Published online Apr 21, 2014. doi: 10.3748/wjg.v20.i15.4316
Revised: December 14, 2013
Accepted: January 19, 2014
Published online: April 21, 2014
Processing time: 176 Days and 14.5 Hours
AIM: To investigate a novel therapeutic strategy to target and suppress c-myc in human cancers using far up stream element (FUSE)-binding protein-interacting repressor (FIR).
METHODS: Endogenous c-Myc suppression and apoptosis induction by a transient FIR-expressing vector was examined in vivo via a HA-tagged FIR (HA-FIR) expression vector. A fusion gene-deficient, non-transmissible, Sendai virus (SeV) vector encoding FIR cDNA, SeV/dF/FIR, was prepared. SeV/dF/FIR was examined for its gene transduction efficiency, viral dose dependency of antitumor effect and apoptosis induction in HeLa (cervical squamous cell carcinoma) cells and SW480 (colon adenocarcinoma) cells. Antitumor efficacy in a mouse xenograft model was also examined. The molecular mechanism of the anti-tumor effect and c-Myc suppression by SeV/dF/FIR was examined using Spliceostatin A (SSA), a SAP155 inhibitor, or SAP155 siRNA which induce c-Myc by increasing FIR∆exon2 in HeLa cells.
RESULTS: FIR was found to repress c-myc transcription and in turn the overexpression of FIR drove apoptosis through c-myc suppression. Thus, FIR expressing vectors are potentially applicable for cancer therapy. FIR is alternatively spliced by SAP155 in cancer cells lacking the transcriptional repression domain within exon 2 (FIR∆exon2), counteracting FIR for c-Myc protein expression. Furthermore, FIR forms a complex with SAP155 and inhibits mutual well-established functions. Thus, both the valuable effects and side effects of exogenous FIR stimuli should be tested for future clinical application. SeV/dF/FIR, a cytoplasmic RNA virus, was successfully prepared and showed highly efficient gene transduction in in vivo experiments. Furthermore, in nude mouse tumor xenograft models, SeV/dF/FIR displayed high antitumor efficiency against human cancer cells. SeV/dF/FIR suppressed SSA-activated c-Myc. SAP155 siRNA, potentially produces FIR∆exon2, and led to c-Myc overexpression with phosphorylation at Ser62. HA-FIR suppressed endogenous c-Myc expression and induced apoptosis in HeLa and SW480 cells. A c-myc transcriptional suppressor FIR expressing SeV/dF/FIR showed high gene transduction efficiency with significant antitumor effects and apoptosis induction in HeLa and SW480 cells.
CONCLUSION: SeV/dF/FIR showed strong tumor growth suppression with no significant side effects in an animal xenograft model, thus SeV/dF/FIR is potentially applicable for future clinical cancer treatment.
Core tip: The authors performed in vivo experiments and included an animal model to examine the Sendai virus/dF/Far Up Stream Element-Binding Protein-Interacting Repressor for cancer gene therapy to minimize side effects for clinical use.
-
Citation: Matsushita K, Shimada H, Ueda Y, Inoue M, Hasegawa M, Tomonaga T, Matsubara H, Nomura F. Non-transmissible Sendai virus vector encoding
c-myc suppressor FBP-interacting repressor for cancer therapy. World J Gastroenterol 2014; 20(15): 4316-4328 - URL: https://www.wjgnet.com/1007-9327/full/v20/i15/4316.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i15.4316
c-Myc plays an essential role in cell proliferation and tumorigenesis. c-myc activation was also shown to be required for skin epidermal and pancreatic beta-cell tumor maintenance in c-MYC-ERTAM transgenic mice[1]. High c-myc expression level in colorectal cancer tissues was associated with poor long-term survival of colorectal cancer patients[2]. The far up stream element (FUSE) is a sequence required for correct expression of the human c-myc gene[3]. The FUSE is located at 1.5 kb upstream of c-myc promoter P1, and binds the FUSE binding protein (FBP), a transcription factor which stimulates c-myc expression in a FUSE-dependent manner[4]. Yeast two-hybrid analysis revealed that FBP binds to a protein that has transcriptional inhibitory activity termed the FBP interacting repressor (FIR). FIR interacts with the central DNA binding domain of FBP[5]. Recently, FIR was found to engage the TFIIH/p89/XPB helicase and repress c-myc transcription by delaying promoter escape[5,6]. Furthermore, exogenous FIR expression represses endogenous c-myc transcription, and drives apoptosis due to the decrease in c-Myc[7]. Although these observations indicate that cancer therapies targeting c-myc suppression by FIR may be a useful strategy, the mechanism of the antitumor effect of FIR should be determined in detail prior to clinical testing. For example, first, FIR is alternatively spliced in colorectal cancer lacking the transcriptional repression domain within exon 2 (FIR∆exon2)[7]. Second, FIR and FIR∆exon2 form a homo- or hetero-dimer, which complexes with SAP155, a subunit of the essential splicing factor 3b (SF3b) subcomplex in the spliceosome, and is required for correct P27Kip1 (P27) pre-mRNA splicing, after which P27 arrests cells in G1[8]. Third, SAP155 is required for correct FIR pre-mRNA splicing and thus the FIR/FIR∆exon2/SAP155 interaction bridged c-myc and p27 expression[9]. Accordingly, SAP155-mediated alternative splicing of FIR serves as a molecular switch for c-myc expression[9]. Finally, spliceostatin A (SSA), a natural SF3b inhibitor, markedly inhibited P27 expression by disrupting its pre-mRNA splicing and reducing cdk2/cyclinE expression[10]. Taken together, these findings suggest that exogenous FIR stimuli potentially affect the FIR/FIR∆exon2/SAP155 interaction which is pivotal for the cell cycle, cancer development and differentiation.
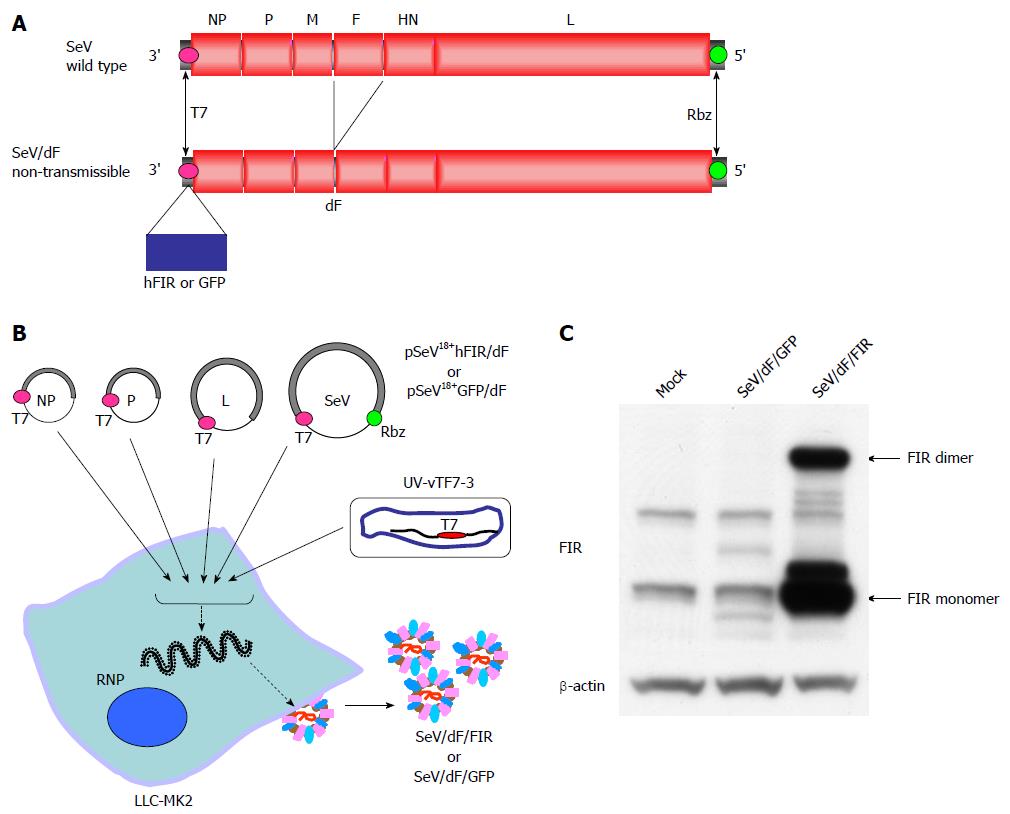
In this study, a fusion gene-deficient human FIR-expressing Sendai virus vector (SeV/dF/FIR) was prepared for future cancer therapy for the following reasons; Sendai virus (SeV), a member of the Paramyxoviridae family, has envelopes and a nonsegmented negative-strand RNA genome. The SeV genome contains six major genes in tandem on a single negative-strand RNA. Three proteins, the nucleoprotein (NP), phosphoprotein (P) and large protein (L; the catalytic subunit of the polymerase) form a ribonucleoprotein complex (RNP) with the SeV RNA. Matrix proteins (M) contribute to the assembly of viral particles, hemagglutinin-neuraminidase (HN) and fusion proteins (F) engage in the attachment of viral particles and infiltration of RNPs into infected cells. Importantly, SeV does not transform cells by integrating its genome into the cellular genome[11]. Therefore, SeV can mediate gene transfer and expression to a cytoplasmic location using cellular tubulin[12], thereby avoiding possible malignant transformation due to the genetic alteration of host cells. These are the safety advantages of SeV. Recently, a novel SeV vector was established where an enhanced green fluorescent protein (EGFP) reporter gene was inserted at the 3’-end of fusion gene-deficient SeV genomic RNA (SeV/dF/EGFP)[13]. This SeV/dF/EGFP is incapable of self-replication, but capable of infecting various cells, including human smooth muscle cells, hepatocytes, and endothelial cells, thus the SeV/dF/EGFP has a broad spectrum of gene transfer activity[9,10]. In this study, SeV/dF/FIR was prepared following the method for SeV/dF/EGFP[12,13]. The validity of SeV/dF/FIR for cancer therapy was examined in animal xenograft models as SeV/dF vectors have been shown to be applicable for clinical use[14-18]. The clinical use of SeV/dF/FIR for cancer therapy is also discussed.
Full-length FIR cDNA (HA-FIR) was cloned into the pCGNM2 vector plasmid to introduce the hemagglutinin (HA)-tag at the amino termini[7]. Full-length FIR cDNA was cloned into the p3xFLAG-CMV-14 vector (Sigma, MO, United States) to introduce the Flag-tag at the amino termini for the selection of FIR-Flag in 293T cells (performed by Dr. T.N.). Plasmids were prepared by CsCl ultra-centrifugation or the Endofree® Plasmid Maxi Kit (Qiagen, MD, United States) and the DNA sequences were verified.
HeLa cells (human cervical squamous cell carcinoma cells), LoVo and SW480 cells (human colon cancer cell lines) and LLC-MK2 a rhesus monkey kidney cell line were purchased from the American Type Culture Collection (Manassas, VA, United States). Yes-5, a human esophageal squamous cell carcinoma cell line was established by Dr Takuo Murakami (Yamaguchi University, Yamaguchi, Japan). All cell lines were cultured at 37 °C in a humidified atmosphere containing 5% CO2. All tumor cell lines, except LLC-MK2 cells [which were maintained in DMEM; Dulbecco’s Modified Eagle’s Medium (Gibco BRL, NY, United States)] were cultured in tissue flasks or Petri dishes containing RPMI-1640 (Gibco, NY, United States) supplemented with 10% heat-inactivated FBS and penicillin (100 units/mL), streptomycin (0.1 mg/mL), and 2 mmol/L glutamine.
Immunocytochemistry was performed as described previously[7]. Protein extraction and immunoblotting are described elsewhere[8,9].
SAP155 siRNA duplexes were purchased from Sigma Aldrich. The target sequences for SAP155 siRNA oligonucleotides were listed previously[8]. Luciferase GL2 duplex was used as a negative control for siRNA targeting 5’-CGTACGCGGAATACTTCGA-3’. Transient transfection of siRNA was carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. The transfected cells were cultured for 72 h at 37 °C in a 5% CO2 incubator.
Apoptotic cells were detected by terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay according to the manufacturer’s instructions (Apoptosis Detection System, Fluorescein, Promega, WI, United States) as described previously[7]. Apoptosis detection by APOPercentage apoptosis assay™ (Funakoshi Co., Ltd., Tokyo, Japan) was performed according to the manufacturer’s instructions[9].
Human FIR cDNA was amplified with a pair of NotI site-tagged primers containing SeV-specific transcriptional regulatory signal sequences, (End and Start, italicized below) 5’-ATTGCGGCCGCCAAGGTTCAATGGCGACGGCGACCATAGC-3’ and 5’-ATTGCGGCCGCGATGAACTTTCACCCTAAGTTTTTCTTACTACGGTCACGCAGAGAGGTCACTGTTATCAAAACGC-3’. The amplified fragment was introduced into the NotI site of the parental SeV vector cDNA, pSeV18+b(+)/dF[15], to generate pSeV18+hFIR/dF. pSeV18+hFIR/dF was transfected to LLC-MK2 cells which were preliminarily infected with psoralen- and long-wave UV-treated vaccinia virus vTF7-3, expressing T7 polymerase. The cells were then washed twice with DMEM, and cultured for 24 h in DMEM containing cytosine β-D-arabinofuranoside (AraC; 40 μg/mL) and trypsin (7.5 μg/mL). LLC-MK2/F7/A cells expressing the F protein were suspended in DMEM containing AraC and trypsin, and layered onto the transfected cells, and cultured at 37 °C for an additional 48 h. The recovered vector in the culture supernatants was propagated using the LLC-MK2/F7/A cells. A GFP expression vector (SeV/dF/GFP) was prepared as previously described[8]. The viral vectors were further amplified by several rounds of propagation. The virus titers of the recovered vectors were determined by their infectivity and expressed using cell-infectious units (CIU). These vectors were frozen at -80 °C until use.
One million LLC-MK2 cells and HeLa cells were seeded in six-well plates and transduced with SeV/dF/GFP when monolayers reached 60%-80% confluence. As the standard inoculation procedure for vaccination, the monolayers were washed twice with PBS and overlaid with serum-free medium containing SeV/dF/GFP at a multiplicity of infection (MOI) of 0, 1, 10, 50, 100, or 300. After incubation at 37 °C for 90 min, non-adsorbed virus was removed, medium containing 10% FBS was added, and the cells were incubated for more than 48 h at 37 °C. The transduction studies were carried out in triplicate for each MOI. Microscopy was used to detect transduced cells by GFP fluorescence. At 72 h after transduction, the GFP-transduced cells were analyzed for GFP expression using a FACS Calibrator (BD Pharmingen, Franklin Lakes, NJ, United States).
The inhibitory effects of viruses on the proliferation of cultured cells were examined using the CellTiter96™ AQueous One Solution Proliferation Assay (Promega, Madison, WI, United States). In brief, five thousand cells were plated in each well on day 0. On day 1, 24 h later, HeLa cells were infected with SeV/dF/FIR or SeV/dF/GFP as the control at 0.1 to 10 MOI, and cultured for 2 d. On day 3, cell viability was quantified by measuring the absorbance at 570 nm after incubation with the tetrazolium compound [3-(4,5-dimethylthiozol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS), and an electron coupling agent, phenazine ethosulfate (PES)] (Promega, Madison, WI, United States) for 4 h. The absorbance at 570 nm was measured by a Mutiabel Counter™, ARVOSX WAIIAC™ (Perkin Elmer, MA, United States). The results are shown as percentages of the uninfected control cells.
Six- to eight-week male immuno-competent Balbc/nu/nu mice were purchased from Clea Japan (Tokyo, Japan) and housed in the Animal Maintenance Facility at Chiba University under specific pathogen-free conditions. All animal experiments were approved by the Committee of the Ethics on Animal Experiments in the Faculty of Medicine, Chiba University and carried out following the Guidelines for Animal Experiments in the Faculty of Medicine, Chiba University, Chiba, Japan and The Law and Notification of the Government. Mouse experiments were carried out at least twice to confirm the results.
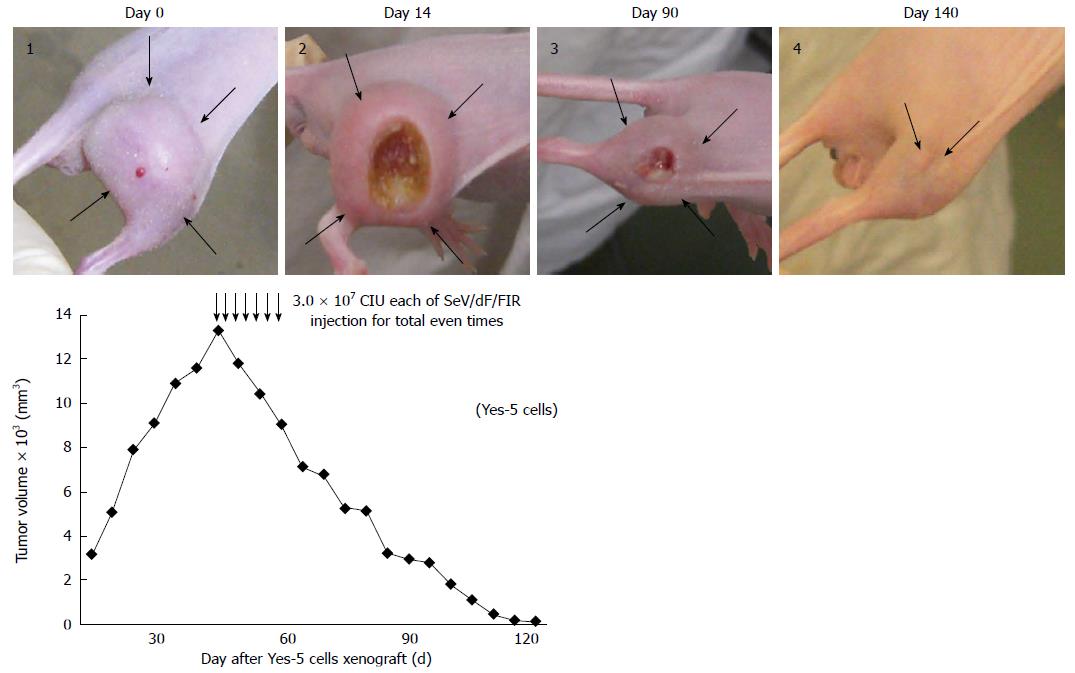
The in vivo inhibition of tumorigenicity of HeLa cells (human cervical squamous cell carcinoma cells) was examined by SeV/dF/FIR or SeV/dF/GFP injection (as the control). 5 × 106 cells/50 μL PBS of HeLa cells were injected under the skin in the right thigh of nude mice (6-wk old males). Tumor growth was observed and the long and short diameter measured for tumor volume calculation. Thirty days after inoculation, tumors grew up to 5-8 mm in diameter in 18 of 18 mice (100%). Tumor size was calculated using the formula, (a × b2)/2, where a and b represent the larger and smaller diameters, respectively, and was monitored every 3 d.
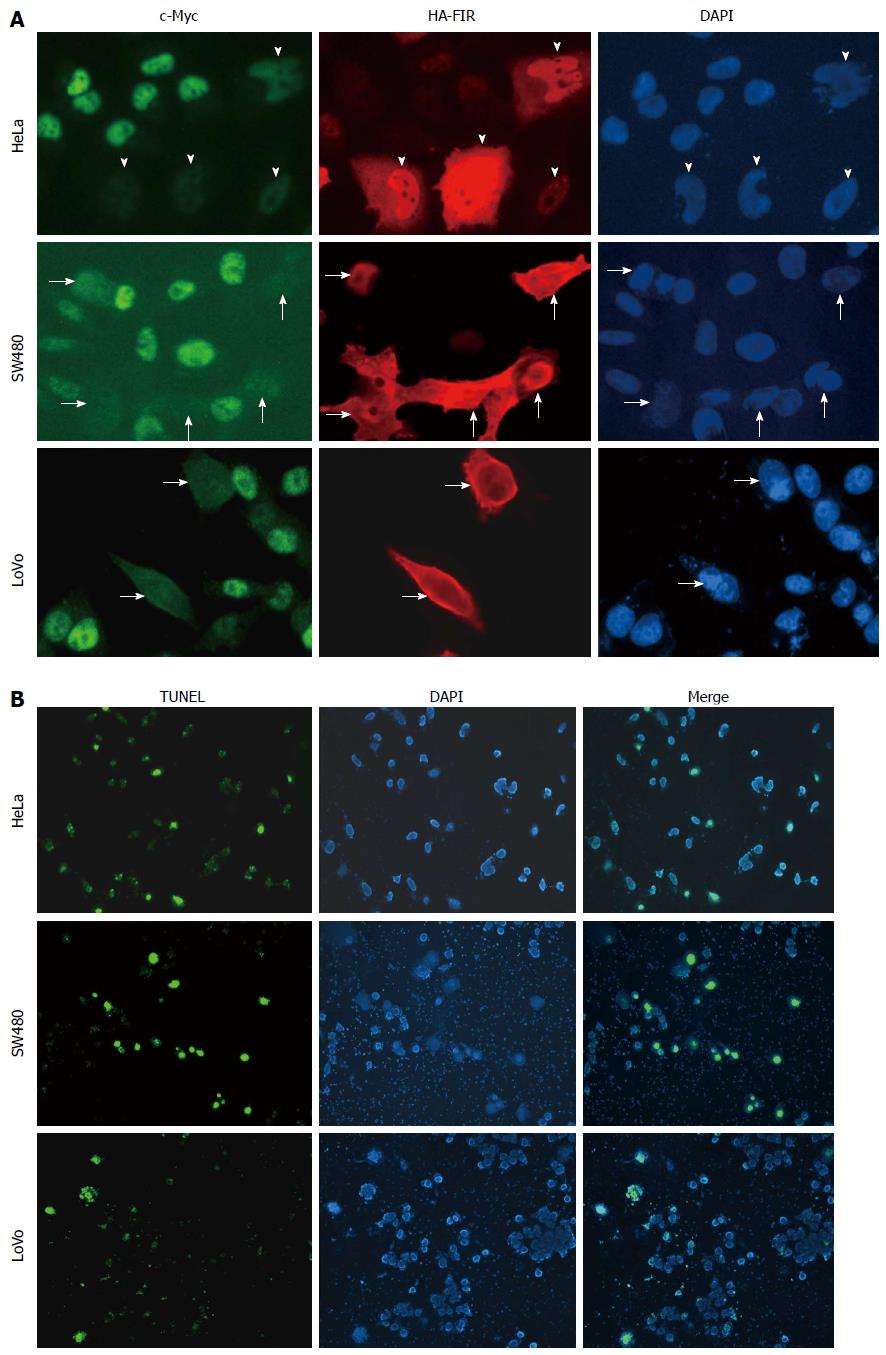
To examine endogenous c-myc gene suppression by FIR, HA-FIR expression plasmids were transfected into HeLa cells, and c-Myc expression was visualized by immunostaining with anti-c-Myc antibodies (Figure 1A; upper panels: HA-FIR is red; c-Myc is green). c-Myc levels were greatly diminished in HA-FIR expressing cells (arrows), demonstrating that FIR represses endogenous c-Myc expression in SW480 (Figure 1A; middle panels) and LoVo cells (Figure 1A; lower panels). After HA-FIR expression plasmids were transfected into HeLa, SW480 or LoVo cells, apoptotic cells were visualized by TUNEL assay. HA-FIR transfected cells were definitively associated with apoptosis (Figure 1B).
SeV/dF/FIR and SeV/dF/GFP were prepared as described in Materials and Methods (Figure 2A, B). SeV/dF/FIR vectors were infected into LLC-MK2 or HeLa cells. FIR protein expression level was examined by western blot with anti-FIR antibody (6B4) (Figure 2C). At least 1 × 1010 CIU of fusion gene-deficient SeV/dF/FIR virus particles were prepared at amplification for use in the experiments.
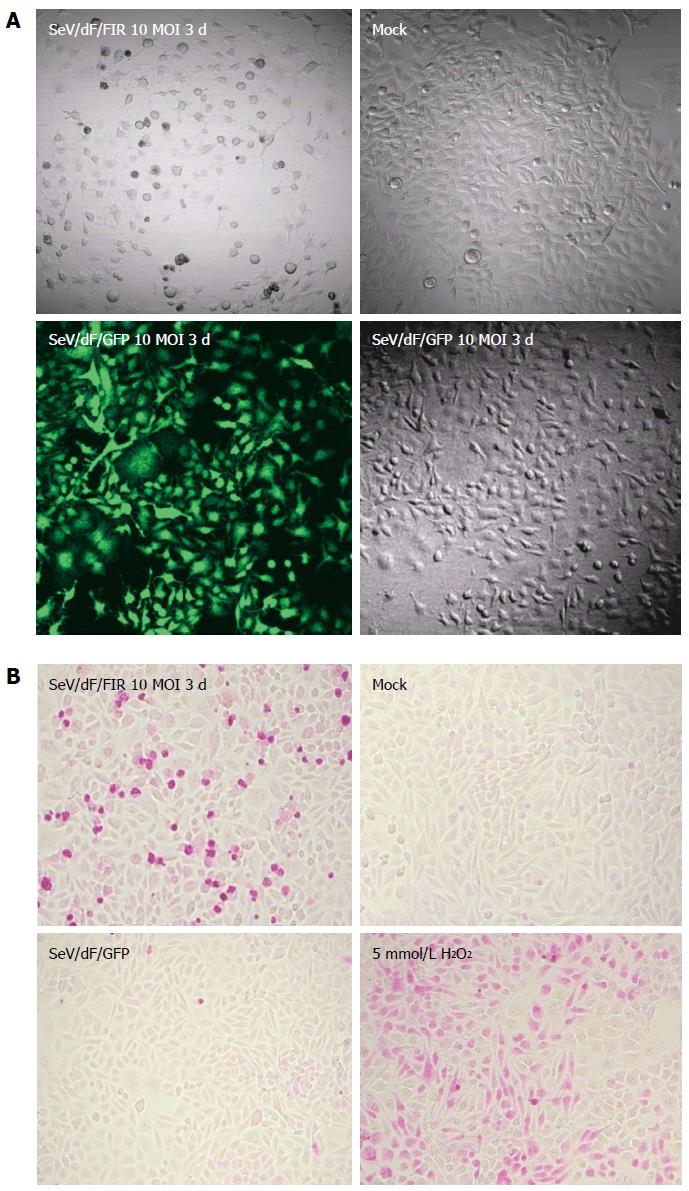
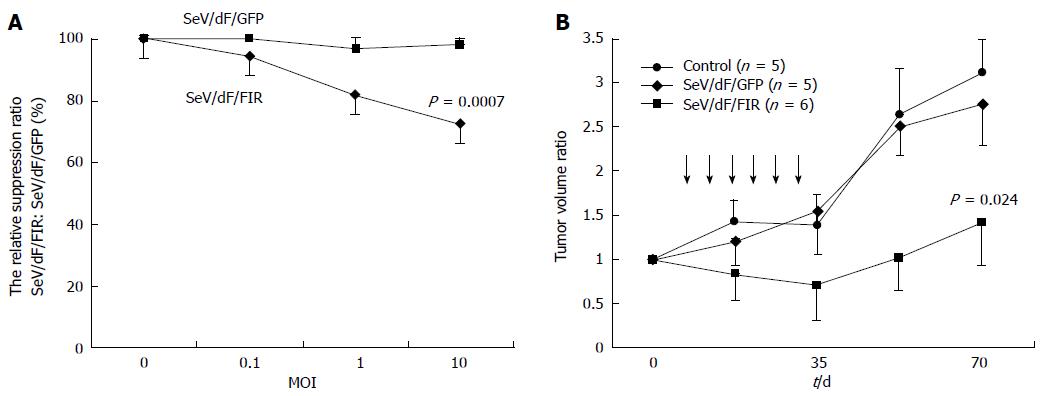
Nine human and five murine tumor cell lines plus non-tumor cells, propagated in vitro were collected, transduced by SeV/dF/GFP, and examined for gene transduction efficiency. Flow cytometric analyses showed dose-dependent GFP expression, and optimal expression was obtained at a MOI of 10-100; > 90% GFP-positive tumor cell lines were detected at a MOI over 10 (Figure 3A and data not shown). Furthermore, SeV/dF/FIR, but not SeV/dF/GFP significantly suppressed HeLa cell (human cervical squamous carcinoma) growth as shown by the APOPercentage assay (Figure 3B), indicating SeV/dF/FIR suppresses tumor cell growth by apoptosis in vivo.
SeV/dF/FIR, but not SeV/dF/GFP, significantly suppressed cell growth in HeLa cells (Figure 4A) and SW480 cells when analyzed by Dunnet’s test for multiple comparisons (Figure 4B). Of note, xenograft tumors 2 cm in diameter disappeared completely following SeV/dF/FIR treatment, indicating that SeV/dF/FIR has immunological effects (Figure 5)[24,25].
If SeV/dF/FIR is to be tested clinically, endogenous FIR-interacting proteins should be identified to avoid unexpected side effects. For this purpose, the FIR-FLAG tag vector was transiently expressed in 293T cells and co-immunoprecipitated with anti-FLAG conjugated beads to detect FIR-binding proteins[19-23] (Table 1). As reported previously, FBP (Far upstream element-binding protein)[26,27], SAP155[28], and SRp54 (splicing factor, arginine/serine rich-12)[28] were identified as candidate FIR-binding proteins. To date, no significant side effects have been observed following SeV/dF/FIR treatment including our study[29].
| Hit preys | |
| CDKN2AIP | CDKN2A interacting protein |
| CDYL | Chromodomain protein, Y chromosome-like |
| DAZAP1 | DAZ associated protein 1 |
| DDX17 | DEAD box polypeptide 17 |
| DDX5 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 |
| ELAVL1 | ELAV-like 1 |
| FAM120A | Oxidative stress-associated Src activator |
| FUBP1|FUBP3|KHSRP | Far upstream element-binding protein family |
| FUBP1|KHSRP | Far upstream element-binding protein family |
| HNRNPA1 | Heterogeneous nuclear ribonucleoprotein A1 |
| HNRNPA1|HNRNPA1L2 | Heterogeneous nuclear ribonucleoprotein; A1 or A1-like |
| HNRNPA1|HNRNPA1L2|LOC402562 | Nuclear ribonucleoprotein A1 family |
| HNRNPA2B1 | Heterogeneous nuclear ribonucleoprotein A2/B1 |
| HNRNPA3 | Heterogeneous nuclear ribonucleoprotein A3 |
| HNRNPAB | Heterogeneous nuclear ribonucleoprotein A/B; isoform a |
| HNRNPAB|HNRNPD | Heterogeneous nuclear ribonucleoprotein; A/B or D |
| HNRNPD | Heterogeneous nuclear ribonucleoprotein D; isoform c |
| HNRNPH1 | Heterogeneous nuclear ribonucleoprotein H1 |
| HNRNPK | Heterogeneous nuclear ribonucleoprotein K |
| HNRNPL | Heterogeneous nuclear ribonucleoprotein L |
| HNRNPM | Heterogeneous nuclear ribonucleoprotein M; isoform a |
| HNRNPR | Heterogeneous nuclear ribonucleoprotein R |
| HNRNPU | Heterogeneous nuclear ribonucleoprotein U; isoform a |
| HNRNPUL1 | Heterogeneous nuclear ribonucleoprotein U-like 1 |
| HNRPDL | Heterogeneous nuclear ribonucleoprotein D-like |
| IFIT5 | Interferon-induced protein with tetratricopeptide repeats 5 |
| IGF2BP1 | Insulin-like growth factor 2 mRNA binding protein 1 |
| IGF2BP1|IGF2BP3 | Insulin-like growth factor 2 mRNA binding protein; 1 or 3 |
| IGF2BP2 | Insulin-like growth factor 2 mRNA binding protein 2 |
| IGF2BP3 | Insulin-like growth factor 2 mRNA binding protein 3 |
| ILF2 | Interleukin enhancer binding factor 2 |
| KHDRBS1 | KH domain containing, RNA binding, signal transduction associated 1 |
| KHDRBS1|KHDRBS2|KHDRBS3 | KH; domain containing, RNA binding, signal transduction associated 1 |
| Or domain-containing, RNA-binding, signal transduction- | |
| KHSRP | KH-type splicing regulatory protein |
| KIAA1967 | p30 DBC protein |
| LARP1 | La related protein |
| LRPPRC | Leucine-rich PPR motif-containing protein |
| LSM12 | LSM12 homolog |
| MAGOH|MAGOHB | Mago-nashi homolog; or B |
| MATR3 | Matrin 3 |
| MOV10 | Mov10, Moloney leukemia virus 10, homolog |
| MSI2 | Musashi 2; isoform a |
| PABP3|PABPC1|PABPC3 | Poly (A) binding protein, cytoplasmic 1 |
| Or poly (A) binding protein, cytoplasmic 3 | |
| PABPC1 | Poly (A) binding protein, cytoplasmic 1 |
| PABPC1|PABPC1L|PABPC5 | Poly (A) binding protein, cytoplasmic 1 or poly(A) -binding protein, |
| Cytoplasmic 1-like or poly (A) binding protein, cytoplasmic 5 | |
| PABPC1|PABPC4 | Poly (A) binding protein, cytoplasmic 1 |
| Or poly A binding protein, cytoplasmic 4 | |
| PABPC1L2B|PABPC4 | Poly (A) binding protein, cytoplasmic 1-like 2B |
| Or poly A binding protein, cytoplasmic 4 | |
| PABPC4 | Poly A binding protein, cytoplasmic 4 |
| PABPC4|PABPC4L | Poly A binding protein, cytoplasmic 4 |
| PABPN1 | Poly (A) binding protein, nuclear 1 |
| PCBP1|PCBP2|PCBP3|PCBP4 | Poly (rC) binding protein; 1 or 2 or 3 or 4 |
| PCBP2|PCBP3 | Poly (rC) binding protein; 2 or 3 |
| PTBP1 | Polypyrimidine tract-binding protein 1; isoform d |
| SF3B1 | Splicing factor 3b, subunit 1 |
| SF3B14 | Splicing factor 3B, 14 kDa subunit |
| SF3B3 | Splicing factor 3b, subunit 3 |
| SF3B4 | Splicing factor 3b, subunit 4 |
| SFPQ | Splicing factor proline/glutamine rich (polypyrimidine tract binding |
| SFRS11 | Splicing factor, arginine/serine-rich 11 (SRp54) |
| SFRS12 | Splicing factor, arginine/serine-rich 12 |
| SNRPD1 | Small nuclear ribonucleoprotein D1 polypeptide 16 kDa |
| SSB | Autoantigen La |
| SYNCRIP | Synaptotagmin binding, cytoplasmic RNA interacting protein |
| THADA | Thyroid adenoma associated |
| TRMT1 | tRNA methyltransferase 1 |
| TROVE2 | TROVE domain family, member 2; isoform 2 |
| XRN2 | 5'-3' exoribonuclease 2 |
| Total hit preys: | 67 |
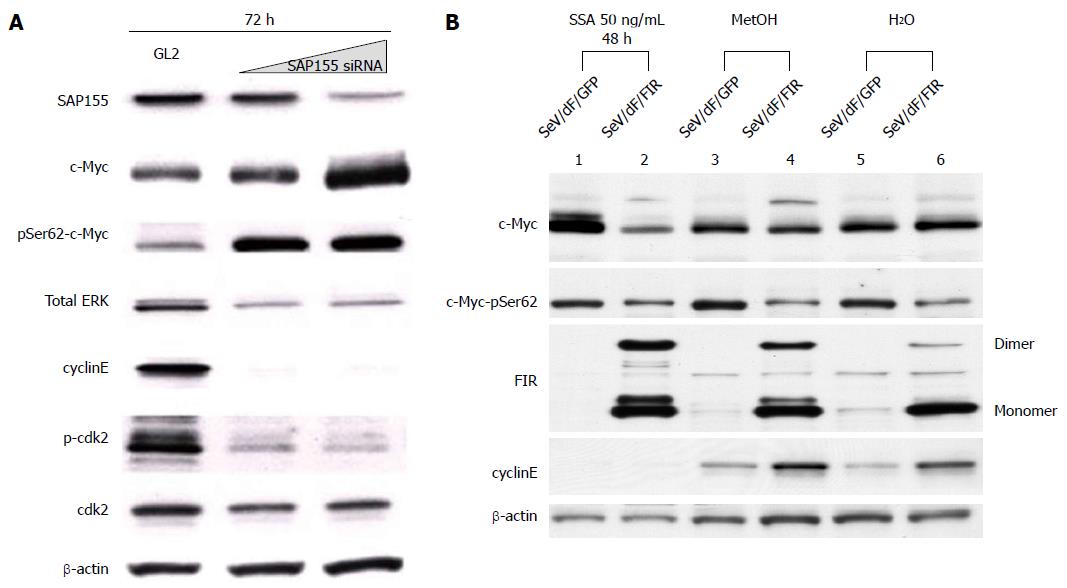
We previously reported that the adenovirus vector encoding FIR∆exon2 (Ad-FIR∆exon2) activates not only c-myc transcription, but also c-Myc protein expression in HeLa cells[8]. However, the extent of c-Myc protein activation by Ad-FIR∆exon2, evaluated by western blot analysis, could not be explained solely by c-myc transcription activation[8]. Therefore, we hypothesized that c-Myc protein should be modified by Ad-FIR∆exon2 to be more stable. Ad-FIR∆exon2 expression leads to increased levels of c-Myc phosphorylated at Ser62 (data not shown), indicating that stable c-Myc protein accumulates in cells[30,31]. As reported previously, SAP155 siRNA inhibited FIR pre-mRNA splicing and generated FIR∆exon2[8,9]. In fact, SAP155 siRNA increased levels of c-Myc phosphorylated at Ser62 and Ad-FIR∆exon2 (Figure 6A). In other words, Ad-FIR∆exon2, which lacks the transcriptional repressor domain, directly or indirectly activated c-myc expression not only through transcription, but also through protein level, suggesting that FIR∆exon2 acts in opposition to the repressor function by FIR[8].
In this study, the effect of SeV/dF/FIR was examined to determine whether it suppresses the increase in c-Myc after SSA treatment. SeV/dF/FIR suppressed SSA-induced c-Myc activation (Figure 6B, compare lane 2 with lane 1), but not basal c-Myc expression (Figure 6B, compare lanes 4 to 3 and 6 to 5, respectively). These results were consistent with previous reports that FIR suppresses activated, but not basal, c-myc transcription[6]. These observations suggest that the increase in c-myc by either SAP155 siRNA or SSA treatment is due to reduced FIR activity, or an increase in the ratio of FIR∆exon2/FIR in HeLa cells. Taken together, these results suggest that SeV/dF/FIR is potentially clinically applicable for cancer therapy as it counteracts SSA-activated c-Myc (Figure 6B, compare lane 2 with lane 1) as well as endogenous c-Myc (Figure 1A).
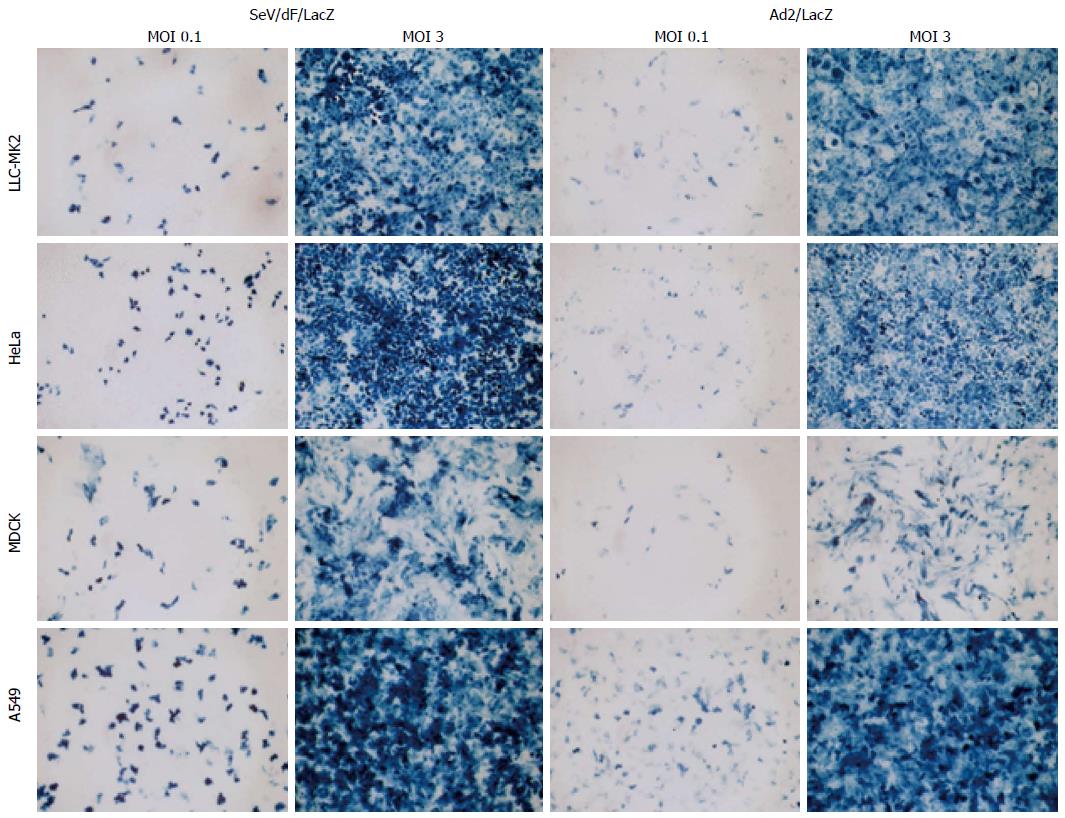
F gene-deficient SeV vectors (SeV/dF) can transduce cells in a wide range of tissues such as respiratory, nervous, muscular, epithelial and immune tissues[11,32-35]. Transduction efficiency to cell lines from various tissues was examined and compared to the adenovirus vector expressing LacZ (Ad5/LacZ) at the same MOI. The transduction efficiency of SeV/dF to those cells was even higher than that of adenovirus vector (Figure 7).
Overexpression of c-Myc has been known to promote cell growth, proliferation and immortalization, whereas a reduction in c-Myc induces apoptosis. The recent genetic construction of the mouse in which the expression of c-myc can be switched on or off in vivo has emphasized the significance of c-Myc expression in tumorigenesis. Ectopic c-myc expression in hematopoietic cells using the tetracycline regulatory system caused malignant T cell lymphomas and acute myeloid leukemia; subsequent inactivation of the transgene caused regression of established tumors[36]. These observations have provided encouragement for the future development of cancer therapies based on targeting individual oncogenes such as c-myc. We have previously reported that FIR strongly represses endogenous c-myc transcription, and induces apoptosis[7] and is thus applicable for cancer treatment. In this study, first, we demonstrated that c-myc suppressor FBP-interacting repressor (FIR) strongly repressed endogenous c-myc transcription and induced apoptosis in SW480, LoVo (human colon cancer cell lines) as well as HeLa cells (human cervical squamous cancer cell line). Second, SeV/dF/FIR showed strong anti-tumor effects in both cultured cells and xenograft tumor growth in an animal model. These results provide new insight into a new therapeutic target for tumor treatment.
What type of suitable vector should be selected and how should FIR expressing vectors be conveyed to cancers? Sendai virus is an RNA virus and exists only in the cytoplasm, hence it is relatively safe as it does not affect chromosomes. In addition, SeV does not transform cells by integrating its genome into the cellular genome, thereby avoiding possible malignant transformation due to the genetic alteration of host cells; this is a safety advantage of SeV. For this reason, we chose SeV and prepared a fusion gene-deficient SeV/dF/FIR vector. The fusion gene-deficient SeV vector cannot transmit to F protein-non-expressing cells as F protein is essential for viral infection. The fusion gene-deficient SeV vector in this study does not require helper virus for reproduction, but is self-replicable in infected cells. Thus, the fusion gene-deficient SeV vector has several advantages over expressing vectors as a gene delivery system for human disease including cancer treatment. First, the fusion gene-deficient SeV vector is not pathogenic in humans. Second, the virus replicates only in the cytoplasm, therefore does not affect chromosome DNA in host cells. Third, SeV vector shows highly efficient gene transfer to a wide spectrum of cells, even to smooth muscle cells, nerve cells, or endothelial cells which are generally difficult to infect. Fourth, the SeV vector shows highly efficient gene transfer to a wide spectrum of cells, even to smooth muscle cells and does not generate wild-type virus in packaging cells. Recently, a gene-deficient SeV (SeV/dF) vector alone demonstrated tumor suppression by activating dendritic cells (DCs)[24], or if granulocyte macrophage colony-stimulating factor was encoded, it produced autologous tumor vaccines[25]. Therefore, the SeV/dF/FIR vector in this study may suppress tumor growth by a dual function through c-Myc suppression of tumor cells and DC activation. Furthermore, SeV/dF/FIR showed a synergistic effect with cisplatin in the treatment of malignant pleural mesothelioma[29]. FIR-binding proteins are basically classified into four categories (Table 1); (1) RNA binding proteins and splicing factors; (2) transcription factors and chromatin remodeling proteins; (3) actin-binding proteins; and (4) signal transduction and protein kinase families. This suggests that FIR potentially engages in some different intracellular events, such as RNA transport, DNA damage repair and pre-mRNA splicing. Accordingly, the side effects of SeV/dF/FIR need to be considered before clinical use, such as pre-mRNA splicing disturbance[8,9], DNA damage repair[37] or intracellular protein transport interference. For clinical safety, SeV/dF/FIR is preferable for local tumor growth control rather than systemic cancer therapy.
Taken together, these findings show that SeV/dF/FIR is a promising approach for cancer gene therapy, although further clinical and basic research are required to explain the precise mechanism of tumor suppression by FIR expressing vectors.
The authors thank to Dr. David Levens (NCI, NIH, United States) for scientific discussions, Dr. Tohru Natsume (Biomedicinal Information Research Center, National Institute of Advanced Industrial Science and Technology, Tokyo, Japan) for FIR-binding proteins analysis, and Dr. Minoru Yoshida (Chemical Genetics Laboratory, RIKEN Advanced Science Institute, Saitama, Japan) for the kind gift of Spliceostatin A (SSA).
Far Up Stream Element-Binding Protein-Interacting Repressor (FIR) is a c-myc transcriptional repressor. Thus, FIR expressing vectors are applicable for cancer therapy. In this study, the authors examined a novel therapeutic strategy to suppress c-myc in human cancers using a fusion gene-deficient Sendai virus (SeV/dF/FIR) which is inherently non-transmissible.
As c-myc transcriptional control is largely unknown, modulation of c-myc regulation by SeV/dF/FIR for cancer therapy should be monitored, strictly and skeptically, from several aspects. This study revealed that SeV/dF/FIR is effective for cancer gene therapy without significant side effects in a xenograft model.
SeV/dF/FIR showed high gene transduction efficiency with significant antitumor effects and apoptosis induction in HeLa and SW480 cells. In the xenograft model, SeV/dF/FIR showed strong suppression of tumor growth with no significant side effects.
SeV/dF/FIR is potentially applicable for future clinical cancer treatment as SeV/dF/FIR suppresses endogenous c-Myc as well as Spliceostatin A (SSA)-activated c-Myc.
FUSE: Far Upstream Element which is required for correct c-myc transcription. FBP: FUSE-Binding protein which has strong transcriptional activity. FIR: FBP interacting repressor which is a critical transcriptional repressor of c-myc gene. SeV: Sendai virus, a member of the Paramyxoviridae family, has envelopes and a nonsegmented negative-strand RNA genome. The SeV genome contains six major genes in tandem on a single negative-strand RNA. DC: Dendritic cell. The gene-deficient SeV (SeV/dF) vector alone demonstrates tumor suppression by activating dendritic cells (DCs).
The authors performed the enthusiastic experiments in vivo and animal model to examine the SeV/dF/FIR for cancer gene therapy to minimize the side effect for the clinical use.
| 1. | Pelengaris S, Khan M, Evan GI. Suppression of Myc-induced apoptosis in beta cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell. 2002;109:321-334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 472] [Cited by in RCA: 501] [Article Influence: 20.9] [Reference Citation Analysis (3)] |
| 2. | Matsushita K, Takenouchi T, Shimada H, Tomonaga T, Hayashi H, Shioya A, Komatsu A, Matsubara H, Ochiai T. Strong HLA-DR antigen expression on cancer cells relates to better prognosis of colorectal cancer patients: Possible involvement of c-myc suppression by interferon-gamma in situ. Cancer Sci. 2006;97:57-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 62] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 3. | Avigan MI, Strober B, Levens D. A far upstream element stimulates c-myc expression in undifferentiated leukemia cells. J Biol Chem. 1990;265:18538-18545. [PubMed] |
| 4. | Bazar L, Meighen D, Harris V, Duncan R, Levens D, Avigan M. Targeted melting and binding of a DNA regulatory element by a transactivator of c-myc. J Biol Chem. 1995;270:8241-8248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 56] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 5. | Michelotti GA, Michelotti EF, Pullner A, Duncan RC, Eick D, Levens D. Multiple single-stranded cis elements are associated with activated chromatin of the human c-myc gene in vivo. Mol Cell Biol. 1996;16:2656-2669. [PubMed] |
| 6. | Liu J, Akoulitchev S, Weber A, Ge H, Chuikov S, Libutti D, Wang XW, Conaway JW, Harris CC, Conaway RC. Defective interplay of activators and repressors with TFIH in xeroderma pigmentosum. Cell. 2001;104:353-363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 92] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 7. | Matsushita K, Tomonaga T, Shimada H, Shioya A, Higashi M, Matsubara H, Harigaya K, Nomura F, Libutti D, Levens D. An essential role of alternative splicing of c-myc suppressor FUSE-binding protein-interacting repressor in carcinogenesis. Cancer Res. 2006;66:1409-1417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 70] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 8. | Matsushita K, Kajiwara T, Tamura M, Satoh M, Tanaka N, Tomonaga T, Matsubara H, Shimada H, Yoshimoto R, Ito A. SAP155-mediated splicing of FUSE-binding protein-interacting repressor serves as a molecular switch for c-myc gene expression. Mol Cancer Res. 2012;10:787-799. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 9. | Matsushita K, Tamura M, Tanaka N, Tomonaga T, Matsubara H, Shimada H, Levens D, He L, Liu J, Yoshida M. Interactions between SAP155 and FUSE-binding protein-interacting repressor bridges c-Myc and P27Kip1 expression. Mol Cancer Res. 2013;11:689-698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 10. | Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, Watanabe H, Kitahara T, Yoshida T, Nakajima H. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol. 2007;3:576-583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 481] [Cited by in RCA: 547] [Article Influence: 28.8] [Reference Citation Analysis (5)] |
| 11. | Li HO, Zhu YF, Asakawa M, Kuma H, Hirata T, Ueda Y, Lee YS, Fukumura M, Iida A, Kato A. A cytoplasmic RNA vector derived from nontransmissible Sendai virus with efficient gene transfer and expression. J Virol. 2000;74:6564-6569. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 195] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 12. | Hirata T, Iida A, Shiraki-Iida T, Kitazato K, Kato A, Nagai Y, Hasegawa M. An improved method for recovery of F-defective Sendai virus expressing foreign genes from cloned cDNA. J Virol Methods. 2002;104:125-133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 20] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 13. | Mitomo K, Griesenbach U, Inoue M, Somerton L, Meng C, Akiba E, Tabata T, Ueda Y, Frankel GM, Farley R. Toward gene therapy for cystic fibrosis using a lentivirus pseudotyped with Sendai virus envelopes. Mol Ther. 2010;18:1173-1182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 95] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 14. | Ueda Y, Hasegawa M, Yonemitsu Y. Sendai virus for cancer immunotherapy. Methods Mol Biol. 2009;515:299-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 15. | Tanaka M, Shimbo T, Kikuchi Y, Matsuda M, Kaneda Y. Sterile alpha motif containing domain 9 is involved in death signaling of malignant glioma treated with inactivated Sendai virus particle (HVJ-E) or type I interferon. Int J Cancer. 2010;126:1982-1991. [PubMed] |
| 16. | Kawaguchi Y, Miyamoto Y, Inoue T, Kaneda Y. Efficient eradication of hormone-resistant human prostate cancers by inactivated Sendai virus particle. Int J Cancer. 2009;124:2478-2487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 64] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 17. | Kinoh H, Inoue M, Komaru A, Ueda Y, Hasegawa M, Yonemitsu Y. Generation of optimized and urokinase-targeted oncolytic Sendai virus vectors applicable for various human malignancies. Gene Ther. 2009;16:392-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 18. | Kinoh H, Inoue M. New cancer therapy using genetically-engineered oncolytic Sendai virus vector. Front Biosci. 2008;13:2327-2334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 19. | Tomonaga T, Levens D. Heterogeneous nuclear ribonucleoprotein K is a DNA-binding transactivator. J Biol Chem. 1995;270:4875-4881. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 152] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 20. | Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, Natsume T, Ohsumi Y, Yoshimori T. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci. 2003;116:1679-1688. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 544] [Cited by in RCA: 606] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 21. | Yanagida M, Hayano T, Yamauchi Y, Shinkawa T, Natsume T, Isobe T, Takahashi N. Human fibrillarin forms a sub-complex with splicing factor 2-associated p32, protein arginine methyltransferases, and tubulins alpha 3 and beta 1 that is independent of its association with preribosomal ribonucleoprotein complexes. J Biol Chem. 2004;279:1607-1614. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 57] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 22. | Natsume T, Yamauchi Y, Nakayama H, Shinkawa T, Yanagida M, Takahashi N, Isobe T. A direct nanoflow liquid chromatography-tandem mass spectrometry system for interaction proteomics. Anal Chem. 2002;74:4725-4733. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 168] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 23. | Komatsu M, Chiba T, Tatsumi K, Iemura S, Tanida I, Okazaki N, Ueno T, Kominami E, Natsume T, Tanaka K. A novel protein-conjugating system for Ufm1, a ubiquitin-fold modifier. EMBO J. 2004;23:1977-1986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 326] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 24. | Komaru A, Ueda Y, Furuya A, Tanaka S, Yoshida K, Kato T, Kinoh H, Harada Y, Suzuki H, Inoue M. Sustained and NK/CD4+ T cell-dependent efficient prevention of lung metastasis induced by dendritic cells harboring recombinant Sendai virus. J Immunol. 2009;183:4211-4219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 1.5] [Reference Citation Analysis (1)] |
| 25. | Inoue H, Iga M, Nabeta H, Yokoo T, Suehiro Y, Okano S, Inoue M, Kinoh H, Katagiri T, Takayama K. Non-transmissible Sendai virus encoding granulocyte macrophage colony-stimulating factor is a novel and potent vector system for producing autologous tumor vaccines. Cancer Sci. 2008;99:2315-2326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 14] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 26. | Liu J, He L, Collins I, Ge H, Libutti D, Li J, Egly JM, Levens D. The FBP interacting repressor targets TFIIH to inhibit activated transcription. Mol Cell. 2000;5:331-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 128] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 27. | Corsini L, Hothorn M, Stier G, Rybin V, Scheffzek K, Gibson TJ, Sattler M. Dimerization and protein binding specificity of the U2AF homology motif of the splicing factor Puf60. J Biol Chem. 2009;284:630-639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 57] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 28. | Page-McCaw PS, Amonlirdviman K, Sharp PA. PUF60: a novel U2AF65-related splicing activity. RNA. 1999;5:1548-1560. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 119] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 29. | Kitamura A, Matsushita K, Takiguchi Y, Shimada H, Tada Y, Yamanaka M, Hiroshima K, Tagawa M, Tomonaga T, Matsubara H. Synergistic effect of non-transmissible Sendai virus vector encoding the c-myc suppressor FUSE-binding protein-interacting repressor plus cisplatin in the treatment of malignant pleural mesothelioma. Cancer Sci. 2011;102:1366-1373. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 30. | Junttila MR, Westermarck J. Mechanisms of MYC stabilization in human malignancies. Cell Cycle. 2008;7:592-596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 115] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 31. | Lee T, Yao G, Nevins J, You L. Sensing and integration of Erk and PI3K signals by Myc. PLoS Comput Biol. 2008;4:e1000013. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 66] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 32. | Yonemitsu Y, Kitson C, Ferrari S, Farley R, Griesenbach U, Judd D, Steel R, Scheid P, Zhu J, Jeffery PK. Efficient gene transfer to airway epithelium using recombinant Sendai virus. Nat Biotechnol. 2000;18:970-973. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 180] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 33. | Inoue M, Tokusumi Y, Ban H, Shirakura M, Kanaya T, Yoshizaki M, Hironaka T, Nagai Y, Iida A, Hasegawa M. Recombinant Sendai virus vectors deleted in both the matrix and the fusion genes: efficient gene transfer with preferable properties. J Gene Med. 2004;6:1069-1081. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 24] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 34. | Shirakura M, Inoue M, Fujikawa S, Washizawa K, Komaba S, Maeda M, Watabe K, Yoshikawa Y, Hasegawa M. Postischemic administration of Sendai virus vector carrying neurotrophic factor genes prevents delayed neuronal death in gerbils. Gene Ther. 2004;11:784-790. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 39] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 35. | Okano S, Yonemitsu Y, Nagata S, Sata S, Onimaru M, Nakagawa K, Tomita Y, Kishihara K, Hashimoto S, Nakashima Y. Recombinant Sendai virus vectors for activated T lymphocytes. Gene Ther. 2003;10:1381-1391. [PubMed] |
| 36. | Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell. 1999;4:199-207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 654] [Cited by in RCA: 676] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 37. | Rahmutulla B, Matsushita K, Satoh M, Seimiya M, Tsuchida S, Kubo S, Shimada H, Otsuka M, Miyazaki M, Nomura F. Alternative splicing of FBP-interacting repressor coordinates c-Myc, P27Kip1/cyclinE and Ku86/XRCC5 expression as a molecular sensor for bleomycin-induced DNA damage pathway. Oncotarget. 2013;December 21. |
P- Reviewer: Takenaga K S- Editor: Ma YJ L- Editor: Webster JR E- Editor: Zhang DN