Published online Aug 21, 2013. doi: 10.3748/wjg.v19.i31.5067
Revised: June 6, 2013
Accepted: June 19, 2013
Published online: August 21, 2013
Processing time: 209 Days and 20.7 Hours
AIM: To study the differential protein profile in serum of hepatitis B patients.
METHODS: Serum samples were obtained from patients with chronic hepatitis B who were receiving peginterferon alfa-2b. The serum samples were subjected to albumin depletion and analyzed by two-dimensional gel electrophoresis (2-DE). Differentially expressed protein spots were identified by electrospray ionization-quadrupole time-of-flight mass spectrometry. Alpha-2-HS-glycoprotein, complement component C3c and CD5 antigen were further analyzed by an enzyme-linked immunosorbent assay and immunonephelometry.
RESULTS: Nineteen patients with HBeAg-positive chronic hepatitis B (CHB) were studied. These patients were followed for at least 1 year after treatment and were classified according to their treatment response: responders (n = 9) and non-responders (n = 10). 2-DE and MS/MS analysis were performed to compare the serum proteins before initiating peginterferon alfa-2b. From the quantitative analysis of the 2-D gel, 7 proteins were detected between the two groups at different levels before treatment. Among these potential candidates, serum levels of alpha-2-HS-glycoprotein, complement component C3c and CD5 antigen-like precursor were further analyzed. In the validation phase, 23 subjects, 9 sustained responders and 14 non-responders, were recruited. Interestingly, the levels of alpha-2-HS-glycoprotein and complement component C3c were elevated in the serum of the non-responders compared to the responders.
CONCLUSION: Serum alpha-2-HS-glycoprotein and complement component C3c may be potential serum biomarkers in predicting the treatment response of peginterferon alfa-2b in patients with CHB prior to treatment.
Core tip: Serum proteins serve as non-invasive biomarkers for several diseases. This is the first report on the potential use of common protein levels in the serum of chronic hepatitis B (CHB) patients to predict treatment responsiveness to peginterferon alfa-2b. We identified 2 potential serum biomarkers, alpha-2-HS-glycoprotein and complement component C3c, that can be used to predict treatment outcome in patients with CHB receiving peginterferon alfa-2b. The identification of these biomarkers prior to treatment is preferable in order to avoid systemic side effects due to interferon therapy.
- Citation: Kuakarn S, SomParn P, Tangkijvanich P, Mahachai V, Thongboonkerd V, Hirankarn N. Serum proteins in chronic hepatitis B patients treated with peginterferon alfa-2b. World J Gastroenterol 2013; 19(31): 5067-5075
- URL: https://www.wjgnet.com/1007-9327/full/v19/i31/5067.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i31.5067
One of the most common health care problems encountered worldwide is hepatitis B virus (HBV) infection which can progress to liver fibrosis, liver cirrhosis and liver cancer (also known as hepatocellular carcinoma). Treatment for chronic hepatitis B includes immunomodulatory agents and antiviral drugs such as nucleoside or nucleotide analogs (NAs). NAs inhibit the replication process of the virus by inhibiting its DNA polymerase[1], whereas immunomodulating therapy mainly includes treatment with interferon-α and pegylated interferon-α. The treatment duration, eradication and lack of drug resistance strains make type 1 interferons ideal for the treatment of chronic HBV[2]. Unfortunately, only 30% of patients will respond to treatment with interferon type 1[3]. The reason for this is because other factors, including the virus and the host, can significantly influence the treatment outcome. Viral factors such as the level of HBV DNA, HBV genotype, levels of hepatitis B surface antigen and hepatitis B core antigen (HBeAg), and HBV viral mutants can affect the outcome of therapy[4,5]. Other factors such as low levels of viral HBV DNA, higher levels of alanine aminotransferase (ALT), older age, being female, and naive to interferon therapy have been shown to be significantly associated with sustainable virological response among HBeAg-positive chronic hepatitis B (CHB) patients[6]. In HBeAg-negative patients, younger age, being female, having high levels of ALT and low levels of viral HBV DNA have been associated with sustainable virological response[7]. In addition, genetic host factors such as human leukocyte antigen (HLA) class II (HLA-DRB1*14 allele), presence of polymorphism A (MxA)-88, levels of interleukin-10 and interleukin-12 have been proposed to predict the patient’s treatment response after therapy[8-10]. The use of biomarkers is invaluable in predicting treatment response as well as being cost-effective in managing patients with CHB.
Various biomarkers for hepatocellular carcinoma (HCC)[11-13], HBV inflammation, HBV liver cirrhosis[14,15], and hepatitis C virus treatment response[16] have been investigated using proteomics, however, there are no predictive data for the treatment of CHB. One report from MA Hui and colleagues identified a potential serum biomarker for detecting changes after treatment, but was not able to predict the treatment outcome prior to treatment[17]. In the present study, albumin and immunoglobulin G (IgG) depleted serum was subjected to 2-dimensional gel electrophoresis and mass spectrometry. We identified 2 potential serum biomarkers, alpha-2-HS-glycoprotein and complement component C3c, and found that they can be used to predict treatment outcome in patients with CHB receiving peginterferon alfa-2b.
Serum samples were obtained from patients with CHB who were followed at the King Chulalongkorn Memorial Hospital. All patients received peginterferon alfa-2b (1.5 mg/kg per week) subcutaneously for 48 wk and their responses to this treatment were assessed. These patients were followed for at least 1 year after treatment and were classified as sustained responders or non-responders. Sustained virological response among HBeAg-positive patients was characterized by undetectable HBeAg, detectable anti-HBe (HBeAg seroconversion) and HBV viral load < 2000 IU/mL 48 wk after treatment[18]. Patients without sustained virological response were classified as non-responders. Serum samples were obtained before initiating peginterferon alfa-2b treatment and at 24 wk after treatment.
Nineteen patients with HBeAg-positive CHB (9 sustained responders and 10 non-responders) were included in the proteomic study before initiating treatment. After 24 wk of treatment, 6 patients (3 sustained responders and 3 non-responders) were included.
Another 23 subjects, 9 sustained responders and 14 non-responders, were enrolled in the validation phase of the proteomic study using an enzyme-linked immunosorbent assay (ELISA) and immunonephelometry.
All studies were approved by the Institutional Review Board, Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand. Informed consent forms were collected from all patients from both phases of the study before any of the procedures were initiated.
Albumin and IgG were removed from the patient’s serum using the ProteoPrep Blue Albumin Depletion Kit (Sigma: PROTBA) according to the company’s protocol. Protein concentrations were measured by the Bio-Rad Bradford total protein assay kit (Biorad Laboratories, Inc., Redmond, WA, United States)[19] using bovine serum albumin (BSA) as the standard curve.
The Immobiline Dry strip (pH 4-7, length 7 cm, Amersham Biosciences, Uppsala, Sweden) was rehydrated with 150 μg protein in 125 μL rehydration buffer containing 9 mol urea, 2% CHAPS, 0.002% w/v bromophenol blue, 0.8% (w/v) DTT, 1% IPG buffer for 14 h at room temperature. Iso-electric focusing (IEF) was performed by IPG ph or IEF apparatus (Amersham Biosciences, Uppsala, Sweden) with a total of 8000 Vhrs. The strip was then equilibrated in equilibration buffer containing 6 mol/L urea, 30% glycerol, 2% SDS, 0.002% bromophenol blue and 50 mmol/L Tris-HCl (pH = 8.8) with 135 mmol/L DTT for 15 min followed by incubation, but replacing with 130 mmol/L iodoacetamide for 15 min. Next, the equilibrated strips were placed on the sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) composed of 12.5% acrylamide and sealed with 0.5% (w/v) agarose. The SDS-PAGE was run on constant electric field, 15 mA per gel, using the SE 260 Mini-Vertical Units (GE Healthcare, Uppsala, Sweden) until the bromophenol blue tracking dye reached the bottom of the gel. Protein spots were stained with Coomassie Brilliant Blue G-250 stain[20]. The stained gels were scanned with an ImageMaster scanner (GE Healthcare; Uppsala, Sweden). Intensity analysis was carried out using the software, Image Master 2D Platinum (GE Healthcare, Uppsala, Sweden).
Differentially expressed protein spots were excised from the 2-DE gels and subjected to in-gel tryptic digestion according to the method modified from Katayama et al[21]. The gel pieces were destained with 50% methanol and 50 mmol ammonium bicarbonate, and dehydrated with 100% acetonitrile (ACN). The gel pieces were reduced and alkylated in 10 mmol/L of DTT and 100 mmol/L iodoacetamide at room temperature for 1 h. They were then dehydrated twice with 100% ACN for 5 min after alkylation. The gel pieces were subsequently digested in 10 µL trypsin (modified porcine trypsin, sequencing grade, Promega, Madison, WI, United States) solution (20 ng in 10 mmol/L ammonium bicarbonate in 50% ACN) and incubated at room temperature overnight. The peptides were extracted twice by adding 30 µL of solution containing 50% ACN and 0.1% formic acid. The extracted solutions were dried in a heat box at 40 °C and kept at -80 °C for further analysis by mass spectrometry. Prior to mass spectrometry analysis, the peptide mixtures were reconstituted in 10 µL of 0.1 % formic acid.
Peptide mixtures were analyzed by ultra-performance liquid chromatography (UPLC) (Ultimate 3000, Dionex, united states) coupled to the micrOTOF-Q II™ ESI-Qq-TOF mass spectrometer (Bruker Daltonics, Germany) equipped with an online nanoESI source. The peptide mixture was injected onto a µ-precolumn cartridge (C18 PepMap; 300 µmol/L × 5 mm; 5 µmol/L particle size) composed of peptides, concentrated and then directly separated using a PepMap100 C18 analytical column (5 µm particle size, with 100 Å pore size). The mobile phase was run for each sample using a linear gradient of 10%-55% of 80% ACN in high performance liquid chromatography (HPLC) water for 30 min, with a hold of 15 min at 90% of 80% ACN in HPLC water, followed by a step to 10% of 80% ACN in HPLC water, hold of 20 min. The Q-TOF instrument was operated in positive ionization mode to switch automatically between MS and MS/MS acquisition. The precursor ion (MS) and fragmentation ion (MS/MS) with a mass range were 400-1600 m/z and 50-3000 m/z, respectively. The source parameters were as follows: capillary 2.0 kV, dry gas 0.3 L/min and dry temperature at 150 °C . The MS and MS/MS spectrometry data were processed using data analysis software (Bruker Daltonics, Germany) and searched against the NCBInr database using the MASCOT search engine. The parameters were identified using the following set up: species-homo sapiens; enzyme-trypsin; allowed up to 1 missed cleavage; fixed modification-carbamidomethylation on cystenine; variable modification-oxidation on methionines. The peptide mass tolerance and fragment mass tolerance were set at 1.2 Da and 0.6 Da, respectively[15]. A probability-based Mowse score of more than 43 was considered significant (P < 0.05).
Validation of the proteomic study was performed in a different population (n = 23) composed of 9 sustained responders and 14 non-responders. ELISA was performed according to the company’s protocol using the Alpha 2 HS Glycoprotein Human ELISA kit (Abcam, Cambridge, United Kingdom) and human CD5 antigen like (CD5L) ELISA kit (Cusabio Biotech., Ltd., China). Complement component C3c was further validated using immunonephelometry and the BN ProSpec system (Siemens Healthcare Diagnostics Products GmbH, Germany).
SPSS version 17.0 (SPSS Inc., Chicago, IL, United States) was used for all statistical analyses. The values of the intensities of the spots are shown as the mean ± SE. Independent sample t test was used to evaluate the baseline characteristics of the patients and compare the intensity data of each matched protein spot between the sustained responders and non-responders; the P value cut-off for the independent sample t test was 0.05. Mann-Whitney U test was used to evaluate the different protein expressions between the two groups 24 wk after treatment. For the Mann-Whitney U test, any proteins identified with P < 0.05 were considered significant. The independent sample t test was performed during the validation phase to compare the different levels of alpha-2-HS-glycoprotein. The Mann-Whitney U test was also performed in the validation phase to compare the different levels of complement component C3c and CD5 antigen like proteins. The Pearson correlation was carried out on age and each protein expression value to determine if age had an influence on the expression of the proteins.
Basic characteristics of the patients are shown in Table 1. The number of men and women, levels of serum ALT and HBV DNA, and presence of HBeAg were comparable between the two groups. However, non-responders were older than the sustained responders.
| Sustained virological | Non-responders | P value | |
| responders (n = 9) | (n = 10) | ||
| Age (yr) | 29.67 ± 8.29 | 36.90 ± 6.40 | 0.047 |
| Sex (male:female) | 7:2 | 8:2 | 0.912 |
| ALT level (U/L) | 103.88 ± 105.04 | 130.50 ± 75.77 | 0.609 |
| HBV DNA (copies/mL) | (11.07 ± 8.52) × 106 | (14.35 ± 8.44) × 106 | 0.511 |
| HBeAg | Positive | Positive | NS |
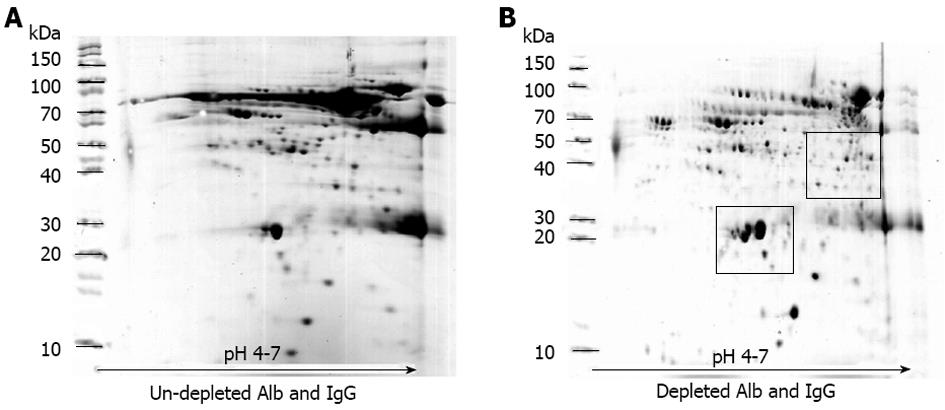
Before performing electrophoresis on the collected serum, the efficiency of the ProteoPrep Blue Albumin Depletion Kit was determined. Figure 1 shows the two representative maps of the serum samples (chronic HBV infection) before and after treatment with the ProteoPrep Blue Albumin Depletion Kit. In the untreated sample, levels of albumin and IgG in serum were approximately 60%-70% and 10%-20%, respectively (Figure 1A). When an equal quantity of protein was pre-treated with ProteoPrep Blue Albumin Depletion Kit, the resolution of the 2D-gels dramatically improved and several spots of other less abundant proteins became visible (Figure 1B).
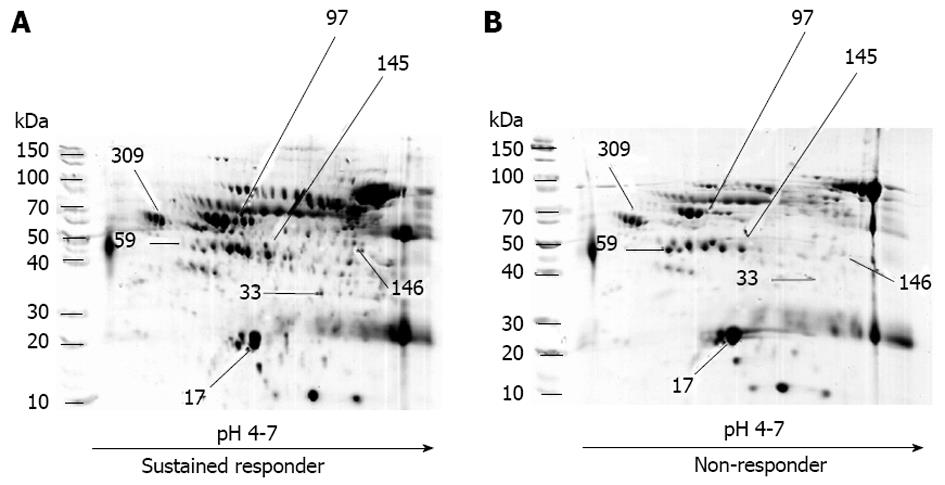
The results of the protein separation by 2D-gel electrophoresis, gel digestion and protein identification by LC/MS/MS are shown in Figure 2. Seven protein spots were detected with various intensities in the patients (Table 2 and Figure 2). Four proteins were significantly detected among the sustained responders: (1) chain A, alpha-1-antitrypsin; (2) albumin, isoform CRA-b; (3) CD5 antigen-like precursor; and (4) albumin. Three proteins were significantly detected among the non-responders: (1) chain A, crystal structure of lipid-free human apolipoprotein A-I; (2) chain C, human complement component C3c; and (3) alpha-2-HS-glycoprotein. Since the sustained responders and non-responders showed significant age differences, the Pearson correlation was used for age and each protein expression value. No significant correlation between age and protein expression was observed; therefore, it is unlikely that age affected protein expression in this study. The identified proteins have the following functions: alpha-1-antitrypsin is a protease inhibitor, serum albumin is a transport and binding protein, alpha-2-HS-glycoprotein is an acute phase response protein, CD5 antigen precursor and complement component C3c are immune protection proteins, and the human apolipoprotein A-I has a role in lipid metabolism
| Spot | Protein | NCBI ID | MS score | %cov | pI | MW | Relative intensity (mean ± SE) | SVR/NR | P value | |
| SVR | NR | |||||||||
| Protease inhibitor | ||||||||||
| 97 | Chain A, alpha-1-antitrypsin | Gi|157831596 | 399 | 64 | 5.37 | 44.28 | 0.2764 ± 0.0327 | 0.4275 ± 0.0609 | 2.34 | 0.038 |
| Transport protein and protein binding | ||||||||||
| 33 | Albumin, isoform | Gi|119626065 | 539 | 38 | 6.96 | 61.12 | 0.0330 ± 0.0121 | 0.0764 ± 0.0144 | 2.31 | 0.033 |
| 146 | Albumin | Gi|332356380 | 243 | 41 | 5.73 | 68.48 | 0.0602 ± 0.0182 | 0.1516 ± 0.0333 | 2.52 | 0.024 |
| Acute phase protein | ||||||||||
| 309 | Alpha-2-HS-glycoprotein | Gi|112910 | 507 | 40 | 5.43 | 40.09 | 0.7165 ± 0.0238 | 0.4782 ± 0.0851 | 0.67 | 0.012 |
| Immunity protection | ||||||||||
| 59 | Chain C, human complement C3c | Gi|78101271 | 513 | 70 | 4.79 | 40.20 | 0.3969 ± 0.0391 | 0.2675 ± 0.0403 | 0.67 | 0.034 |
| 145 | CD5 antigen-like precursor | Gi|5174411 | 443 | 67 | 5.28 | 39.60 | 0.1086 ± 0.0192 | 0.1903 ± 0.0197 | 1.75 | 0.009 |
| Lipid metabolism | ||||||||||
| 17 | Chain A, crystal structure of lipid-free human apoliprotein A-I | Gi|90108664 | 1347 | 78 | 5.27 | 28.06 | 9.4993 ± 0.5044 | 6.0364 ± 1.0047 | 0.64 | 0.005 |
In the validation phase, nine and 14 patients with sustained virological response and nonresponders were enrolled, respectively. The sex ratio, levels of serum and HBV DNA, and HBeAg were not significantly different, but there was a significant difference in age between the groups (Table 3).
| Sustained virological responders | Non-responders | P value | |
| Validation phase | (n = 9) | (n = 14) | |
| Age (yr) | 29.56 ± 2.78 | 37.71 ± 1.97 | 0.023 |
| Sex (male:female) | 7:2 | 11:3 | 0.966 |
| ALT level (U/L) | 106.88 ± 36.26 | 130.50 ± 30.93 | 0.644 |
| HBV DNA (copies/mL) | (11.06 ± 2.84) × 106 | (13.12 ± 2.45) × 106 | 0.593 |
| HBeAg | Positive | Positive | NS |
| 24 wk | (n = 3) | (n = 3) | |
| Age (yr) | 29.33 ± 6.89 | 35.67 ± 6.06 | NS |
| Sex (male:female) | Male | Male | NS |
| ALT level (U/L) | 52.33 ± 13.78 | 253.67 ± 187.74 | NS |
| HBV DNA (copies/mL) | (6.70 ± 6.65) × 106 | (13.65 ± 6.35) × 106 | NS |
| HBeAg | Positive | Positive | NS |
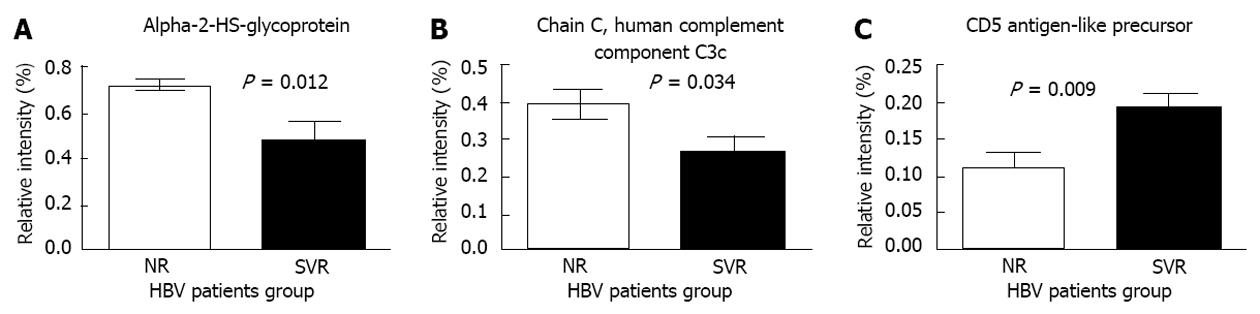
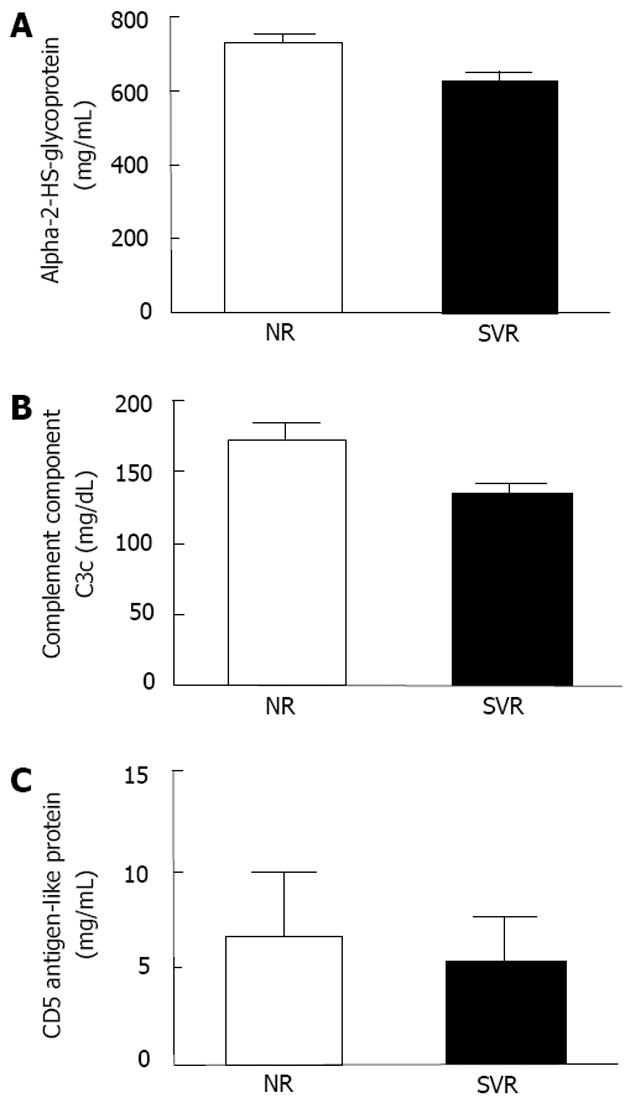
We selected 3 proteins (alpha-2-HS-glycoprotein, complement component C3c, and CD5 antigen-like proteins) that were significantly different between the 2 groups, and have functions related to immune response for further validation (Figure 3). According to the ELISA results, the serum levels of alpha-2-HS-glycoprotein were significantly elevated among the non-responders when compared to the sustained responders at baseline (Figure 4A). Similarly, the immunonephelometry results showed that serum levels of complement component C3c were significantly elevated in the non-responders when compared to the sustained responders at baseline (Figure 4B), however, the serum levels of CD5 antigen like proteins were comparable between the two groups (Figure 4C). No significant correlation between age and protein expression was observed.
A total of 6 patients, divided into 2 groups, were included in the analysis 24 wk post-treatment. The clinical data were not significantly different between the 2 groups (Table 3). The samples were separated by 2D-gel electrophoresis, and identified by LC/MS/MS. Thirteen protein spots were significantly changed at 24 wk after treatment among the patients in the sustained response group, whereas 6 were found in the non-responders (Table 4). Interestingly, all 13 proteins in the sustained response group which were increased at 24 wk were composed of cholesterol metabolites, proteins from the acute phase response, protease inhibitors, transport proteins and immune protection. Of these, alpha-2-HS-glycoprotein was higher than the baseline level in the responder group. In the non-responder group, levels of proapoliprotein, chain A of human antithrombin III complex and alpha-1-B-glycoprotein were higher than the baseline level, whereas levels of albumin and alpha-2-HS-glycoprotein decreased 24 wk post-treatment.
| Spot | Protein Name | NCBI ID | MW/pI | No. of match peptide | MS score | Fold change | ∆ relative intensity | Biological function |
| Sustained virological response | ||||||||
| 38 | Proapolipoprotein | Gi|178775 | 28.94/5.50 | 63 | 798 | ↑ 5.17 | 0.1525a | Cholesterolmetabolism |
| 26 | Chain A, the structure of pentameric human serum amyloid P | Gi|576259 | 23.36/6.12 | 12 | 174 | ↑ 6.65 | 0.0783a | Acute phase protein |
| 86 | Alpha-2-HS-glycoprotein | Gi|112910 | 40.098/5.43 | 32 | 494 | ↑ 2.39 | 0.3773a | |
| 309 | Alpha-2-HS-glycoprotein | Gi|112910 | 40.09/5.43 | 29 | 507 | ↑ 2.04 | 0.4987a | |
| 171 | Chain A, the intact and cleaved III complex as a model for serpin-proteinase interaction | Gi|999513 | 49.35/5.95 | 37 | 396 | ↑ 9.66 | 0.1481b | |
| 173 | Chain A, the intact and cleaved III complex as a model for serpin-proteinase interaction | Gi|999513 | 49.35/5.95 | 26 | 210 | ↑ 8.01 | 0.0694a | Protease inhibitor |
| 149 | Serotransferin precursor | Gi|4557871 | 79.28/6.81 | 58 | 650 | ↑ 3.43 | 0.1165a | |
| 201 | PRO2619 | Gi|11493459 | 58.51/5.96 | 36 | 278 | ↑ 4.95 | 0.0594a | Transport protein and proteinbinding |
| 207 | Albumin | Gi|332356380 | 68.48/5.73 | 48 | 781 | ↑ 6.97 | 0.1653a | |
| 280 | Albumin | Gi|332356380 | 68.48/5.73 | 31 | 201 | ↑ 7.33 | 0.1206a | |
| 202 | CD5 antigen-like precursor | Gi|5174411 | 39.60/5.28 | 11 | 70 | ↑ absent at baseline | 0.0348a | |
| 229 | Ig J-chain | Gi|532598 | 16.04/4.62 | 3 | 42 | ↑ absent at baseline | 0.0923b | Immunity protection |
| 299 | Immunoglobulin light | Gi|218783338 | 24.16/5.95 | 26 | 747 | ↑ absent at base line | 0.1709b | |
| Non-responder | ||||||||
| 20 | Proapolipoprotein | Gi|178775 | 28.94/5.45 | 63 | 798 | ↑ absent at base line | 0.2383b | Cholesterol metabolism |
| 171 | Chain A, the intact and cleaved III complex as a model for serpine-proteinase interaction | Gi|999513 | 49.35/5.95 | 37 | 396 | ↑ 2.39 | 0.0896a | Acute phase protein |
| 4 | Albumin isoform | Gi|119626066 | 27.67/6.39 | 20 | 660 | ↓ 0.04 | -0.2832a | Transport protein and protein binding |
| 264 | Serum albumin | Gi|62113341 | 71.09/5.85 | 21 | 115 | ↓ absent at 24 wk | -0.1196a | |
| 123 | Alpha-1-B-glycoprotein | Gi|69990 | 52.47/5.69 | 27 | 247 | ↑ 1.38 | 0.1610a | Serum protein |
| 310 | Alpha-2-HS-glycoprotein | Gi|112910 | 40.09/5.43 | 30 | 475 | ↓ 0.46 | -0.4119a | Acute phase protein |
The proteomic approach is usually used to analyze protein expression. It can be used with various specimens such as tissue, serum, plasma or body fluids. For this study, serum samples were used to identify potential biomarkers that can be further applied to predict the outcome of CHB therapy. Serum was selected because the collection process is non-invasive and proteins from the liver are secreted into the serum. Therefore, serum is an ideal specimen to screen for new proteins or biomarkers. In addition, serum proteomics can be used to detect post-translational modified proteins. 2-DE was used to separate and identify the proteins between 10-200 kDa. The high sensitivity and high throughput of mass spectrometry has resulted in the detection of several new biomarkers in ovarian cancer, prostate cancer, breast cancer and hepatocellular carcinoma. However, it should be noted that 2-DE does have limitations. 2-DE cannot detect low abundant proteins because high abundant proteins such as albumin and IgG can suppress the detection of low abundant proteins. To overcome this obstacle, albumin and IgG were removed from the serum samples before electrophoresis using the ProteoPrep Blue Albumin Depletion Kit. Albumin (-45 mg/mL) and IgG (-10 mg/mL) are the two major protein components of serum, representing 60%-70% and 10%-20% of the total serum protein, respectively[22]. When the high abundant proteins were depleted from the serum, this allowed the low abundant protein spots to become visible. However, it is also possible to miss certain low abundant proteins when the high abundant proteins are removed[23]. The reason for this is that low abundant proteins sometimes bind themselves to high abundant proteins. Hence, only albumin and IgG were removed in order to prevent the loss of other important proteins[24]. As expected, in the untreated sample, albumin dominated the gel, obscuring signals from other less abundant proteins. When an equal quantity of protein was pre-treated with the ProteoPrep Blue Albumin Depletion Kit, the resolution of the 2D-gels significantly improved. This process cannot completely eliminate all albumin, but can eliminate enough to allow several protein spots to be clearly visible. Using these techniques, a total of seven protein spots were found to be differentially expressed in the serum of CHB patients, 9 sustained responders and 10 non-responders, before starting peginterferon alfa-2b treatment. Interestingly, four of the proteins were higher in the sustained responders. Of these proteins, only 3 were involved in immune responses: alpha-2-HS-glycoprotein, complement component C3c and CD5 antigen-like proteins. We further validated the proteomic results using the sensitive ELISA and nephelometry assay. In the validation phase of the study, the serum levels of alpha-2-HS-glycoprotein and complement component C3c were significantly higher in the non-responders when compared to the sustained responders. However, the level of CD5 antigen-like protein was not statistically different between the two groups. Since the band intensity of this protein was lower than the other proteins and the level was detected only in the validation phase of the study when the abundant proteins were not depleted, it is possible that the level of CD5 antigen-like protein may have altered when albumin and IgG were removed.
Based on these findings, the authors believe that alpha-2-HS-glycoprotein and complement component C3c are potential serum pre-treatment biomarkers in predicting sustained virological response in CHB patients treated with peginterferon alfa-2b. In this study, the levels of serum alpha-2-HS-glycoprotein were elevated in the non-responders before treatment initiation, whereas the levels were much lower in the sustained responders. In addition, we performed a subsequent serum proteomic analysis in a subset of samples at 24 wk after peginterferon alfa-2b treatment. Serum alpha-2-HS-glycoprotein was also up-regulated in the sustained virological responders, but downregulated in the non-responders. This finding is consistent with the results reported earlier by Ma et al[17]. However, a previous study only detected changes after treatment, but was not able to predict the treatment outcome prior to treatment. The identification of biomarkers prior to treatment is preferable in this case to avoid systemic side effects due to interferon therapy.
Alpha-2-HS-glycoprotein is a high abundant protein produced by the liver and osteoblasts and is usually concentrated in the mineralized tissues. This protein belongs to the cystatin super family[25]. Several studies have reported various levels of alpha-2-HS-glycoprotein in patients with liver diseases[26-28]. Some have found low levels of serum alpha-2-HS-glycoprotein in patients with acute drug-induced hepatitis, alcoholic hepatitis, chronic autoimmune hepatitis, primary biliary cirrhosis, fatty liver and HCC[26,28]. Aside from its multiple functions and ability to affect metabolic diseases, tumor and sepsis[29,30], Dai et al[31] reported that it was an independent marker for liver injury and a prognostic marker for CHB. They suggested that the protein may decrease liver inflammation by inhibiting the release of inflammatory factors from activated peripheral blood mononuclear cells[31]. Similarly, Patel et al[32] found reduced levels of alpha-2-HS-glycoprotein in chronic hepatitis C patients identified as sustained virological responders before initiating treatment. The reduced levels of alpha-2-HS-glycoprotein in chronic hepatitis C[32] and B patients identified as sustained virological responders before treatment suggest that this protein may potentially be used as a biomarker in predicting the outcome of peginterferon alfa-2b treatment.
Another protein detected in this study was complement component C3c which is the degradation product of complement C3. This protein is important for both the acquired and innate immune systems, especially against microbial infection as it switches the cellular responses from cell death to opsonization[33]. 80%-90% of complement is produced by hepatocytes and is associated with the pathogenesis of many chronic human diseases such as autoimmune diseases, complement-mediated hemolytic anemia, vascular and liver diseases[33,34]. Interestingly, low levels of C3 fragments were detected in HBV and HCC patients compared to normal, healthy controls[11]. According to the results obtained from the screening and validation phases of this study, the expression and levels of complement component C3c were elevated in non-responders before treatment. Thus, complement component C3c may be a possible biomarker in predicting the outcome of peginterferon alfa-2b treatment in patients with CHB.
In conclusion, alpha-2-HS-glycoprotein and complement component C3c proteins were elevated in non-responders indicating that these proteins may be potential biomarkers in predicting the response to peginterferon alfa-2b treatment in patients with CHB prior to treatment. However, external validation is needed to assess the clinical applicability of these two proteins as predictors of anti-HBV treatment outcome.
The treatment duration, eradication and lack of drug resistance strains make type 1 interferons ideal for the treatment of chronic hepatitis B virus. Unfortunately, only 30% of patients will respond to treatment with interferon type 1. To date, there is no effective predictor of interferon responsiveness.
The authors identified 2 potential serum biomarkers, alpha-2-HS-glycoprotein and complement component C3c, and determined that they can be used to predict treatment outcome in patients with chronic hepatitis B (CHB) receiving peginterferon alfa-2b.
Serum proteins serve as non-invasive biomarkers for several diseases. This is the first report of the potential use of common protein levels in the serum of CHB patients to predict responsiveness to peginterferon alfa before treatment. The identification of biomarkers prior to treatment is preferable in this case to avoid systemic side effects due to interferon therapy.
These 2 serum proteins can be added to the list of potential biomarkers to predict responsiveness to peginterferon alfa. However, these findings require further validated using a larger sample size and other independent studies.
The authors examined the differential protein profile in serum of hepatitis B patients before treatment with peginteferon-2b. From the quantitative analysis of the 2D-gel and validation step using Enzyme-linked immunosorbent assay and immunonephelometry, serum levels of alpha-2-HS-glycoprotein and complement component C3c were elevated in the serum of the non-responders compared to the responders. This indicated that these 2 serum proteins may be potential serum biomarkers in predicting the treatment response in CHB patients on peginterferon alfa-2b therapy.
| 1. | Nguyen T, Locarnini S. Hepatitis: Monitoring drug therapy for hepatitis B--a global challenge? Nat Rev Gastroenterol Hepatol. 2009;6:565-567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 2. | Liu CJ, Kao JH. Pegylated interferons for the treatment of chronic hepatitis B. Recent Pat Antiinfect Drug Discov. 2006;1:85-94. [PubMed] |
| 3. | Leemans W, Janssen HL, de Man R. Future prospectives for the management of chronic hepatitis B. World J Gastroenterol. 2007;13:2554-2567. [PubMed] |
| 4. | Kao JH. Role of viral factors in the natural course and therapy of chronic hepatitis B. Hepatol Int. 2007;1:415-430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 79] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 5. | Wong GL, Chan HL. Predictors of treatment response in chronic hepatitis B. Drugs. 2009;69:2167-2177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 6. | Buster EH, Hansen BE, Lau GK, Piratvisuth T, Zeuzem S, Steyerberg EW, Janssen HL. Factors that predict response of patients with hepatitis B e antigen-positive chronic hepatitis B to peginterferon-alfa. Gastroenterology. 2009;137:2002-2009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 301] [Cited by in RCA: 330] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 7. | Bonino F, Marcellin P, Lau GK, Hadziyannis S, Jin R, Piratvisuth T, Germanidis G, Yurdaydin C, Diago M, Gurel S. Predicting response to peginterferon alpha-2a, lamivudine and the two combined for HBeAg-negative chronic hepatitis B. Gut. 2007;56:699-705. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 212] [Cited by in RCA: 215] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 8. | Huang YX, Ma LN, Chen XY, Li Z, Huang YL, Shen CL, Ma B. Genetic polymorphisms of MxA protein and eIF-2a-reg2 and their responses to interferon treatment in patients with chronic hepatitis B. Zhonghua Ganzangbing Zazhi. 2007;15:187-191. [PubMed] |
| 9. | Han YN, Yang JL, Zheng SG, Tang Q, Zhu W. Relationship of human leukocyte antigen class II genes with the susceptibility to hepatitis B virus infection and the response to interferon in HBV-infected patients. World J Gastroenterol. 2005;11:5721-5724. [PubMed] |
| 10. | Wu JF, Wu TC, Chen CH, Ni YH, Chen HL, Hsu HY, Chang MH. Serum levels of interleukin-10 and interleukin-12 predict early, spontaneous hepatitis B virus e antigen seroconversion. Gastroenterology. 2010;138:165-72.e1-3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 98] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 11. | Steel LF, Shumpert D, Trotter M, Seeholzer SH, Evans AA, London WT, Dwek R, Block TM. A strategy for the comparative analysis of serum proteomes for the discovery of biomarkers for hepatocellular carcinoma. Proteomics. 2003;3:601-609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 119] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 12. | Feng JT, Liu YK, Song HY, Dai Z, Qin LX, Almofti MR, Fang CY, Lu HJ, Yang PY, Tang ZY. Heat-shock protein 27: a potential biomarker for hepatocellular carcinoma identified by serum proteome analysis. Proteomics. 2005;5:4581-4588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 157] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 13. | Yang MH, Tyan YC, Jong SB, Huang YF, Liao PC, Wang MC. Identification of human hepatocellular carcinoma-related proteins by proteomic approaches. Anal Bioanal Chem. 2007;388:637-643. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 14. | He QY, Lau GK, Zhou Y, Yuen ST, Lin MC, Kung HF, Chiu JF. Serum biomarkers of hepatitis B virus infected liver inflammation: a proteomic study. Proteomics. 2003;3:666-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 125] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 15. | Marrocco C, Rinalducci S, Mohamadkhani A, D’Amici GM, Zolla L. Plasma gelsolin protein: a candidate biomarker for hepatitis B-associated liver cirrhosis identified by proteomic approach. Blood Transfus. 2010;8 Suppl 3:s105-s112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 16. | Paradis V, Asselah T, Dargere D, Ripault MP, Martinot M, Boyer N, Valla D, Marcellin P, Bedossa P. Serum proteome to predict virologic response in patients with hepatitis C treated by pegylated interferon plus ribavirin. Gastroenterology. 2006;130:2189-2197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 17. | Ma H, Wang J, Guo F, Wei L. α-2-HS-glycoprotein is a potential marker predicting hepatitis B e antigen seroconversion in patients with chronic hepatitis B during treatment with pegylated interferon alpha-2b. Sci China Life Sci. 2011;54:39-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 18. | Tangkijvanich P, Komolmit P, Mahachai V, Sa-nguanmoo P, Theamboonlers A, Poovorawan Y. Low pretreatment serum HBsAg level and viral mutations as predictors of response to PEG-interferon alpha-2b therapy in chronic hepatitis B. J Clin Virol. 2009;46:117-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 47] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 19. | Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248-254. [PubMed] |
| 20. | Candiano G, Bruschi M, Musante L, Santucci L, Ghiggeri GM, Carnemolla B, Orecchia P, Zardi L, Righetti PG. Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis. 2004;25:1327-1333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1437] [Cited by in RCA: 1457] [Article Influence: 69.4] [Reference Citation Analysis (0)] |
| 21. | Katayama H, Nagasu T, Oda Y. Improvement of in-gel digestion protocol for peptide mass fingerprinting by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2001;15:1416-1421. [PubMed] |
| 22. | Rengarajan K, de Smet MD, Wiggert B. Removal of albumin from multiple human serum samples. Biotechniques. 1996;20:30-32. [PubMed] |
| 23. | Lu Y, Liu J, Lin C, Wang H, Jiang Y, Wang J, Yang P, He F. Peroxiredoxin 2: a potential biomarker for early diagnosis of hepatitis B virus related liver fibrosis identified by proteomic analysis of the plasma. BMC Gastroenterol. 2010;10:115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 37] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 24. | Echan LA, Tang HY, Ali-Khan N, Lee K, Speicher DW. Depletion of multiple high-abundance proteins improves protein profiling capacities of human serum and plasma. Proteomics. 2005;5:3292-3303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 270] [Cited by in RCA: 254] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 25. | Dziegielewska KM, Møllgård K, Reynolds ML, Saunders NR. A fetuin-related glycoprotein (alpha 2HS) in human embryonic and fetal development. Cell Tissue Res. 1987;248:33-41. [PubMed] |
| 26. | Yilmaz Y, Yonal O, Kurt R, Ari F, Oral AY, Celikel CA, Korkmaz S, Ulukaya E, Ozdogan O, Imeryuz N. Serum fetuin A/α2HS-glycoprotein levels in patients with non-alcoholic fatty liver disease: relation with liver fibrosis. Ann Clin Biochem. 2010;47:549-553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 51] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 27. | Kalabay L, Gráf L, Vörös K, Jakab L, Benko Z, Telegdy L, Fekete B, Prohászka Z, Füst G. Human serum fetuin A/alpha2HS-glycoprotein level is associated with long-term survival in patients with alcoholic liver cirrhosis, comparison with the Child-Pugh and MELD scores. BMC Gastroenterol. 2007;7:15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 28. | Kalabay L, Jakab L, Prohászka Z, Füst G, Benkö Z, Telegdy L, Lörincz Z, Závodszky P, Arnaud P, Fekete B. Human fetuin/alpha2HS-glycoprotein level as a novel indicator of liver cell function and short-term mortality in patients with liver cirrhosis and liver cancer. Eur J Gastroenterol Hepatol. 2002;14:389-394. [PubMed] |
| 29. | Sari I, Kebapcilar L, Taylan A, Bilgir O, Kozaci DL, Yildiz Y, Yuksel A, Gunay N, Akkoc N. Fetuin-A and interleukin-18 levels in ankylosing spondylitis. Int J Rheum Dis. 2010;13:75-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 30. | Ix JH, Sharma K. Mechanisms linking obesity, chronic kidney disease, and fatty liver disease: the roles of fetuin-A, adiponectin, and AMPK. J Am Soc Nephrol. 2010;21:406-412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 243] [Cited by in RCA: 273] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 31. | Dai XH, Zhang P, Xiao MF, Zhou RR, Zhang BX, Hu GS, Huang ZB, Fan XG. Protective Role of α2HS-Glycoprotein in HBV-Associated Liver Failure. Int J Mol Sci. 2011;12:3846-3856. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 32. | Patel K, Lucas JE, Thompson JW, Dubois LG, Tillmann HL, Thompson AJ, Uzarski D, Califf RM, Moseley MA, Ginsburg GS. High predictive accuracy of an unbiased proteomic profile for sustained virologic response in chronic hepatitis C patients. Hepatology. 2011;53:1809-1818. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 31] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 33. | Qin X, Gao B. The complement system in liver diseases. Cell Mol Immunol. 2006;3:333-340. [PubMed] |
P- Reviewer Genaro MSD S- Editor Huang XZ L- Editor Webster JR E- Editor Li JY