:1553-1566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 208] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 120. | Suzuki M, Mimuro H, Kiga K, Fukumatsu M, Ishijima N, Morikawa H, Nagai S, Koyasu S, Gilman RH, Kersulyte D. Helicobacter pylori CagA phosphorylation-independent function in epithelial proliferation and inflammation. Cell Host Microbe. 2009;5:23-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 220] [Cited by in RCA: 275] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 121. | Lu HS, Saito Y, Umeda M, Murata-Kamiya N, Zhang HM, Higashi H, Hatakeyama M. Structural and functional diversity in the PAR1b/MARK2-binding region of Helicobacter pylori CagA. Cancer Sci. 2008;99:2004-2011. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 37] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 122. | Murata-Kamiya N, Kurashima Y, Teishikata Y, Yamahashi Y, Saito Y, Higashi H, Aburatani H, Akiyama T, Peek RM, Azuma T. Helicobacter pylori CagA interacts with E-cadherin and deregulates the beta-catenin signal that promotes intestinal transdifferentiation in gastric epithelial cells. Oncogene. 2007;26:4617-4626. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 324] [Cited by in RCA: 379] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 123. | Wallace JL, Cucala M, Mugridge K, Parente L. Secretagogue-specific effects of interleukin-1 on gastric acid secretion. Am J Physiol. 1991;261:G559-G564. [PubMed] |
| 124. | Beales IL, Calam J. Interleukin 1 beta and tumour necrosis factor alpha inhibit acid secretion in cultured rabbit parietal cells by multiple pathways. Gut. 1998;42:227-234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 211] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 125. | Noach LA, Bosma NB, Jansen J, Hoek FJ, van Deventer SJ, Tytgat GN. Mucosal tumor necrosis factor-alpha, interleukin-1 beta, and interleukin-8 production in patients with Helicobacter pylori infection. Scand J Gastroenterol. 1994;29:425-429. [PubMed] |
| 126. | Basso D, Scrigner M, Toma A, Navaglia F, Di Mario F, Rugge M, Plebani M. Helicobacter pylori infection enhances mucosal interleukin-1 beta, interleukin-6, and the soluble receptor of interleukin-2. Int J Clin Lab Res. 1996;26:207-210. [PubMed] |
| 127. | El-Omar EM, Carrington M, Chow WH, McColl KE, Bream JH, Young HA, Herrera J, Lissowska J, Yuan CC, Rothman N. Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature. 2000;404:398-402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1690] [Cited by in RCA: 1688] [Article Influence: 64.9] [Reference Citation Analysis (3)] |
| 128. | Chiurillo MA, Moran YH, Cañas M, Valderrama EJ, Armanie E. Infection with specific Helicobacter pylori-cag pathogenicity island strains is associated with interleukin-1B gene polymorphisms in Venezuelan chronic gastritis patients. Dig Dis Sci. 2011;56:449-456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 129. | Zambon CF, Basso D, Navaglia F, Belluco C, Falda A, Fogar P, Greco E, Gallo N, Rugge M, Di Mario F. Pro- and anti-inflammatory cytokines gene polymorphisms and Helicobacter pylori infection: interactions influence outcome. Cytokine. 2005;29:141-152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 166] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 130. | Queiroz DM, Saraiva IE, Rocha GA, Rocha AM, Gomes LI, Melo FF, Bittencourt PF. IL2-330G polymorphic allele is associated with decreased risk of Helicobacter pylori infection in adulthood. Microbes Infect. 2009;11:980-987. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 131. | Kirkegaard K, Taylor MP, Jackson WT. Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nat Rev Microbiol. 2004;2:301-314. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 313] [Cited by in RCA: 351] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 132. | Orvedahl A, Levine B. Eating the enemy within: autophagy in infectious diseases. Cell Death Differ. 2009;16:57-69. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 168] [Cited by in RCA: 169] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 133. | Raju D, Hussey S, Ang M, Terebiznik MR, Sibony M, Galindo-Mata E, Gupta V, Blanke SR, Delgado A, Romero-Gallo J. Vacuolating cytotoxin and variants in Atg16L1 that disrupt autophagy promote Helicobacter pylori infection in humans. Gastroenterology. 2012;142:1160-1171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 184] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 134. | Vinall LE, King M, Novelli M, Green CA, Daniels G, Hilkens J, Sarner M, Swallow DM. Altered expression and allelic association of the hypervariable membrane mucin MUC1 in Helicobacter pylori gastritis. Gastroenterology. 2002;123:41-49. [PubMed] |
| 135. | Carvalho F, Seruca R, David L, Amorim A, Seixas M, Bennett E, Clausen H, Sobrinho-Simões M. MUC1 gene polymorphism and gastric cancer--an epidemiological study. Glycoconj J. 1997;14:107-111. [PubMed] |
| 136. | Silva F, Carvalho F, Peixoto A, Teixeira A, Almeida R, Reis C, Bravo LE, Realpe L, Correa P, David L. MUC1 polymorphism confers increased risk for intestinal metaplasia in a Colombian population with chronic gastritis. Eur J Hum Genet. 2003;11:380-384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 18] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 137. | Nakayama J, Yeh JC, Misra AK, Ito S, Katsuyama T, Fukuda M. Expression cloning of a human alpha1, 4-N-acetylglucosaminyltransferase that forms GlcNAcalpha1--& gt; 4Galbeta--& gt; R, a glycan specifically expressed in the gastric gland mucous cell-type mucin. Proc Natl Acad Sci USA. 1999;96:8991-8996. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 80] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 138. | Zheng Z, Jia Y, Hou L, Persson C, Yeager M, Lissowska J, Chanock SJ, Blaser M, Chow WH, Ye W. Genetic variation in a4GnT in relation to Helicobacter pylori serology and gastric cancer risk. Helicobacter. 2009;14:120-125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 139. | Clausen H, Hakomori S. ABH and related histo-blood group antigens; immunochemical differences in carrier isotypes and their distribution. Vox Sang. 1989;56:1-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 381] [Cited by in RCA: 363] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 140. | Henry S, Oriol R, Samuelsson B. Lewis histo-blood group system and associated secretory phenotypes. Vox Sang. 1995;69:166-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 136] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 141. | Kelly RJ, Rouquier S, Giorgi D, Lennon GG, Lowe JB. Sequence and expression of a candidate for the human Secretor blood group alpha(1,2)fucosyltransferase gene (FUT2). Homozygosity for an enzyme-inactivating nonsense mutation commonly correlates with the non-secretor phenotype. J Biol Chem. 1995;270:4640-4649. [PubMed] |
| 142. | Magalhães A, Gomes J, Ismail MN, Haslam SM, Mendes N, Osório H, David L, Le Pendu J, Haas R, Dell A. Fut2-null mice display an altered glycosylation profile and impaired BabA-mediated Helicobacter pylori adhesion to gastric mucosa. Glycobiology. 2009;19:1525-1536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 91] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 143. | Lindén S, Mahdavi J, Semino-Mora C, Olsen C, Carlstedt I, Borén T, Dubois A. Role of ABO secretor status in mucosal innate immunity and H. pylori infection. PLoS Pathog. 2008;4:e2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 131] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 144. | Heneghan MA, Moran AP, Feeley KM, Egan EL, Goulding J, Connolly CE, McCarthy CF. Effect of host Lewis and ABO blood group antigen expression on Helicobacter pylori colonisation density and the consequent inflammatory response. FEMS Immunol Med Microbiol. 1998;20:257-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 37] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 145. | Ley RE. Obesity and the human microbiome. Curr Opin Gastroenterol. 2010;26:5-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 558] [Cited by in RCA: 533] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 146. | Karlsson FH, Tremaroli V, Nookaew I, Bergström G, Behre CJ, Fagerberg B, Nielsen J, Bäckhed F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1766] [Cited by in RCA: 2239] [Article Influence: 172.2] [Reference Citation Analysis (0)] |
| 147. | Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol. 2012;9:599-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 771] [Cited by in RCA: 917] [Article Influence: 65.5] [Reference Citation Analysis (0)] |
P- Reviewers: Crabtree JE, Linden SK, Tamara V S- Editor: Ma YJ L- Editor: A E- Editor: Liu XM