Published online Nov 7, 2013. doi: 10.3748/wjg.v19.i41.7032
Revised: September 1, 2013
Accepted: September 15, 2013
Published online: November 7, 2013
Processing time: 114 Days and 19.3 Hours
Except for the most organized mature hepatocytes, liver stem/progenitor cells (LSPCs) can differentiate into many other types of cells in the liver including cholangiocytes. In addition, LSPCs are demonstrated to be able to give birth to other kinds of extra-hepatic cell types such as insulin-producing cells. Even more, under some bad conditions, these LSPCs could generate liver cancer stem like cells (LCSCs) through malignant transformation. In this review, we mainly concentrate on the molecular mechanisms for controlling cell fates of LSPCs, especially differentiation of cholangiocytes, insulin-producing cells and LCSCs. First of all, to certificate the cell fates of LSPCs, the following three features need to be taken into account to perform accurate phenotyping: (1) morphological properties; (2) specific markers; and (3) functional assessment including in vivo transplantation. Secondly, to promote LSPCs differentiation, systematical attention should be paid to inductive materials (such as growth factors and chemical stimulators), progressive materials including intracellular and extracellular signaling pathways, and implementary materials (such as liver enriched transcriptive factors). Accordingly, some recommendations were proposed to standardize, optimize, and enrich the effective production of cholangiocyte-like cells out of LSPCs. At the end, the potential regulating mechanisms for generation of cholangiocytes by LSPCs were carefully analyzed. The differentiation of LSPCs is a gradually progressing process, which consists of three main steps: initiation, progression and accomplishment. It’s the unbalanced distribution of affecting materials in each step decides the cell fates of LSPCs.
Core tip: After liver stem/progenitor cells (LSPCs) are isolated by different groups from both fetal and adult livers, it is urgent to decide the cell fates of LSPCs. Especially, it is found that the core issue for LSPCs application lies in their accurate differentiation. Because there are lots of literatures concentrating on self-renewal and hepatic differentiation of LSPCs, in this review, we mainly summarize the molecular mechanisms for controlling other cell fates of LSPCs, especially differentiation into cholangiocytes. For biliary differentiation, we propose that it is a gradually progressing process consisting of three main steps: initiation, progression and accomplishment.
- Citation: Liu WH, Ren LN, Chen T, Liu LY, Tang LJ. Stages based molecular mechanisms for generating cholangiocytes from liver stem/progenitor cells. World J Gastroenterol 2013; 19(41): 7032-7041
- URL: https://www.wjgnet.com/1007-9327/full/v19/i41/7032.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i41.7032
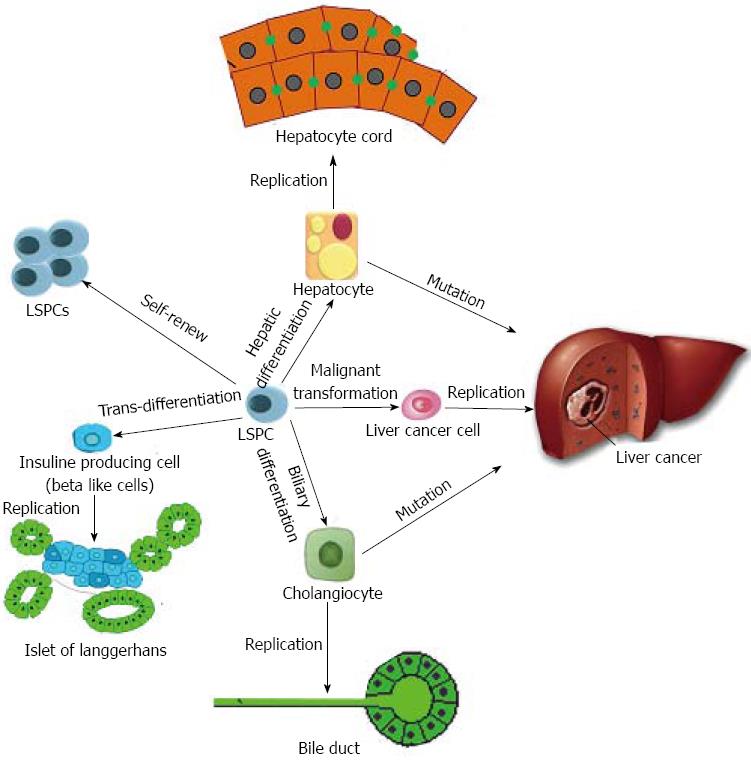
Liver stem/progenitor cells (LSPCs) possess high proliferative capacity and low immunogenicity and are robust in the face of cryopreservation or ischemic injury: properties that could enhance their engraftment within a recipient liver. Because of this, these LSPCs are very promising for the treatment of end-stage liver disease[1]. A series of animal models transplanted with LSPCs have been established, and several clinical trials have been reported. In one animal model, transplanted rat embryonic day (ED) 14 fetal liver stem/progenitor cells (FLSPCs) differentiated into the two mature epithelial cell phenotypes in the liver, i.e., hepatocytes and cholangiocytes, and long-term, in vivo functional reconstitution of the liver tissue was achieved (Figure 1)[2,3]. The important recent progress is the use of human FLSPCs engrafted into naturally derived scaffolds to create a liver-like tissue in vitro[4]. However promising LSPCs are for cell therapy or tissue engineering, the fundamental purpose lies in generating mature, functional cells[5,6].
LSPCs constitute approximately 0.5%-2.5% of liver parenchyma at all donor ages. The self-renewal capacity of LSPCs is demonstrated by their phenotypic stability after expansion for > 150 population doublings in a serum-free, defined medium, with a doubling time of approximately 36 h[7]. In fetal liver, LSPCs are commonly called hepatoblasts[2,3]. In some studies, some groups have used other terms than hepatoblasts to represent the cell populations with stem properties in fetal liver, such as embryonic hepatic stem cells or fetal liver stem-like epithelia. Thus, it would be more appropriate to denote these cells as the “FLSPCs”, and we will adopt this description in this review. In adult liver, LSPCs are generally referred to as oval cells (OCs), with scant, lightly basophilic cytoplasm and pale blue-staining nuclei[8]. The appearance of OCs has been reported in rat livers treated with hepatotoxins, such as 2-acetylaminofluorene, combined with partial hepatectomy (PHx) and D-galactosamine[9,10]. However, in addition to OCs, small hepatocytes (SHs) are also well known in adult liver, and they are better suited to the appellation “progenitor” cells. As it is not an easy task to distinguish stem cells from progenitors because of the difficulty of proving the unlimited self-renewal activity of stem cells in many situations, we use the term “adult liver stem/progenitor cells (ALSPCs)” to describe such cells, including both OCs and SHs in this review article. In the field of liver biology, the definitions of “LSPCs” include the following: (1) cells responsible for normal tissue turnover; (2) cells that regenerate liver after PHx; (3) cells responsible for progenitor-dependent regeneration; (4) transplantable liver repopulating cells; and (5) cells that adopt hepatocyte and bile duct phenotypes in vitro.
Currently, researchers are working hard to characterize, localize and isolate LSPCs, though this has been difficult because of the lack of specific markers[11]. To avoid the restriction of lacking specific markers, Liu et al[12,13] have tried other strategies for isolating LSPCs. Based on the concept that stem cells have specific physical and morphological properties, Liu et al[12] isolated FLSPCs by a percoll continuous gradient centrifugation-centered three-step method. Because stem cells have specific functional characteristics, such as excluding biological vital dyes such as Hoechst 33342, Liu et al[13] obtained ALSPCs by side population (SP) enrichment. Although many groups have isolated LSPCs using various strategies, LSPCs for disease application remain far off. The core issue is how to manipulate LSPC differentiation, which is essential for both cell therapy and liver regeneration[14,15]. Thus, to guarantee the efficiency and security of LSPC-based therapy for liver diseases, it is important to clarify the strategies and related mechanisms for proper differentiation of LSPCs.
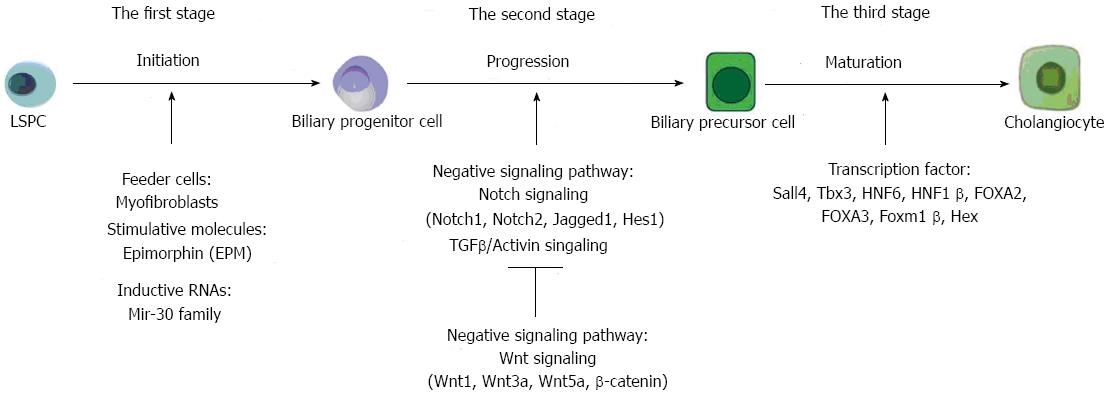
In recent years, many studies have shed light on the tangle of regulatory mechanisms that govern the complex process of LSPC differentiation, but an overall understanding remains a challenge. Here, we review the current understanding of the exact mechanisms related to the differentiation of LSPCs, especially toward cholangiocytic differentiation. We divided the process of LSPC differentiation into three stages (Figure 2). The first stage is the onset of differentiation, when LSPCs are induced to mature into certain cell types. In this stage, the lineage-specific cytokines/growth factors (GFs), their (relative) doses and order of application are crucial for directing the lineage specification of the LSPCs[16]. The second stage is the acceleration of differentiation, when LSPCs are quickly progressing through the process. Many developmental regulatory signaling pathways, including the Wnt, Notch, bone morphogenetic protein and fibroblast growth factor pathways, may play a role in directing the cell fates of LSPCs[17]. The third stage is to guarantee the accomplishment of differentiation. In this stage, transcription factors are vital to make cells express lineage-specific markers[5].
LSPCs can differentiate into a wide range of cell types, including hepatocytes, cholangiocytes, pancreatic cells and intestinal epithelial cells (Figure 1)[18]. However, in this review, we focus on cholangiocytic specification from LSPCs. To ascertain the cell fates of LSPCs, the following three features inherent to LSPC transitions must be taken into account for accurate phenotyping: (1) the differentiation of LSPCs toward a specific lineage often involves uncontrolled processes, resulting in a heterogeneous cell population; (2) the differentiation into mature cells is a steady process; and (3) the ultimate proof of functional cell behavior is in vivo transplantation of ex vivo generated LSPC-based mature cells into immunodeficient animal models with liver injury[19,20].
As LSPCs differentiate into cholangiocytes, the cells grow in size to > 12 μm and display a keystone morphology with cholangiocyte-type epithelial polarity. These cells are concentrically layered to form a cyclic structure or arranged in lines to form ductal plates. Under the electron microscope, these cells acquired the classic cholangiocyte features of small numbers of organelles and many primary cilia on their surface.
Aside from morphological identification, the analytical work is limited to the elucidation of (1) cholangiocytic RNA transcripts via (quantitative) reverse transcriptase polymerase chain reaction and (2) cholangiocytic proteins by immunofluorescence. During the process of LSPC differentiation into cholangiocytes, cells transition from the expression of early biliary markers (such as Sox9, which is a representative transcriptional factor expressed in biliary precursor cells), to the expression of mid-stage biliary markers (such as the cytokine CK19 and E-cadherin), and then mature biliary markers (such as CK7)[21]. In addition, gamma-glutamyl transpeptidase (GGT), a major enzyme of glutathione homeostasis, is often used as a biliary marker to follow the differentiation of LSPCs[22]. Furthermore, multidrug resistance-associated protein 3[23] and secretin receptors[24] are also found to be expressed in cholangiocytes.
Although the induced differentiation of cholangiocytes has been performed, the functional examination of LSPC-derived cholangiocytes is very scarce. Thus, the in vivo identification of induced cholangiocytes is essential, and to some extent it can be considered the “gold standard” of certifying the cell fates of LSPCs[25]. LSPC-derived cholangiocytes in vivo should be able to replace injured cholangiocytes or lost bile duct cells.
The components of the stem-cell microenvironment regulating differentiation include distinct cell-cell interactions and paracrine signals, which comprise both soluble and extracellular matrix factors, as well as the three-dimensional architecture, which shapes and dictates the delivery of these cues. It is reported that mature stellate cells and/or myofibroblasts resulted in differentiation of LSPCs into cholangiocytes[26]. These feeder cells control the cell fates of LSPCs through either paracrine signaling pathways or cell-cell interaction[27-30]. Thus, if the paracrine signals produced by the feeders are replaced with similar components, the same induced differentiation of LSPCs could be achieved. There are feeder-free conditions that yield equivalent results, consisting of the embedding of LSPCs into hydrogels containing type I collagen (60%) and Matrigel (40%) with modified Kubota's medium for cholangiocytes. It is also demonstrated that the murine FLSPC cell line, hepatoblast cell line-3, can be induced to differentiate toward cholangiocyte by plating in Matrigel[31]. Furthermore, Matrigel-coated films are also widely used for manipulating LSPCs. Although PLL-terminal t-(poly-l-lysine/poly-l-glutamic acid) (PLL/PLGA) films are less favorable for stem cell cultures than PLGA-terminal t-(PLL/PLGA) films, the cell fates of LSPCs are correlated with the film thickness on both types of film, with differentiation favored on the thinner films[32].
Recent evidence has shown that expression of miRNAs can regulate the divergent differentiation pathways of stem cells[33]. Therefore, Liu et al[13] reasoned that miRNAs could be responsible for regulating cell fate decisions in LSPCs by regulating the cells’ responses to ubiquitous GFs. It was found that the miR-23b cluster, including miR-23b, miR-27b, and miR-24-1 and miR-10a, miR-26a and miR-30a, was highly expressed in LSPCs[34]. MiR-23b cluster repressed bile duct gene expression in LSPCs while promoting their growth; low levels of the miR-23b miRNAs were needed in cholangiocytic differentiation and bile duct formation[34].
The process of intrahepatic bile duct (IHBD) formation from LSPCs involves cholangiocyte differentiation (lineage specification) and morphogenesis of ductal structures[21]. Understanding how LSPCs can generate differentiated bile ducts is crucial for studies on epithelial morphogenesis and for development of cell therapies for hepatobiliary diseases. Many groups[35-37] have demonstrated that, during in vivo liver development and in vitro differentiation, LSPCs located around the portal vein first develop as biliary precursor cells and then generate cholangiocytes. Nevertheless, the molecular mechanisms behind these events have yet to be fully elucidated. It is shown that Wnt and Notch signaling are active in the adult human liver to drive proliferation and differentiation of LSPCs into the hepatocyte or cholangiocyte lineages[38]. The Notch pathway is triggered by expression of the Notch ligand Jagged1 by myofibroblasts, thereby promoting biliary differentiation of LSPCs, and the enhancement of Wnt3a expression in macrophages after uptake of hepatocyte debris and paracrine activation of Wnt signaling in neighboring LSPCs specifies hepatocytic differentiation[39,40]. The opposing roles of Wnt and Notch signals in cholangiocyte fate determination in the LSPCs are described below. The molecules responsible for differentiation of LSPCs into cholangiocytes are also discussed in this section (Figure 2).
When the LSPCs are cultured in Matrigel, they are likely to differentiate into cholangiocytes. Recently, the key stimulator has been found. Epimorphin/syntaxin 2 (EPM) is a highly conserved and very abundant protein involved in epithelial morphogenesis in various epithelial organs[41], and in the liver, it is exclusively expressed on the surface of hepatic stellate cells and myofibroblasts[42]. Biliary differentiation markers elevated by EPM include Yp, Cx43, aquaporin-1, CK19 and GGT[41]. Moreover, the signaling pathway of EPM was analyzed by focal adhesion kinase (FAK), extracellular regulated kinase 1/2 (ERK1/2) and RhoA. Most importantly, RhoA was found to be necessary for EPM-induced activation of FAK and ERK1/2 and bile duct formation. In addition, EPM regulated GGT IV and GGT V expression differentially, and this was possibly mediated by C/EBPβ. Taken together, these data demonstrated that EPM regulates biliary differentiation of LSPCs through effects on RhoA and C/EBPβ, implicating a dual aspect of this morphoregulator in bile duct epithelial morphogenesis. In another study, it was reported that EPM selectively induced bile duct formation through upregulation of CK19 expression and suppression of hepatocyte nuclear factor (HNF) 3α and HNF6[43]. These results demonstrate a new biophysical action of EPM in bile duct formation, during which the determination of LSPCs play a crucial role. MiRNAs could also initiate biliary differentiation of LSPCs. In the previous section, we described the requirement for miR-23b miRNAs in growing hepatocytes to repress bile duct genes and repress tumor growth factor (TGF) β signaling. There has also been another report providing evidence that miR-30 family miRNAs were required for complete bile duct formation to repress hepatocyte genes[44].
Bile ducts are formed only around the portal side, suggesting that region-specific signals induce cholangiocytes from LSPCs. Two signaling pathways, TGFβ/Activin[21,45] and Notch[46,47], are specifically activated in LSPCs near the portal vein. Although differentiation of LSPCs to cholangiocytes by TGFβ and Notch signaling occurs in mid-gestation, surprisingly, LSPCs can be induced to differentiate into cholangiocytes and form ectopic duct structures in the parenchyma upon Notch activation after birth[48]. That is, the Notch pathway plays an essential role in the morphogenesis of bile duct structures[49]. Indeed, conditional knockout of Recombination signal binding protein Jκ, an essential downstream signal component of the Notch receptor, results in a reduced number of cholangiocytes at ED 16.5, confirming a role for this signaling pathway in cholangiocyte cell fate specification[48]. In general, Notch signaling is likely to play the most important role in controlling biliary differentiation of LSPCs.
A study using an in vitro culture of FLSPCs has shown that activation of the Notch signaling pathway promotes LSPC differentiation into the cholangiocyte lineage by coordinating a network of LETFs including HNF1α/β, HNF4α and C/EBPα[46]. Among multiple Notch signaling components, Notch1, Notch2 and Jagged1, Hes1 are widely accepted as essential for promoting bile duct differentiation[49-51], while Notch3 and Jagged2 play key roles in hepatic differentiation[52]. Lacking Hes1, a target of the Notch signaling, ductal plate formation occurs normally, but the subsequent remodeling and tubular structure formation is completely blocked[53]. In humans, mutations in Jagged1, a ligand for the Notch receptors, are associated with Alagille syndrome, an autosomal dominant disorder characterized by multiple developmental defects including neonatal cholestasis caused by a paucity of IHBD[54-56]. In addition, another form of Alagille syndrome has been found to be caused by mutations in the Notch2 gene[57].
TGFβ is necessary for the formation of bile ducts[58]. The inhibition of TGFβ signaling allows LSPCs to undergo normal hepatocyte differentiation[45]. Wnt signaling is also involved in regulating biliary epithelial cell fate. The addition of Wnt3a in ex vivo culture experiments supports biliary epithelial cell differentiation of FLSPCs[59]. However, as to Wnt5a, a non-canonical Wnt ligand, in vitro differentiation assays showed that Wnt5a-mediated signaling in FLSPCs suppresses biliary differentiation through the activation of phosphorylated Calcium/calmodulin-dependent protein kinase II[60]. Similarly, in the absence of Wnt1 signaling, LSPCs failed to differentiate into hepatocytes and underwent atypical ductular hyperplasia, exhibiting epithelial metaplasia and mucin production[61,62]. Furthermore, the inhibition of β-catenin, a core component of canonical Wnt signaling, prevents LSPCs from expressing biliary markers[63].
In brief, Notch signaling promotes LSPCs differentiation into the biliary epithelial lineage and concurrently inhibits hepatic differentiation by reducing the expression of hepatic genes. In contrast, Wnt signaling is more likely to aid in promoting hepatic differentiation and repressing biliary differentiation. The unbalanced activation of Wnt and Notch signaling pathways influences the cell fates of LSPCs.
With regard to the molecular mechanisms involved in cholangiocytic differentiation, several transcription factors have been implicated, including Sal-like 4 (Sall4), T-box transcription factor 3 (Tbx3), the Onecut transcription factor HNF6 and HNF1β, HES1, FOXA2, FOXA3, forkhead Box (Fox) m1β (Foxm1β), and Hex[36,37,64-68]. Sall4 is expressed in LSPCs but not in mature liver cells. The expression level of Sall4 gradually falls during liver development. Sall4 has been shown to play a role in regulating the lineage commitment of LSPCs by inhibiting their differentiation into hepatocytes while driving differentiation toward cholangiocytes[36]. When bile duct-like structures were induced by collagen gel-embedded culture conditions, overexpression of Sall4 markedly augmented the size and number of CK19+ branching structures. These results suggest that Sall4 plays a crucial role in controlling the lineage commitment of LSPCs not only by inhibiting their differentiation into hepatocytes but also by driving their differentiation toward cholangiocytes[36]. Tbx3 also contributes to the hepato-biliary lineage decision[37,69]. Tbx3 functions to maintain expression of the hepatocyte transcription factors HNF4α and C/EBPα while suppressing expression of the cholangiocyte transcription factors HNF6 and HNF1β[69]. In addition, as a direct and critical target of HNF6, HNF1β shows a decisive effect in bile duct development[65].
Although organ-specific stem cells possess plasticity that permits differentiation along new lineages, production of endocrine pancreas and insulin-secreting beta cells from stem cells has not been fully demonstrated. The liver and pancreas share a common developmental origin, and a bipotential precursor cell population for these organs has been identified within the embryonic endoderm[70]. Consistent with these facts, many studies have demonstrated that LSPCs can be converted to insulin-producing cells by stable expression of pancreatic duodenal homeobox 1 (Pdx1) or its super-active form (Pdx1-VP16) or to functional pancreatic beta-cell-like cells, and/or islet-like cell clusters containing other pancreatic lineages under certain other conditions[71-74]. The most common condition under which LSPCs are induced to differentiate into insulin-producing cells is a high-glucose environment[75]. In addition, there are studies indicating an efficient chemical protocol for differentiating LSPCs into functional insulin-producing cells using small molecules, and they represent a promising LSPC-based treatment for diabetes mellitus. When ALSPCs were incubated with a combination of 5 mmol/L sodium butyrate and 1 nmol/L betacellulin, most of the cells were converted into morphologically beta cell-like cells. An immunoreactive pancreatic polypeptide, somatostatin, and insulin were detected in sodium butyrate and betacellulin-treated ALSPCs[72]. Based on induction by a combination of 5-aza-2’-deoxycytidine, trichostatin A, retinoic acid and a mix of insulin, transferrin and selenite, LSPCs could also trans-differentiate into beta-like cells[76]. Furthermore, transduction of pancreatic transcription factors, such as Pdx1, Neurogenin3, NeuroD and MafA, can induce the formation of ectopic islet-like cells and the production of insulin in ALSPCs[77,78]. Stepwise differentiation from LSPCs into functional insulin-secreting cells will identify key steps in beta-cell development and may yet prove useful for transplantation therapy for diabetic patients[79].
Stem cells have potential for therapy of liver diseases, but they may also be involved in the formation of cancer[80]. At present, it is widely accepted that cancer arises from the malignant transformation of stem cells[81,82] because these are the only cells that persist sufficiently long to acquire the required number of genetic changes. Specifically, LSPCs are hypothesized to be the precursors for a subset of liver cancer[83,84]. Presently, accumulating evidence supports the above notion as follows[85]: (1) similar signaling pathways may regulate self-renewal in LSPCs and liver cancer cells; and (2) liver cancer contains rare cells with stem cell-like properties, which may derive from malignant transformation of LSPCs. Herein, we propose that liver cancer stem like cells (LCSCs) might arise from LSPCs it would facilitate our understanding of stem-cell origin of liver cancer. It has been demonstrated that deletion of p53 from LSPCs is sufficient to induce tumor formation[86]. Recently, through loss expression of Tg737, You et al[87] successfully induced FLSPCs to malignantly transform into LCSCs. These LCSCs from LSPCs could generate liver cancer after transplantation into immuno-insufficient mice. In addition to gene manipulation, dysregulated miRNAs may also initiate malignant transformation. To find the possible target miRNAs, Liu et al[13] compared the miRNA profiles between LSPCs and LCSCs. As a result, Liu et al[13] found 78 miRNAs were dysregulated, including miR-200a (the most down-regulated miRNA in LCSCs) and miR-181 (the most greatly upregulated miRNA in LCSCs)[13]. After inhibition of miR-200a in LSPCs, Liu et al[13] found that cells displayed malignant properties such as unlimited proliferation and strong metastasis. A novel regulatory link between miR-181 and LCSCs was proven by a study from another group[88]. They found that miR-181 could induce LSPCs’ malignant transformation by directly targeting hepatic transcriptional regulators of differentiation (for example, caudal type homeobox transcription factor 2 and GATA binding protein 6) and an inhibitor of Wnt/beta-catenin signaling (nemo-like kinase).
Despite uncertainty surrounding the mechanism underlying the role of LSPCs in liver regeneration[89], there is great hope for the use of these cells in liver-based therapies[90]. First, LSPCs can be used for the treatment of inherited end-stage liver disease. Second, they can also serve as a source of cells for cell transplantation in acquired liver diseases such as acute failure due to toxic or viral injury. Third, because LSPCs can be expanded in vitro to a desired extent, they can be used to populate liver assist devices or artificial livers based on bioengineered matrices. Lastly, they can be used as targets for gene therapies in primary liver diseases or diseases where extra-hepatic manifestations arise from abnormal gene expression or defective protein production in the liver. Considering the strong proliferative potential and amenability for in vitro manipulation, LSPCs may be attractive candidates for liver disease treatment. In addition, LSPCs may be useful for cell therapy to treat diabetic patients, given their potential to be effectively reprogrammed toward pancreatic lineages[91]. Furthermore, the development of such protocols would reduce the likelihood of malignant transformation upon transplantation.
Although LSPCs are promising for the future use in many fields, the accurate control of cell fates of LSPCs is far from accomplishment. Thus, it is necessary to clear the mechanisms for LSPCs differentiation and build standardisation of the production of functional cholangiocytes from LSPCs. Here, we want to list several directions that may help to guide future research of LSPCs differentiation: (1) The knowledge of biliary development and liver regeneration can best provide detailed information for in vitro cholangiocytic differention of LSPCs. It is a good choice to thoroughly investigate the molecular basis of biliary development during the period from fetal liver to adult liver; (2) LSPCs react differently to stimulative materials at different stages. The dosage, timing, and combinations of materials should thus be fine-tuned according to the differentiated stage LSPCs located. Hence, it is important to figure out what state LSPCs are presented; (3) The molecular mechanism in each step of cholangiocytic differentiation from LSPCs is essential for cell-based therapies. Both positive and negative factors responsible for the initiation, progression and maturation of cholangiocytic differentiation should be specially considered; and (4) Although we divide the process of cholangiocytic differentiation into three stages, it is actually a continuous evolving process. That is to say, it should be kept in mind that many key factors may not only take effect in some stage of cholangiocytic differentiation from LSPCs.
| 1. | Flohr TR, Bonatti H, Brayman KL, Pruett TL. The use of stem cells in liver disease. Curr Opin Organ Transplant. 2009;14:64-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 30] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 2. | Oertel M, Menthena A, Chen YQ, Teisner B, Jensen CH, Shafritz DA. Purification of fetal liver stem/progenitor cells containing all the repopulation potential for normal adult rat liver. Gastroenterology. 2008;134:823-832. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 100] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 3. | Oertel M, Menthena A, Chen YQ, Shafritz DA. Properties of cryopreserved fetal liver stem/progenitor cells that exhibit long-term repopulation of the normal rat liver. Stem Cells. 2006;24:2244-2251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 29] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 4. | Baptista PM, Siddiqui MM, Lozier G, Rodriguez SR, Atala A, Soker S. The use of whole organ decellularization for the generation of a vascularized liver organoid. Hepatology. 2011;53:604-617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 493] [Cited by in RCA: 482] [Article Influence: 32.1] [Reference Citation Analysis (0)] |
| 5. | Lemaigre FP. Mechanisms of liver development: concepts for understanding liver disorders and design of novel therapies. Gastroenterology. 2009;137:62-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 180] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 6. | Tanaka M, Itoh T, Tanimizu N, Miyajima A. Liver stem/progenitor cells: their characteristics and regulatory mechanisms. J Biochem. 2011;149:231-239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 98] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 7. | Schmelzer E, Zhang L, Bruce A, Wauthier E, Ludlow J, Yao HL, Moss N, Melhem A, McClelland R, Turner W. Human hepatic stem cells from fetal and postnatal donors. J Exp Med. 2007;204:1973-1987. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 485] [Cited by in RCA: 444] [Article Influence: 23.4] [Reference Citation Analysis (13)] |
| 8. | Farber E. Similarities in the sequence of early histological changes induced in the liver of the rat by ethionine, 2-acetylamino-fluorene, and 3’-methyl-4-dimethylaminoazobenzene. Cancer Res. 1956;16:142-148. [PubMed] |
| 9. | Shafritz DA, Oertel M, Menthena A, Nierhoff D, Dabeva MD. Liver stem cells and prospects for liver reconstitution by transplanted cells. Hepatology. 2006;43:S89-S98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 99] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 10. | Bird TG, Lorenzini S, Forbes SJ. Activation of stem cells in hepatic diseases. Cell Tissue Res. 2008;331:283-300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 125] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 11. | Itoh , Tm Miyajima. Liver regeneration by stem/progenitor cells. Hepatology. 2013;Oct 1; Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 116] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 12. | Liu WH, Li R, Dou KF. Convenient and efficient enrichment of the CD133+ liver cells from rat fetal liver cells as a source of liver stem/progenitor cells. Stem Cell Rev. 2011;7:94-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 13. | Liu WH, Tao KS, You N, Liu ZC, Zhang HT, Dou KF. Differences in the properties and mirna expression profiles between side populations from hepatic cancer cells and normal liver cells. PLoS One. 2011;6:e23311. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 15. | Navarro-Alvarez N, Soto-Gutierrez A, Kobayashi N. Hepatic stem cells and liver development. Methods Mol Biol. 2010;640:181-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 16. | Moore KA, Lemischka IR. Stem cells and their niches. Science. 2006;311:1880-1885. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1139] [Cited by in RCA: 1072] [Article Influence: 53.6] [Reference Citation Analysis (0)] |
| 17. | Theise ND. Gastrointestinal stem cells. III. Emergent themes of liver stem cell biology: niche, quiescence, self-renewal, and plasticity. Am J Physiol Gastrointest Liver Physiol. 2006;290:G189-G193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 46] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 18. | He ZP, Tan WQ, Tang YF, Feng MF. Differentiation of putative hepatic stem cells derived from adult rats into mature hepatocytes in the presence of epidermal growth factor and hepatocyte growth factor. Differentiation. 2003;71:281-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 19. | Aurich I, Mueller LP, Aurich H, Luetzkendorf J, Tisljar K, Dollinger MM, Schormann W, Walldorf J, Hengstler JG, Fleig WE. Functional integration of hepatocytes derived from human mesenchymal stem cells into mouse livers. Gut. 2007;56:405-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 245] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 20. | Brulport M, Schormann W, Bauer A, Hermes M, Elsner C, Hammersen FJ, Beerheide W, Spitkovsky D, Härtig W, Nussler A. Fate of extrahepatic human stem and precursor cells after transplantation into mouse livers. Hepatology. 2007;46:861-870. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 73] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 21. | Antoniou A, Raynaud P, Cordi S, Zong Y, Tronche F, Stanger BZ, Jacquemin P, Pierreux CE, Clotman F, Lemaigre FP. Intrahepatic bile ducts develop according to a new mode of tubulogenesis regulated by the transcription factor SOX9. Gastroenterology. 2009;136:2325-2333. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 319] [Cited by in RCA: 291] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 22. | Holic N, Suzuki T, Corlu A, Couchie D, Chobert MN, Guguen-Guillouzo C, Laperche Y. Differential expression of the rat gamma-glutamyl transpeptidase gene promoters along with differentiation of hepatoblasts into biliary or hepatocytic lineage. Am J Pathol. 2000;157:537-548. [PubMed] |
| 23. | Kojima H, Nies AT, König J, Hagmann W, Spring H, Uemura M, Fukui H, Keppler D. Changes in the expression and localization of hepatocellular transporters and radixin in primary biliary cirrhosis. J Hepatol. 2003;39:693-702. [PubMed] |
| 24. | Renzi A, DeMorrow S, Onori P, Carpino G, Mancinelli R, Meng F, Venter J, White M, Franchitto A, Francis H. Modulation of the biliary expression of arylalkylamine N-acetyltransferase alters the autocrine proliferative responses of cholangiocytes in rats. Hepatology. 2013;57:1130-1141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 42] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 25. | Zhang H, Liu Z, Li R, Wang D, Liu W, Li J, Yu H, Zhang F, Dou K. Transplantation of embryonic small hepatocytes induces regeneration of injured liver in adult rat. Transplant Proc. 2009;41:3887-3892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 26. | Wang Y, Yao HL, Cui CB, Wauthier E, Barbier C, Costello MJ, Moss N, Yamauchi M, Sricholpech M, Gerber D. Paracrine signals from mesenchymal cell populations govern the expansion and differentiation of human hepatic stem cells to adult liver fates. Hepatology. 2010;52:1443-1454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 80] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 27. | Talbot NC, Caperna TJ. A feeder-cell independent subpopulation of the PICM-19 pig liver stem cell line capable of long-term growth and extensive expansion. Cytotechnology. 2013;Feb 9; Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 28. | Ishii T, Yasuchika K, Ikai I. Hepatic differentiation of embryonic stem cells by murine fetal liver mesenchymal cells. Methods Mol Biol. 2013;946:469-478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 29. | Talbot NC, Caperna TJ, Garrett WM. Growth and Development Symposium: Development, characterization, and use of a porcine epiblast-derived liver stem cell line: ARS-PICM-19. J Anim Sci. 2013;91:66-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 30. | Xi J, Wang Y, Zhang P, He L, Nan X, Yue W, Pei X. Human fetal liver stromal cells that overexpress bFGF support growth and maintenance of human embryonic stem cells. PLoS One. 2010;5:e14457. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 20] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 31. | Ader T, Norel R, Levoci L, Rogler LE. Transcriptional profiling implicates TGFbeta/BMP and Notch signaling pathways in ductular differentiation of fetal murine hepatoblasts. Mech Dev. 2006;123:177-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 48] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 32. | Tsai HA, Shen CN, Chang YC. Use of surface properties to control the growth and differentiation of mouse fetal liver stem/progenitor cell colonies. Biomacromolecules. 2012;13:3483-3493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 33. | Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125-1129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 896] [Cited by in RCA: 985] [Article Influence: 54.7] [Reference Citation Analysis (0)] |
| 34. | Rogler CE, Levoci L, Ader T, Massimi A, Tchaikovskaya T, Norel R, Rogler LE. MicroRNA-23b cluster microRNAs regulate transforming growth factor-beta/bone morphogenetic protein signaling and liver stem cell differentiation by targeting Smads. Hepatology. 2009;50:575-584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 163] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 35. | Meng F, Francis H, Glaser S, Han Y, DeMorrow S, Stokes A, Staloch D, Venter J, White M, Ueno Y. Role of stem cell factor and granulocyte colony-stimulating factor in remodeling during liver regeneration. Hepatology. 2012;55:209-221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 48] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 36. | Oikawa T, Kamiya A, Kakinuma S, Zeniya M, Nishinakamura R, Tajiri H, Nakauchi H. Sall4 regulates cell fate decision in fetal hepatic stem/progenitor cells. Gastroenterology. 2009;136:1000-1011. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 88] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 37. | Suzuki A, Sekiya S, Büscher D, Izpisúa Belmonte JC, Taniguchi H. Tbx3 controls the fate of hepatic progenitor cells in liver development by suppressing p19ARF expression. Development. 2008;135:1589-1595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 103] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 38. | Spee B, Carpino G, Schotanus BA, Katoonizadeh A, Vander Borght S, Gaudio E, Roskams T. Characterisation of the liver progenitor cell niche in liver diseases: potential involvement of Wnt and Notch signalling. Gut. 2010;59:247-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 160] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 39. | Boulter L, Govaere O, Bird TG, Radulescu S, Ramachandran P, Pellicoro A, Ridgway RA, Seo SS, Spee B, Van Rooijen N. Macrophage-derived Wnt opposes Notch signaling to specify hepatic progenitor cell fate in chronic liver disease. Nat Med. 2012;18:572-579. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 577] [Cited by in RCA: 622] [Article Influence: 44.4] [Reference Citation Analysis (0)] |
| 40. | Diehl AM. Neighborhood watch orchestrates liver regeneration. Nat Med. 2012;18:497-499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 41. | Jia Y, Yao H, Zhou J, Chen L, Zeng Q, Yuan H, Shi L, Nan X, Wang Y, Yue W. Role of epimorphin in bile duct formation of rat liver epithelial stem-like cells: involvement of small G protein RhoA and C/EBPβ. J Cell Physiol. 2011;226:2807-2816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 42. | Miura K, Nagai H, Ueno Y, Goto T, Mikami K, Nakane K, Yoneyama K, Watanabe D, Terada K, Sugiyama T. Epimorphin is involved in differentiation of rat hepatic stem-like cells through cell-cell contact. Biochem Biophys Res Commun. 2003;311:415-423. [PubMed] |
| 43. | Zhou J, Zhao L, Qin L, Wang J, Jia Y, Yao H, Sang C, Hu Q, Shi S, Nan X. Epimorphin regulates bile duct formation via effects on mitosis orientation in rat liver epithelial stem-like cells. PLoS One. 2010;5:e9732. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 44. | Hand NJ, Master ZR, Eauclaire SF, Weinblatt DE, Matthews RP, Friedman JR. The microRNA-30 family is required for vertebrate hepatobiliary development. Gastroenterology. 2009;136:1081-1090. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 107] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 45. | Clotman F, Jacquemin P, Plumb-Rudewiez N, Pierreux CE, Van der Smissen P, Dietz HC, Courtoy PJ, Rousseau GG, Lemaigre FP. Control of liver cell fate decision by a gradient of TGF beta signaling modulated by Onecut transcription factors. Genes Dev. 2005;19:1849-1854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 259] [Cited by in RCA: 278] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 46. | Tanimizu N, Miyajima A. Notch signaling controls hepatoblast differentiation by altering the expression of liver-enriched transcription factors. J Cell Sci. 2004;117:3165-3174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 207] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 47. | Sparks EE, Huppert KA, Brown MA, Washington MK, Huppert SS. Notch signaling regulates formation of the three-dimensional architecture of intrahepatic bile ducts in mice. Hepatology. 2010;51:1391-1400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 104] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 48. | Zong Y, Panikkar A, Xu J, Antoniou A, Raynaud P, Lemaigre F, Stanger BZ. Notch signaling controls liver development by regulating biliary differentiation. Development. 2009;136:1727-1739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 328] [Cited by in RCA: 354] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 49. | Lozier J, McCright B, Gridley T. Notch signaling regulates bile duct morphogenesis in mice. PLoS One. 2008;3:e1851. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 95] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 50. | Tchorz JS, Kinter J, Müller M, Tornillo L, Heim MH, Bettler B. Notch2 signaling promotes biliary epithelial cell fate specification and tubulogenesis during bile duct development in mice. Hepatology. 2009;50:871-879. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 97] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 51. | Geisler F, Nagl F, Mazur PK, Lee M, Zimber-Strobl U, Strobl LJ, Radtke F, Schmid RM, Siveke JT. Liver-specific inactivation of Notch2, but not Notch1, compromises intrahepatic bile duct development in mice. Hepatology. 2008;48:607-616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 165] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 52. | Zhang W, Li W, Liu B, Wang P, Li W, Zhang H. Efficient generation of functional hepatocyte-like cells from human fetal hepatic progenitor cells in vitro. J Cell Physiol. 2012;227:2051-2058. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 53. | Kodama Y, Hijikata M, Kageyama R, Shimotohno K, Chiba T. The role of notch signaling in the development of intrahepatic bile ducts. Gastroenterology. 2004;127:1775-1786. [PubMed] |
| 54. | Crosnier C, Attié-Bitach T, Encha-Razavi F, Audollent S, Soudy F, Hadchouel M, Meunier-Rotival M, Vekemans M. JAGGED1 gene expression during human embryogenesis elucidates the wide phenotypic spectrum of Alagille syndrome. Hepatology. 2000;32:574-581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 99] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 55. | Li L, Krantz ID, Deng Y, Genin A, Banta AB, Collins CC, Qi M, Trask BJ, Kuo WL, Cochran J. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat Genet. 1997;16:243-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 932] [Cited by in RCA: 889] [Article Influence: 30.7] [Reference Citation Analysis (0)] |
| 56. | Oda T, Elkahloun AG, Pike BL, Okajima K, Krantz ID, Genin A, Piccoli DA, Meltzer PS, Spinner NB, Collins FS. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat Genet. 1997;16:235-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 857] [Cited by in RCA: 771] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 57. | McDaniell R, Warthen DM, Sanchez-Lara PA, Pai A, Krantz ID, Piccoli DA, Spinner NB. NOTCH2 mutations cause Alagille syndrome, a heterogeneous disorder of the notch signaling pathway. Am J Hum Genet. 2006;79:169-173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 582] [Cited by in RCA: 510] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 58. | Clotman F, Lemaigre FP. Control of hepatic differentiation by activin/TGFbeta signaling. Cell Cycle. 2006;5:168-171. [PubMed] |
| 59. | Hussain SZ, Sneddon T, Tan X, Micsenyi A, Michalopoulos GK, Monga SP. Wnt impacts growth and differentiation in ex vivo liver development. Exp Cell Res. 2004;292:157-169. [PubMed] |
| 60. | Kiyohashi K, Kakinuma S, Kamiya A, Sakamoto N, Nitta S, Yamanaka H, Yoshino K, Fujiki J, Murakawa M, Kusano-Kitazume A. Wnt5a signaling mediates biliary differentiation of fetal hepatic stem/progenitor cells in mice. Hepatology. 2013;57:2502-2513. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 61. | Coleman WB, McCullough KD, Esch GL, Faris RA, Hixson DC, Smith GJ, Grisham JW. Evaluation of the differentiation potential of WB-F344 rat liver epithelial stem-like cells in vivo. Differentiation to hepatocytes after transplantation into dipeptidylpeptidase-IV-deficient rat liver. Am J Pathol. 1997;151:353-359. [PubMed] |
| 62. | Williams JM, Oh SH, Jorgensen M, Steiger N, Darwiche H, Shupe T, Petersen BE. The role of the Wnt family of secreted proteins in rat oval “stem” cell-based liver regeneration: Wnt1 drives differentiation. Am J Pathol. 2010;176:2732-2742. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 31] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 63. | Monga SP, Monga HK, Tan X, Mulé K, Pediaditakis P, Michalopoulos GK. Beta-catenin antisense studies in embryonic liver cultures: role in proliferation, apoptosis, and lineage specification. Gastroenterology. 2003;124:202-216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 169] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 64. | Clotman F, Lannoy VJ, Reber M, Cereghini S, Cassiman D, Jacquemin P, Roskams T, Rousseau GG, Lemaigre FP. The onecut transcription factor HNF6 is required for normal development of the biliary tract. Development. 2002;129:1819-1828. [PubMed] |
| 65. | Coffinier C, Gresh L, Fiette L, Tronche F, Schütz G, Babinet C, Pontoglio M, Yaniv M, Barra J. Bile system morphogenesis defects and liver dysfunction upon targeted deletion of HNF1beta. Development. 2002;129:1829-1838. [PubMed] |
| 66. | Hunter MP, Wilson CM, Jiang X, Cong R, Vasavada H, Kaestner KH, Bogue CW. The homeobox gene Hhex is essential for proper hepatoblast differentiation and bile duct morphogenesis. Dev Biol. 2007;308:355-367. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 136] [Cited by in RCA: 130] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 67. | Strazzabosco M. Foxa1 and Foxa2 regulate bile duct development in mice. J Hepatol. 2010;52:765-767. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 68. | Li Z, White P, Tuteja G, Rubins N, Sackett S, Kaestner KH. Foxa1 and Foxa2 regulate bile duct development in mice. J Clin Invest. 2009;119:1537-1545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 111] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 69. | Lüdtke TH, Christoffels VM, Petry M, Kispert A. Tbx3 promotes liver bud expansion during mouse development by suppression of cholangiocyte differentiation. Hepatology. 2009;49:969-978. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 87] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 70. | Deutsch G, Jung J, Zheng M, Lóra J, Zaret KS. A bipotential precursor population for pancreas and liver within the embryonic endoderm. Development. 2001;128:871-881. [PubMed] |
| 71. | Feng RQ, Du LY, Guo ZQ. In vitro cultivation and differentiation of fetal liver stem cells from mice. Cell Res. 2005;15:401-405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 72. | Yamada S, Terada K, Ueno Y, Sugiyama T, Seno M, Kojima I. Differentiation of adult hepatic stem-like cells into pancreatic endocrine cells. Cell Transplant. 2005;14:647-653. [PubMed] |
| 73. | Tang DQ, Lu S, Sun YP, Rodrigues E, Chou W, Yang C, Cao LZ, Chang LJ, Yang LJ. Reprogramming liver-stem WB cells into functional insulin-producing cells by persistent expression of Pdx1- and Pdx1-VP16 mediated by lentiviral vectors. Lab Invest. 2006;86:83-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 59] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 74. | Zalzman M, Gupta S, Giri RK, Berkovich I, Sappal BS, Karnieli O, Zern MA, Fleischer N, Efrat S. Reversal of hyperglycemia in mice by using human expandable insulin-producing cells differentiated from fetal liver progenitor cells. Proc Natl Acad Sci USA. 2003;100:7253-7258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 194] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 75. | Yang L, Li S, Hatch H, Ahrens K, Cornelius JG, Petersen BE, Peck AB. In vitro trans-differentiation of adult hepatic stem cells into pancreatic endocrine hormone-producing cells. Proc Natl Acad Sci USA. 2002;99:8078-8083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 374] [Cited by in RCA: 364] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 76. | Liu J, Liu Y, Wang H, Hao H, Han Q, Shen J, Shi J, Li C, Mu Y, Han W. Direct differentiation of hepatic stem-like WB cells into insulin-producing cells using small molecules. Sci Rep. 2013;3:1185. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 36] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 77. | Li H, Li X, Lam KS, Tam S, Xiao W, Xu R. Adeno-associated virus-mediated pancreatic and duodenal homeobox gene-1 expression enhanced differentiation of hepatic oval stem cells to insulin-producing cells in diabetic rats. J Biomed Sci. 2008;15:487-497. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 78. | Wang AY, Ehrhardt A, Xu H, Kay MA. Adenovirus transduction is required for the correction of diabetes using Pdx-1 or Neurogenin-3 in the liver. Mol Ther. 2007;15:255-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 84] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 79. | Zaret KS. Genetic programming of liver and pancreas progenitors: lessons for stem-cell differentiation. Nat Rev Genet. 2008;9:329-340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 210] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 80. | Rountree CB, Mishra L, Willenbring H. Stem cells in liver diseases and cancer: recent advances on the path to new therapies. Hepatology. 2012;55:298-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 78] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 81. | Teng IW, Hou PC, Lee KD, Chu PY, Yeh KT, Jin VX, Tseng MJ, Tsai SJ, Chang YS, Wu CS. Targeted methylation of two tumor suppressor genes is sufficient to transform mesenchymal stem cells into cancer stem/initiating cells. Cancer Res. 2011;71:4653-4663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 80] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 82. | Vander Griend DJ, Karthaus WL, Dalrymple S, Meeker A, DeMarzo AM, Isaacs JT. The role of CD133 in normal human prostate stem cells and malignant cancer-initiating cells. Cancer Res. 2008;68:9703-9711. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 168] [Cited by in RCA: 185] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 83. | Rountree CB, Ding W, Dang H, Vankirk C, Crooks GM. Isolation of CD133+ liver stem cells for clonal expansion. J Vis Exp. 2011;pii: 3183. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 84. | Cardinale V, Carpino G, Reid LM, Gaudio E, Alvaro D. Notch2 signaling and undifferentiated liver cancers: evidence of hepatic stem/progenitor cell origin. Hepatology. 2013;58:1188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 85. | Peng N, Li L, Cai X, Tan S, Wu T. Liver stem/progenitor cells in the canals of Hering: cellular origin of hepatocellular carcinoma with bile duct tumor thrombi? Stem Cell Rev. 2010;6:579-584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 11] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 86. | Katz SF, Lechel A, Obenauf AC, Begus-Nahrmann Y, Kraus JM, Hoffmann EM, Duda J, Eshraghi P, Hartmann D, Liss B. Disruption of Trp53 in livers of mice induces formation of carcinomas with bilineal differentiation. Gastroenterology. 2012;142:1229-1239.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 74] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 87. | You N, Liu W, Zhong X, Ji R, Zhang M, You H, Dou K, Tao K. Tg737 inhibition results in malignant transformation in fetal liver stem/progenitor cells by promoting cell-cycle progression and differentiation arrest. Mol Carcinog. 2012;51:659-673. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 88. | Ji J, Yamashita T, Budhu A, Forgues M, Jia HL, Li C, Deng C, Wauthier E, Reid LM, Ye QH. Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells. Hepatology. 2009;50:472-480. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 423] [Cited by in RCA: 439] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 89. | Sancho-Bru P. [Therapeutic possibilities of stem cells in the treatment of liver diseases]. Gastroenterol Hepatol. 2011;34:701-710. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 90. | Heng BC, Yu H, Yin Y, Lim SG, Cao T. Factors influencing stem cell differentiation into the hepatic lineage in vitro. J Gastroenterol Hepatol. 2005;20:975-987. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 91. | Russo FP, Parola M. Stem and progenitor cells in liver regeneration and repair. Cytotherapy. 2011;13:135-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 37] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
P- Reviewer: Larentzakis A S- Editor: Cui XM L- Editor: A E- Editor: Wu HL