Published online Feb 14, 2013. doi: 10.3748/wjg.v19.i6.909
Revised: December 4, 2012
Accepted: December 27, 2012
Published online: February 14, 2013
Processing time: 213 Days and 22.5 Hours
AIM: To give a comprehensive report of E-cadherin gene (CDH1) variations in a population at a high risk for gastric cancer (GC).
METHODS: The samples consisted of 178 men and 58 women with a mean age of 62.3 ± 9.4 years and an age range of 30-84 years. A total of 240 cancer-free controls were recruited (mean age of 61.8 ± 10.1 years, age range of 26-82 years). Samples were screened for CDH1 germline mutations by high-resolution melting analysis or directly sequencing. Luciferase reporter assay, RNA splicing assay and bioinformatic analysis were used to evaluate the effect of mutations.
RESULTS: Four novel CDH1 sequence alterations were identified in GC patients including a G>T transition 49 bp before the start codon; a three-nucleotide deletion, c.44_46del TGC; one missense mutation, c.604G>A (V202I); and one variation in the intron, c.1320+7A>G. In addition, polymorphism frequencies were observed for CDH1-164delT, -161C>A, -73A>C, c.48+6C>T, c.48+62_48+63delinsCGTGCCCCAGCCC, c.894C>T (A298A), c.1224G>A (A408A), c.1888C>G (L630V), c.2076T>C (A692A), and c.2253C>T (N751N) which is similar to the data reported in http://www.ncbi.nlm.nih.gov/projects/SNP/. RNA splicing analysis suggested that the c.1320+7A>G and c.1224G>A variations did not affect exon splicing ability. Luciferase reporter assay demonstrated that the c.-49T variation might be helpful for E-cadherin transcription, though the increase in transcription activity is limited (only 33%). SIFT score and PolyPhen analysis both demonstrated that the L630V missense mutation probably damages protein function, while the V202I variant does not.
CONCLUSION: This study reveals novel mutations in sporadic GC patients which had been poorly investigated for susceptibility genes.
-
Citation: Chen QH, Deng W, Li XW, Liu XF, Wang JM, Wang LF, Xiao N, He Q, Wang YP, Fan YM. Novel
CDH1 germline mutations identified in Chinese gastric cancer patients. World J Gastroenterol 2013; 19(6): 909-916 - URL: https://www.wjgnet.com/1007-9327/full/v19/i6/909.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i6.909
Gastric cancer (GC) is one of the most common malignancies worldwide and the leading cancer in East Asian countries[1]. There are two histopathological types of gastric cancer, differentiated and undifferentiated[2], or intestinal and diffuse, respectively[3]. Genetic factors are important for the etiology of GC. The E-cadherin gene (CDH1), a calcium-dependent transmembrane glycoprotein, is critical for epithelial architecture, intercellular adhesion, and cell invasion[4]. E-cadherin consists of a large extracellular domain composed of five repeat domains and smaller transmembrane and cytoplasmic domains[5]. Mutations in the CDH1 gene and perturbation of E-cadherin expression are the most frequent genetic alterations in hereditary diffuse gastric cancer (HDGC)[6,7]. The CDH1 germline mutation spectrum is heterogeneous and includes point mutations, small deletions, and insertions distributed along the entire coding sequence[8-10]. In CDH1 germline variation carriers, the lifetime penetrance is estimated to be approximately 70%[11]. The identification of CDH1 mutations offers the opportunity for the development of cancer risk-reduction strategies for unaffected at-risk individuals. About 90% of gastric carcinoma presents a sporadic setting and only 10% shows a familial cluster; among this group, about 15% are considered as hereditary syndromes, such as the HDGC. For sporadic GC, germline CDH1 mutations are seldom reported.
In this study, we carried out a comprehensive screen of CDH1 germline mutations in 236 Chinese GC patients (175 sporadic cases and 61 cases with hereditary predisposition) (Table 1) and identified four novel germline CDH1 mutations in sporadic GC patients. In addition, the CDH1 polymorphism frequencies in Chinese GC patients and controls were determined. Furthermore, functional assays were carried out to evaluate the impact of the novel mutations identified.
| Variables | Cases | Controls | P value |
| Number | 236 | 240 | |
| Age (yr) | 0.997 | ||
| ≤ 49 | 19 (8.1) | 19 (7.9) | |
| 50-59 | 65 (27.5) | 65 (27.1) | |
| 60-69 | 94 (39.8) | 98 (40.8) | |
| ≥ 70 | 58 (24.6) | 58 (24.2) | |
| Gender | 0.915 | ||
| Male | 178 (75.4) | 180 (75.0) | |
| Female | 58 (24.6) | 60 (25.0) | |
| Family history | |||
| Familial recurrence for gastric cancer1 | 6 (2.5) | ||
| Low familial recurrence for gastric cancer2 | 39 (16.5) | ||
| Young age (< 50 yr) of sporadic disease | 16 (6.8) | ||
| Old age (≥ 50 yr) of sporadic disease | 175 (74.2) | ||
| Histologic grade3 | |||
| Poorly differentiated adenocarcinoma | 64 (39.5) | ||
| Moderately differentiated adenocarcinoma | 69 (42.6) | ||
| Well differentiated adenocarcinoma | 29 (17.9) | ||
| Depth invasion (pT)34 | |||
| pT1 | 12 (7.4) | ||
| pT2 | 32 (19.8) | ||
| pT3 | 109 (67.3) | ||
| pT4 | 9 (5.5) | ||
| Lymph node involvement (pN)34 | |||
| pN0 | 35 (21.6) | ||
| pN1 | 3 (1.9) | ||
| pN2 | 65 (40.1) | ||
| pN3 | 59 (36.4) | ||
| Distant metastasis (M)34 | |||
| M0 | 161 (99.4) | ||
| M1 | 1 (0.6) | ||
| TNM stage34 | |||
| Stage I | 8 (4.9) | ||
| Stage II | 39 (24.1) | ||
| Stage III | 115 (71.0) |
Gastric cancer patients from the East District of China having disease onset between January 1 and December 31, 2008, in whom tumors had been confirmed using histology, were investigated. The samples consisted of 178 men and 58 women with a mean age of 62.3 ± 9.4 years and an age range of 30-84 years. A total of 240 cancer-free controls were recruited (mean age of 61.8 ± 10.1 years, age range of 26-82 years) (Table 1). Details regarding the following information are summarized in Table 1: gastric cancer family history, age of onset and histological and tumor-node-metastasis (TNM) staging classifications. Informed consent, according to the Ethics Committee of the Medical School of Nanjing University, was obtained from all subjects who underwent genetic testing.
Genomic DNA was extracted from peripheral blood leukocytes using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Mutation screening of CDH1 exons 2-16 and neighboring intronic sequences was performed using polymerase chain reaction (PCR) and high-resolution melting analysis using a LightScanner system (Idaho technology, Salt Lake City, UT, United States). The samples that presented abnormal profiles were sequenced on an ABI 3130-Avant automated sequencer (Applied Biosystems, Foster City, CA, United States). The region around the transcription start site (TSS) of the CDH1 gene (From promoter region to intron 1 of CDH1 gene) was genotyped using PCR and directly sequenced on the ABI 3130-Avant automated sequencer. Primer sequences and PCR conditions are available upon request.
A dual-luciferase reporter assay system (http://www.promega.com) was used to examine the effects of novel sequence variation in the promoter region on the transcriptional activity of CDH1. Briefly, DNA fragments around the TSS (-345 to 271 bp) were amplified by PCR using genomic DNA containing either the particular variant sequence or CDH1 wild type as a template. The amplified fragments were designed to contain the region possessing basal promoter activity. The following primer sequences were used: 5’-ATGCCTCGAGCCATCTCCAAAACGAACAAAC-3’ (forward) and 5’-ATGCAAGCTTGAAGGGAAGCGGTGACGAC-3’ (reverse), which include the restriction sites (underlined) for XhoI and HindIII, respectively. The PCR products were digested with XhoI and HindIII and subsequently cloned into the pGL3-basic vector carrying the firefly luciferase gene (Promega). The nucleotide sequence of the fragment inserted into each plasmid was confirmed by DNA sequencing. Plasmids were then transiently transfected into Hela cells using the Lipofect transfection reagent (Tiangen Biotech, Beijing, Co., Ltd., China). All plasmids were co-transfected with the renilla luciferase gene containing the pRL-CMV plasmid (Promega) as an internal standard. Cell extracts were prepared, and luciferase activity was measured by a luminometer instrument (Promega) using the dual-luciferase reporter assay system (Promega). The transcriptional activity in each cell extract was determined from the level of firefly luciferase after normalization to renilla luciferase activity. Four independent experiments were performed using DNA from plasmid preparations.
Because it has been recognized that DNA sequence variants localized in exon-intron boundaries could be pathogenic by affecting exon definition and the splicing of pre-mRNA[12,13], we used RNA splicing assay to evaluate the variant located at the 5’ terminal of intron 9. Total RNA from frozen tumor tissue and paired normal tissue was extracted using RNAiso Plus (TaKaRa Biotechnology [(Dalian) Co., Ltd.]. Reverse transcription (RT)-PCR was performed in 2 steps. First strand cDNA synthesis was performed using PrimerScript RT reagent Kit (TaKaRa) with random DNA hexamers and oligo-dT primer according to the manufacturer’s protocol. cDNA was amplified in the region of exons 7-10. Primer sequences were 5’-GGACCGAGAGAGTTTCCCTACG-3’ (sense) and 5’-GTTATTTTCTGTTCCATAAATG-3’ (antisense). PCR conditions were as follows: 35 cycles at 94 °C for 30 s, 58 °C for 30 s and 72 °C for 30 s, followed by a final extension at 72 °C for 5 min. Agarose gel electrophoresis was carried out using 2% gels run at 100 V for 40 min. The purified amplification products were sequenced on the ABI 3130-Avant automated sequencer.
The impact of amino acid allelic variants on protein structure/function can be predicted via analysis of multiple sequence alignments and protein 3D-structures. The sorting intolerant from tolerant (SIFT) algorithm and Polymorphism Phenotyping (PolyPhen) were adopted.
SIFT is a program that predicts the effect of amino acid substitutions on protein function based on sequence conservation during evolution and the nature of the amino acids substituted in a gene of interest[14]. The SIFT score was calculated online (http://sift.jcvi.org/). If the value is less than 0.05, the amino acid substitution is predicted as intolerant, while those with a value greater than or equal to 0.05 are classified as tolerated.
PolyPhen is an automatic tool for prediction of the possible impact of an amino acid substitution on the structure and function of a human protein based on straightforward physical and comparative considerations[15] (http://genetics.bwh.harvard.edu/pph/). Each of the two amino acid residues [the original residue and the single-nucleotide polymorphism (SNP)] was entered and the difference between the position-specific independent counts (PSIC) scores of the two residues was computed. The higher a PSIC score difference is, the higher the functional impact a particular amino acid substitution is likely to have. A PSIC score difference of 1.5 and above is considered to be damaging.
χ2 tests or Fisher’s exact tests were used to compare the distribution of variables between cases and controls. Luciferase activities were compared using Student’s unpaired t test. All statistical tests were two-sided, with a P value of 0.05 considered to be significant, using SPSS software (version 16).
The study was comprised of 236 gastric cancer cases and 240 cancer-free controls. There were no significant differences in the distributions of age or gender between the cases and controls (P = 0.997 and 0.915, respectively) (Table 1). The majority of studied cases were sporadic; approximately 20% had a family history of cancer. The tumor type was assessed in 162 cases, and more than 80% of the cases had poorly differentiated or moderately differentiated adenocarcinoma; more than 70% of the cases were in TNM Stage III (Table 1).
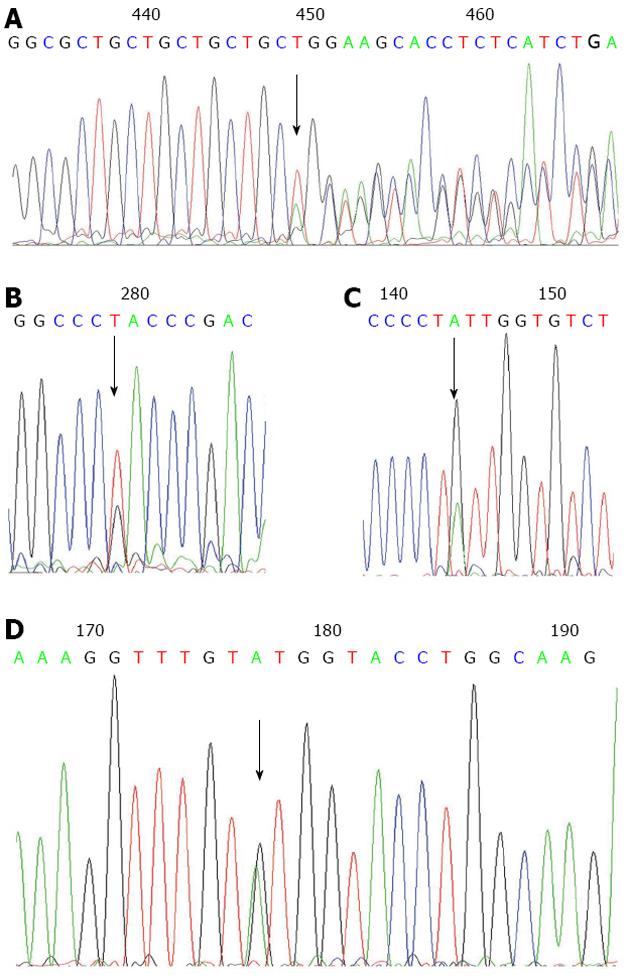
Four novel CDH1 germline variations were identified in gastric cancer patients. One of the variants was located in the CDH1 5’UTR (c.-49 G>T) and is seemed to be a polymorphism since it is found in both the cases and controls. The other three were only detected in the GC cases and not seen in the controls. One was a missense mutation in the coding region [c.604G>A (V202I)], one was a three-nucleotide deletion in exon 1 (c.44_46del TGC) and the other was an intronic variation (c.1320+7A>G) (Figure 1 and Table 2). In addition, ten CDH1 polymorphisms (and their frequencies) were observed. The polymorphisms frequencies are similar to the data available at the SNP website (http://www.ncbi.nlm.nih.gov/projects/SNP/) (Table 3).
| Gene location | Sequence variant | Consequence | Genotype | ||
| Gastric cancer patients (n = 236) | Control (n = 240) | P value | |||
| 5’UTR | c.-49 G>T | Substitution at the 5’UTR | 4 (1.7) | 3 (1.3) | 0.723 |
| Exon 1 | c.44_46delTGC | Loss of the 15th code (Leu) | 2 (0.85) | 0 (0.0) | 0.245 |
| Exon 5 | c.604G>A | Missense V202I | 1 (0.4) | 0 (0.0) | 0.496 |
| Intron 9 | c.1320+7 A>G | Substitution of invariant A | 1 (0.4) | 0 (0.0) | 0.496 |
| Gene location | Sequence variant | Condon | MAF in 236 GC patients | MAF in 240 controls | Reported MAF1 |
| Promoter | -164del T(g.4837delT) | - | 0.004del T | 0.000del T | rs5030658: NA |
| Promoter | -161C>A(g.4840C>A) | - | 0.242A | 0.227A | rs16260: 0.227A |
| Promoter | -73A>C (g.4928A>C) | - | 0.129C | 0.146C | rs28372783: 0.040C |
| Intron 1 | c.48+6C>T | - | 0.280C | 0.271C | rs3743674: 0.214C |
| Intron1 | c.48+62_48+63delinsCGTGCCCCAGCCC | - | 0.280del2 | 0.271del2 | rs3833051: NA |
| Exon 7 | c.894C>T | A298A | 0.002T | 0.000T | rs139110184: NA |
| Exon 9 | c.1224G>A | A408A | 0.002A | 0.000A | rs200161607: 0.001A |
| Exon 12 | c.1888C>G | L630V | 0.004G | 0.004G | rs2276331: 0.002G |
| Exon 13 | c.2076T>C | A692A | 0.377T | 0.404T | rs1801552: 0.307T |
| Exon 14 | c.2253C>T | N751N | 0.089T | 0.102T | rs33964119: 0.058T |
The cases carrying novel CDH1 mutations were all sporadic GC patients, with poorly differentiated or moderately differentiated adenocarcinoma (Table 4). The case with CDH1 c.604G>A (V202I) mutation harbors the MLH1 c.2101C>A (Q701K) mutation as well[16].
| ID code | Age onset (yr) | Gender | Family history | Histologic grade | Depth invasion (pT) | Lymph node involvement (pN) | Distant metastasis (M) | CDH1 variants |
| G45 | 66 | Female | Sporadic | Poorly differentiated adenocarcinoma | pT3 | pN3 | M0 | c.44_46delTGC |
| G68 | 51 | Male | Sporadic | NA | NA | NA | NA | c.44_46delTGC |
| G1501 | 58 | Male | Sporadic | Moderately differentiated adenocarcinoma | pT2 | pN3 | M0 | c.604G>A (V202I) |
| G26 | 56 | Male | Sporadic | Moderately differentiated adenocarcinoma | pT3 | pN2 | M0 | c.1320+7 A>G |
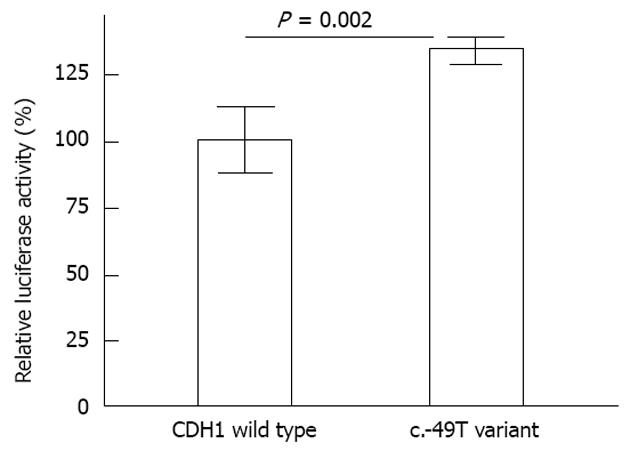
c.-49T variation contribute a slightly higher promoter activity of the CDH1 gene than the wild type: The c.-49T alteration was near the TSS of the CDH1 gene (49 bp before the start codon, and +76 bp relative to the TSS). To examine the potential effect of the c.-49T variation on E-cadherin gene transcription, a 616-bp promoter of the E-cadherin gene (-345 to 271) carrying either the G or T allele was inserted upstream of the luciferase gene in the pGL3 promoterless enhancer plasmid vector. The activity of the E-cadherin G/T promoter-luciferase reporter gene constructs was assessed using transient transfection assays in Hela cells. As shown in Figure 2, slightly higher luciferase activities were observed for the pGL-T construct compared with the pGL-G (wild type) construct. The average activity of the promoter having the c.-49T variation was 133.7% (P = 0.002) relative to the CDH1 wild type promoter.
c.44_46del TGC variant causes the loss of one leucine in the signal peptide region of the E-cadherin protein: Codon sequence analysis demonstrated that the three-nucleotide deletion c.44_46del TGC causes the loss of a single amino acid [the 15th codon (Leucine)] in exon 1 of CDH1, which is in the signal peptide region of the E-cadherin protein.
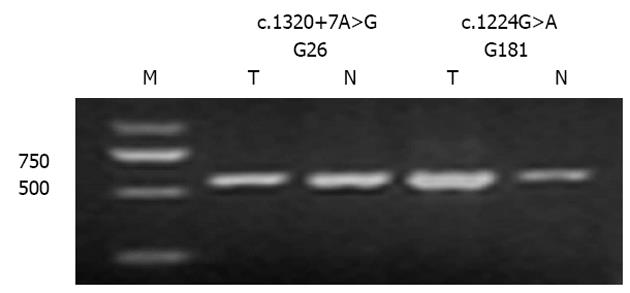
Intron variation (c.1320+7A>G) and the silent mutation [c.1224G>A (A408A)] do not induce CDH1 splicing defects: To evaluate functional consequence of those novel mutations detected on pre-mRNA splicing, we analyzed cDNA produced in vivo from tissues retrieved from the patients harboring the novel mutations. The PCR fragment generated using primers flanking the silent mutation [c.1224G>A (A408A)] and the one intron variation (c.1320+7A>G) indicated normal-sized mRNA. This result demonstrated that a splicing defect was not likely to occur as a consequence of these mutations (Figure 3).
c.1888C>G (L630V) missense mutation might impair E-cadherin protein function while the V202I mutation does not: Two CDH1 missense mutations were detected. Both the SIFT score and the PolyPhen analysis demonstrated that the L630V variant was sorted as being intolerant, suggesting that this amino acid substitution is predicted to damage protein function. The other variant, V202I, of CDH1 was sorted as tolerant (Table 5).
| Sequence variant | Structural alteration | SIFT scores | PSIC score difference | Prediction |
| c.604G>A | V202I | 0.20 | 0.381 | Benign |
| c.1888C>G | L630V | 0.02 | 1.748 | Probably damaging |
The discovery of genetic variants responsible for the pathogenesis of gastric cancer is important in understanding this disease. Although screening of CDH1 germline mutations in hereditary GC has been fairly well established, the report of CDH1 germline mutations in sporadic GC is limited. Bacani et al[17] identified a germline deletion (nt41delT) in a 30-year-old sporadic GC patient and suggested that 2%-3% of cases of early-onset gastric cancer in North America may be owing to high-risk genetic mutations. Garziera et al[18] reported a germline missense mutation in CDH1 exon 6, c. 820 G>A (G274S) in one sporadic Italian gastric cancer patient. Here, we report a population-based study of GC to determine the role of germline mutations in a population at a high risk for GC. The majority of studied cases were sporadic. We have studied all of the coding and promoter core regions of the most important gene implicated in GC and have identified four novel CDH1 sequence variants distributed along the entire coding sequence and the non-coding regions in the CDH1 gene. This is consistent with previously published reports[19], suggesting that CDH1 mutations have arisen without mutational hotspots.
The c.-49 G>T transition was detected in 4/236 (1.7%) of GC patients and 3/240 (1.3%) of cancer-free controls; however, these differences did not achieve significance (P = 0.723, Table 2). One patient with this variant had low familial recurrence for gastric cancer, but IHC showed normal CDH1 protein expression. Previous studies have demonstrated that a fragment spanning -399 to +31 bp relative to the TSS of the CDH1 gene possesses basal promoter activity[20]. As this variant is around the TSS of the CDH1 gene, a luciferase reporter assay was carried out. This in vitro assay showed that the c.-49T promoter had 33% higher activity than the promoter containing the c.-49G (Figure 2). So this polymorphism might be helpful for E-cadherin transcription, though the increase in transcription activity is limited (only 33%).
The c.44_46del TGC variant was detected in two sporadic GC patients of 51 years and 66 years and was not detected in the 240 controls. One patient’s pathological data was available, which showed a poorly differentiated adenocarcinoma (Table 4). To the best of our knowledge, this variant has never been reported in the open access mutation database and literatures. As the parents of the probands are not available for mutation analyzing, we are not certain whether it’s a de novo mutation. This variant seems to be a rare variant with an allele frequency in GC patients of 0.85% and can even be considered as a kind of mutation hotspot, as it has been detected in two patients with no relationship. Recently rare variants have been reported in several diseases, include cancer. The identified rare variants often have functional effects on protein-protein interactions. Further, rare variants might confer a stronger increase in disease risk than common variants and may make a substantial contribution to the multifactorial inheritance of common chronic diseases[21-24]. Codon sequence analysis demonstrated that the c.44_46del TGC variant causes the loss of a single amino acid [the 15th codon (Leucine)] in exon 1 of CDH1 which is in the signal peptide region of the E-cadherin protein. This amino acid loss might have effect on E-cadherin protein, and further functional analysis should be carried out to investigate associations of the variant with phenotype.
Growing evidence has shown that rare single base substitutions localized in exon-intron boundaries can disrupt one of the cis-transcriptional elements known as exonic splicing enhancers and affect normal pre-mRNA splicing[12,13]. Therefore, it appears reasonable to verify the effect of variants at the mRNA level. The sequence alteration c.1320+7A>G, located in an exon-intron boundary, was detected in a 56-year-old GC patient and not in the 240 controls (Table 2). RNA splicing assay demonstrated that this variation did not affect exon splicing ability (Figure 3) and might be rare polymorphism.
The CDH1 molecule consists of five tandemly repeated extracellular domains (EC1-EC5, containing exons 4-13), each about 110 amino acids in length. This large extracellular domain is responsible for Ca2+ binding and is important for cell-cell adhesion. The NH2-terminal EC1 domain is required for lateral E-cadherin dimerization contributing to the intercellular junction[25-27]. A novel missense mutation, V202I, is located in the middle of EC1. While EC1 shows remarkably high conservation between various species[28], SIFT and PolyPhen analyses both showed that V202I might be a tolerant variation (Table 5). The GC patient with this variation carried the MLH1 c.2101C>A (Q701K) mutation as well. IHC analysis in the index patient demonstrated a loss of MLH1 protein and normal expression of MSH2 and E-cadherin[16]; therefore, we suggest that the MLH1 c.2101 C>A (Q701K) mutation, and not the CDH1 c.604G>A (V202I) variation, might be the cause of GC in this patient.
In addition, frequencies of CDH1 polymorphisms in Chinese GC patients and controls were reported which were similar to those reported at http://www.ncbi.nlm.nih.gov/projects/SNP/ (Table 3). It needs to say something about the rare polymorphism, c.1888C>G (L630V). The SIFT score of the CDH1 variants and the PolyPhen analysis both showed that the L630V variant probably damages protein function (Table 5). However, data from case-control analysis did not support an effect of this L630V variant (Table 3). Accordingly, the pathogenic role of this polymorphism remains elusive and in vitro approaches should be performed to elucidate its function.
In conclusion, this study reveals novel mutations in sporadic GC patients in China, a high-incidence country for GC. Though the pathogenic role of the variants remains still uncertain, our findings display the necessity to scan germline CDH1 variants in sporadic gastric cancer population.
Although E-cadherin gene (CDH1) germline mutations are well implicated in hereditary diffuse gastric cancer, the report of CDH1 germline mutations in sporadic gastric cancer (GC) is limited.
Here, the authors report a population-based study of GC to determine the role of CDH1 germline mutations in a population at a high risk for GC. The majority of studied cases were sporadic.
The authors have studied all of the coding and promoter core regions of CDH1 and identified four novel CDH1 sequence variants, including one transition near the transcription start site, one three-nucleotide deletion in code region, one missense mutation, and one variation in exon-intron boundary. Three of the four variants were detected only in sporadic GC patients and not in the 240 cancer-free controls. Though the functional significance of the variants remains still uncertain, this study reveals novel mutations in sporadic GC patients which had been poorly investigated for susceptibility genes
The findings display the necessity to scan germline CDH1 variants in sporadic gastric cancer population.
High-resolution melting analysis, is a high-throughput single-nucleotide polymorphism genotyping technology based on the analysis of the melting profile of polymerase chain reaction products.
In this study, the authors identified 4 novel CDH1 germline mutations in different patients harbouring GC with a sporadic setting. The identification of mutations represents an important discovery, to assess the cancer risk for the novel generations.
| 1. | Ushijima T, Sasako M. Focus on gastric cancer. Cancer Cell. 2004;5:121-125. [PubMed] |
| 2. | Nakamura K, Sugano H, Takagi K. Carcinoma of the stomach in incipient phase: its histogenesis and histological appearances. Gann. 1968;59:251-258. [PubMed] |
| 3. | Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31-49. [PubMed] |
| 4. | Takeichi M. Cadherin cell adhesion receptors as a morphogenetic regulator. Science. 1991;251:1451-1455. [PubMed] |
| 5. | Ringwald M, Schuh R, Vestweber D, Eistetter H, Lottspeich F, Engel J, Dölz R, Jähnig F, Epplen J, Mayer S. The structure of cell adhesion molecule uvomorulin. Insights into the molecular mechanism of Ca2+-dependent cell adhesion. EMBO J. 1987;6:3647-3653. [PubMed] |
| 6. | Pedrazzani C, Corso G, Marrelli D, Roviello F. E-cadherin and hereditary diffuse gastric cancer. Surgery. 2007;142:645-657. [PubMed] |
| 7. | Corso G, Marrelli D, Pascale V, Vindigni C, Roviello F. Frequency of CDH1 germline mutations in gastric carcinoma coming from high- and low-risk areas: metanalysis and systematic review of the literature. BMC Cancer. 2012;12:8. [PubMed] |
| 8. | Brooks-Wilson AR, Kaurah P, Suriano G, Leach S, Senz J, Grehan N, Butterfield YS, Jeyes J, Schinas J, Bacani J. Germline E-cadherin mutations in hereditary diffuse gastric cancer: assessment of 42 new families and review of genetic screening criteria. J Med Genet. 2004;41:508-517. [PubMed] |
| 9. | Kaurah P, MacMillan A, Boyd N, Senz J, De Luca A, Chun N, Suriano G, Zaor S, Van Manen L, Gilpin C. Founder and recurrent CDH1 mutations in families with hereditary diffuse gastric cancer. JAMA. 2007;297:2360-2372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 332] [Cited by in RCA: 327] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 10. | Shah MA, Salo-Mullen E, Stadler Z, Ruggeri JM, Mirander M, Pristyazhnyuk Y, Zhang L. De novo CDH1 mutation in a family presenting with early-onset diffuse gastric cancer. Clin Genet. 2012;82:283-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 24] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 11. | Pharoah PD, Guilford P, Caldas C. Incidence of gastric cancer and breast cancer in CDH1 (E-cadherin) mutation carriers from hereditary diffuse gastric cancer families. Gastroenterology. 2001;121:1348-1353. [PubMed] |
| 12. | Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1559] [Cited by in RCA: 1649] [Article Influence: 68.7] [Reference Citation Analysis (0)] |
| 13. | Tournier I, Vezain M, Martins A, Charbonnier F, Baert-Desurmont S, Olschwang S, Wang Q, Buisine MP, Soret J, Tazi J. A large fraction of unclassified variants of the mismatch repair genes MLH1 and MSH2 is associated with splicing defects. Hum Mutat. 2008;29:1412-1424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 138] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 14. | Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812-3814. [PubMed] |
| 15. | Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894-3900. [PubMed] |
| 16. | Zhi W, Xue B, Wang L, Xiao N, He Q, Wang Y, Fan Y. The MLH1 2101C& gt; A (Q701K) variant increases the risk of gastric cancer in Chinese males. BMC Gastroenterol. 2011;11:133. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 17. | Bacani JT, Soares M, Zwingerman R, di Nicola N, Senz J, Riddell R, Huntsman DG, Gallinger S. CDH1/E-cadherin germline mutations in early-onset gastric cancer. J Med Genet. 2006;43:867-872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 33] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 18. | Garziera M, De Re V, Geremia S, Seruca R, Figueiredo J, Melo S, Simões-Correia J, Caggiari L, De Zorzi M, Canzonieri V. A novel CDH1 germline missense mutation in a sporadic gastric cancer patient in north-east of Italy. Clin Exp Med. 2012;Apr 29; Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 19. | Blair V, Martin I, Shaw D, Winship I, Kerr D, Arnold J, Harawira P, McLeod M, Parry S, Charlton A. Hereditary diffuse gastric cancer: diagnosis and management. Clin Gastroenterol Hepatol. 2006;4:262-275. [PubMed] |
| 20. | Giroldi LA, Bringuier PP, de Weijert M, Jansen C, van Bokhoven A, Schalken JA. Role of E boxes in the repression of E-cadherin expression. Biochem Biophys Res Commun. 1997;241:453-458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 107] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 21. | Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695-701. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 875] [Cited by in RCA: 837] [Article Influence: 46.5] [Reference Citation Analysis (0)] |
| 22. | Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A. Finding the missing heritability of complex diseases. Nature. 2009;461:747-753. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5794] [Cited by in RCA: 5927] [Article Influence: 348.6] [Reference Citation Analysis (1)] |
| 23. | Gorlov IP, Gorlova OY, Sunyaev SR, Spitz MR, Amos CI. Shifting paradigm of association studies: value of rare single-nucleotide polymorphisms. Am J Hum Genet. 2008;82:100-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 249] [Cited by in RCA: 261] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 24. | Fearnhead NS, Winney B, Bodmer WF. Rare variant hypothesis for multifactorial inheritance: susceptibility to colorectal adenomas as a model. Cell Cycle. 2005;4:521-525. [PubMed] |
| 25. | Parisini E, Higgins JM, Liu JH, Brenner MB, Wang JH. The crystal structure of human E-cadherin domains 1 and 2, and comparison with other cadherins in the context of adhesion mechanism. J Mol Biol. 2007;373:401-411. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 95] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 26. | Nelson WJ. Regulation of cell-cell adhesion by the cadherin-catenin complex. Biochem Soc Trans. 2008;36:149-155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 292] [Cited by in RCA: 315] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 27. | Shi Q, Maruthamuthu V, Li F, Leckband D. Allosteric cross talk between cadherin extracellular domains. Biophys J. 2010;99:95-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 35] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
P- Reviewer Marrelli D S- Editor Zhai HH L- Editor A E- Editor Li JY