Published online May 28, 2013. doi: 10.3748/wjg.v19.i20.3090
Revised: March 7, 2013
Accepted: April 9, 2013
Published online: May 28, 2013
Processing time: 133 Days and 11.7 Hours
AIM: To develop a real-time polymerase chain reaction (PCR) method to detect and quantify Campylobacter jejuni (C. jejuni) from stool specimens.
METHODS: Primers and a probe for real-time PCR were designed based on the specific DNA sequence of the hipO gene in C. jejuni. The specificity of the primers and probe were tested against a set of Campylobacter spp. and other enteric pathogens. The optimal PCR conditions were determined by testing a series of conditions with standard a C. jejuni template. The detection limits were obtained using purified DNA from bacterial culture and extracted DNA from the stool specimen. Two hundred and forty-two specimens were analyzed for the presence of C. jejuni by direct bacterial culture and real-time PCR.
RESULTS: The optimal PCR system was determined using reference DNA templates, 1 × uracil-DNA glycosylase, 3.5 mmol/L MgCl2, 1.25 U platinum Taq polymerase, 0.4 mmol/L PCR nucleotide mix, 0.48 μmol/L of each primer, 0.2 μmol/L of probe and 2 μL of DNA template in a final volume of 25 μL. The PCR reaction was carried as follows: 95 °C for 4 min, followed by 45 cycles of 10 s at 95 °C and 30 s at 59 °C. The detection limit was 4.3 CFU/mL using purified DNA from bacterial culture and 103 CFU/g using DNA from stool specimens. Twenty (8.3%, 20/242) C. jejuni strains were isolated from bacterial culture, while 41 (16.9%, 41/242) samples were found to be positive by real-time PCR. DNA sequencing of the PCR product indicated the presence of C. jejuni in the specimen. One mixed infection of C. jejuni and Salmonella was detected in one specimen and the PCR test for this specimen was positive.
CONCLUSION: The sensitivity of detection of C. jejuni from stool specimens was much higher using this PCR assay than using the direct culture method.
Core tip: In the present study, we developed an effective real-time polymerase chain reaction method based on the specific DNA sequence of the hipO gene in Campylobacter jejuni (C. jejuni). The detection limit of this assay is 4.3 CFU/mL. A study of 242 clinical stool specimens from diarrheal patients indicated that this method is more sensitive than direct bacterial culture for the identification of C. jejuni from stool specimens.
-
Citation: Zhang MJ, Qiao B, Xu XB, Zhang JZ. Development and application of a real-time polymerase chain reaction method for
Campylobacter jejuni detection. World J Gastroenterol 2013; 19(20): 3090-3095 - URL: https://www.wjgnet.com/1007-9327/full/v19/i20/3090.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i20.3090
Campylobacter spp., Salmonella spp., Yersinia spp., Shigella spp., and Escherichia coli (E. coli) O157 are the leading causes of human bacterial gastroenteritis worldwide[1]. Campylobacter jejuni (C. jejuni) is the main species of Campylobacter that affects humans[2,3]. Campylobacter infections have been investigated and followed with considerable interest since the early 1980s in China, and there have been numerous reports on C. jejuni infection in patients with diarrhea. However, reports and the frequency of isolation of C. jejuni from diarrheal patients have decreased since the late 1990s. Improvement in sanitary conditions may explain this decline in numbers; however, the difficulty in detecting this fastidious pathogen might be another contributor to the decreased number of reports and frequency of isolation. Our recent pilot study revealed that the ratio of isolation of this pathogen from stool specimens of diarrheal patients was considerably different among surveillance spots in different laboratories (2%-15%, unpublished). Sensitive and accurate detection of this pathogen is important, both for the treatment of patients and for prompt epidemiological investigation.
With the development of genomic DNA sequencing, online databases, and bioinformatic analyses, nucleic acid-based methods, particularly polymerase chain reaction (PCR) methods, have become promising tools for the rapid, reliable, and sensitive detection and diagnosis of pathogenic infection[4-8]. In this study, we developed a real-time PCR assay to detect C. jejuni. We then combined this PCR assay with DNA sequence analysis and compared this method with the direct culture method to detect C. jejuni in stool specimens obtained from patients with diarrhea. The results obtained in this study provide a proof of concept for PCR-based detection of C. jejuni infection in patients with diarrhea, may lead to the development of a pre-screening approach for isolating C. jejuni.
Two hundred and forty-two stool specimens were collected from diarrheal patients (with ages ranging from 6 to 72 years) in the outpatient facilities of 11 hospitals. The specimens were transferred to a laboratory within 4 h for bacterial culture. The remainder of the samples was frozen at -80 °C for DNA extraction. A sterile cotton swab was twisted in each stool sample and streaked directly on selective Skirrow agar plates consisting of Columbia agar base (CM331, Oxoid, Basingstoke, United Kingdom), selective supplement (SR0069, Oxoid), and 5% defibrinated sheep blood. The plates were incubated in a microaerobic atmosphere (5% O2, 10% CO2 and 85% N2) at 42 °C for 48 h. Suspected colonies were picked and identified by gram staining, oxidase and catalase tests, and hippurate hydrolysis analysis, according to our previous report[9]. All specimens were also examined for the presence of Shigella spp., Salmonella spp., Yersinia spp. and E. coli O157 by culture on xylose lysine deoxycholate agar, deoxycholate hydrogen sulfide lactose agar, cefsulodin-irgasan-novobiocin agar, and sorbitol MacConkey agar, respectively. Suspected colonies were identified biochemically using API 20E strips, and the cultures were confirmed up to the species level by further serotype or PCR analysis, as previously described[10,11].
Reference genomic DNA was extracted from the Campylobacter isolates cultured in this study by using a QIAamp DNA mini kit (Qiagen, Düsseldorf, Germany). DNA templates from other reference pathogens were gifts from Drs. Huaiqi Jing and Biao Kan. The reference DNA templates are shown in Table 1.
| Bacteria | Strain name |
| Campylobacter jejuni | NCTC11168 |
| Campylobacter jejuni | 81116 |
| Campylobacter jejuni | ATCC33560 |
| Campylobacter jejuni sub doylei | ATCC49349 |
| Campylobacter jejuni | ICDCCJ07001 |
| Campylobacter coli | ATCC33559 |
| Campylobacter coli | WHO-10.2 |
| Campylobacter fetus | ATCC27374 |
| Campylobacter lari | ATCC35221 |
| Escherichia coli O157:H7 | Isolate-1 |
| Enterotoxic Escherichia coli | ETEC10407 |
| Enteroaggregative Escherichia coli | EAECO42 |
| Enteroinvasive Escherichia Coli | EIEC44825 |
| Enteropathogenic Escherichia coli | Isolate-2 |
| Salmonella Serovar typhi | CT18 |
| Typhoid-paratyphoid A | Isolate-3 |
| Salmonella typhimurium | Isolate-4 |
| Cholera O1 | Isolate-5 |
| Shigella | Isolate-6 |
| Salmonella enteritidis | Isolate-7 |
| Yersinia enterocolitica O3 | 52203 |
| Vibrio parahemolyticus | Isolate-8 |
| Enterococcus | Isolate-9 |
| Listeria monocytogenes | Isolate-10 |
Specific DNA fragments of the hipO gene were compared using BLASTn and Vector NTI suite 6.0 software. The primers and probe sets were designed and synthesized by Shanghai Huirui Biotechnology Co., Ltd. The sequences of the primers and probe used in this study were: hipO-F, 5’-CGGATAGTTATAGTATTGAAGTTATTGG-3’, hipO-R, 5’-GAAGCAGCATAAATAGGATCTTTTG-3’, and hipO-P, 5’-FAM-TTCTGGAGCACTTCCATGACCACC-BHQ1-3’.
The optimal PCR conditions were determined by testing a series of conditions with standard C. jejuni template. The standard PCR curve was constructed using genomic DNA from C. jejuni strain NCTC11168. The detection limits of this assay from pure culture and stool specimens were determined using 10-fold dilutions of quantified reference C. jejuni genomic DNA templates (1 × 100 to 1 × 106 CFU/mL) and the same amount of bacteria inoculated into stool specimens that were previously confirmed to contain no Campylobacter. The specificity of this assay was verified using genomic DNA from other enteric pathogens. The C. jejuni-inoculated samples were also cultured to compare the detection limits.
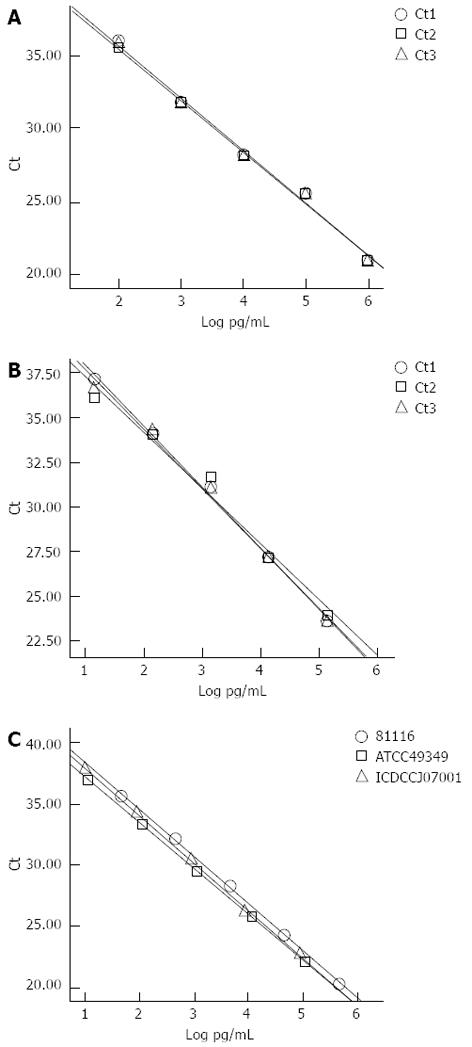
To evaluate the reproducibility of the assay, five serial dilutions with three replicates per concentration were performed for two separate C. jejuni genomic DNA templates (NCTC11168 and ATCC33560). The serial dilutions were also performed on three different C. jejuni strains (81116, ATCC49349 and ICDCCJ07001) simultaneously. The curves were constructed on the basis of the log pg/mL of the genomic DNA and threshold cycle (Ct). Reproducibility was assessed using the SD of the Ct value and the SD of the slope of each dilution curve, respectively.
After direct culture, the stool specimens were stored at -80 °C until DNA extraction using a QIAamp DNA stool mini kit (Qiagen), which was performed in accordance with the manufacturer’s instructions. Conventional PCR with the universal primers targeting conserved bacterial 16s rRNA gene sequences was carried out to evaluate the validity of the template used in this study. Real-time PCR was performed in a 25-μL volume with 2 μL of purified DNA obtained directly from stool specimens. All the samples with Ct < 35 were considered positive. Each of the positive PCR products, except for the ones from the culture-positive specimens, was confirmed with conventional PCR using the same primers, and the PCR products were purified and sequenced. Online sequence BLAST was performed using the NCBI BLASTN suite (http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&BLAST_PROGRAMS=megaBlast&PAGE_TYPE=BlastSearch&SHOW_DEFAULTS=on&LINK_LOC=blasthome).
Twenty C. jejuni (8.3%, 20/242) isolates, 34 (14%, 34/242) Salmonella isolates, and one (0.4%, 1/242) Shigella isolate were obtained by direct plating of the 242 stool specimens. Salmonella was the most frequently isolated pathogenic genus from the stool specimens and C. jejuni was the second most frequently isolated. No Yersinia or E. coli O157 isolates were found. In another laboratory, serotyping methods were used to further identify Salmonella and Shigella colonies.
The optimal PCR system was determined using reference DNA templates, 1 × uracil-DNA glycosylase, 3.5 mmol/L MgCl2, 1.25 U platinum Taq polymerase, 0.4 mmol/L PCR nucleotide mix, 0.48 μmol/L of each primer, 0.2 μmol/L of probe, and 2 μL of DNA template in a final volume of 25 μL run with the following parameters: 95 °C for 4 min, followed by 45 cycles of 10 s at 95 °C and 30 s at 59 °C. Fluorescence signals were measured at the end of the annealing step of every cycle. PCR products were verified using DNA electrophoresis through 2% agarose gels. The size of the PCR product was consistent with the expected size (85 bp) in each case. The specificity of the primers and probe were tested against Campylobacter spp. and other enteric pathogens. Only C. jejuni showed positive reactions. All the other templates gave negative results, including other Campylobacter spp.
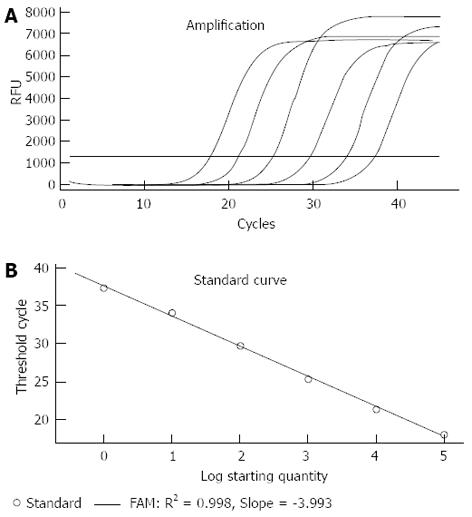
Serial dilutions from 100 to 106 CFU/ml of the target template were subjected to the real-time PCR assay (Figure 1A). The standard curve based on the dilutions of genomic DNA showed a linear relationship between Log CFU/mL and threshold cycles (Ct; Figure 1B). Copy numbers could be obtained by the following equation using the standard curve: Y(Ct) = -3.993X (log CFU/mL) + 37.51 (R2 = 0.998). When Ct = 35, the detection limit of the PCR assay for pure culture was approximately 4.3 CFU/mL. However, when Ct = 35, the detection limit for C. jejuni in the stool specimens was 103 CFU/g, and we could not obtain a linear relationship between log CFU/g and Ct using serial bacterial inoculation in different stool specimens. The curves constructed from the serial dilution replicates of the individual C. jejuni templates and those generated from the three different C. jejuni templates are shown in Figure 2. The variation of the Ct value and the slopes of the curves indicated that this assay could be stably reproduced.
The detection limit from both bacterial culture and real-time PCR (with Ct < 35) for the inoculated stool samples was 103 CFU/g; therefore, a Ct value of ≤ 35 was considered positive for the real-time PCR assay in this study. Forty-one (16.9%, 41/242) samples were found to be positive by the real-time PCR method. All culture-positive specimens showed positive PCR results (Ct < 34). DNA sequencing of the positive PCR products for the culture-negative samples indicated the presence of C. jejuni. The samples containing Salmonella and Shigella isolates tested negative by real-time PCR (Ct > 40), except for one sample that contained both C. jejuni and Salmonella (Ct = 27).
C. jejuni is one of the major causes of food borne disease worldwide[12]. It is an important pathogen of acute bacterial diarrhea in humans and has been associated with the development of Guillain-Barré syndrome, a post-infectious polyneuropathy[13,14]. Routine detection of Campylobacter species in the clinical setting is currently based on culture-based detection and subsequent phenotypic identification. Its detection is difficult because of its special growth requirements, low infectious doses, and potential for entering a viable, but not cultivable, state[15,16]. The traditional “gold standard” diagnostic methods for Campylobacter infection include bacterial culture and culture-based biochemical tests. These methods are both time-consuming and laborious. In addition to the fastidious culture requirements, the discrimination and differentiation of Campylobacter spp. are complicated and error prone. For example, distinguishing C. jejuni from C. coli is usually done on the basis of the biochemistry test for hippurate hydrolysis. Only C. jejuni gives a positive reaction, but previous studies have found that 10% of C. jejuni isolates failed to hydrolyze hippurate under laboratory conditions[17,18]. The accurate identification of Campylobacter up to species level is an essential prerequisite for many epidemiological studies.
In the present study, we developed a real-time PCR assay based on the specific DNA fragment of the hipO gene from C. jejuni, which could identify C. jejuni directly and differentiate it from other pathogens, especially from other Campylobacter spp., in stool specimens from diarrheal patients. The specificity of the PCR system was verified with 27 reference enteric pathogens (Table 1). The detection limit of this PCR assay is less effective for stool samples than for pure culture. Loss of template during DNA extraction or inhibitors present in the stool specimens could reduce the efficiency[19]. Two hundred and forty-two stool specimens from patients with diarrhea were tested by the direct culture and by the PCR assay. All the culture-positive samples gave positive PCR results and samples containing other pathogens tested negative by PCR (Ct > 40). The DNA sequence of the positive PCR products indicated the presence of C. jejuni in the specimens. The sensitivity of this PCR assay (100%, 41/41) was much higher than that of direct bacterial culture (49%, 20/41). The result from the mixed infection sample indicated that this PCR assay is considerably accurate. The results presented in this study contrast with those reported by certain previous studies, in which PCR results were found to have the same or less sensitivity compared with the direct culture method for the detection of C. jejuni from stool specimens[20,21]. However, Maher et al[22] used a DNA probe-based PCR assay and were able to identify 94% (17/18) and 32% (35/109) of Campylobacter infections from culture-positive and culture-negative stool specimens, respectively. Furthermore, Bessède et al[23] proved the weakness of the culture methods compared with PCR and immunoenzymatic methods of detection of Campylobacter. Notably, many aspects of the culture-based method, such as the choice of the selective media used for culture, need to be improved[24]. Although the sensitivity of the PCR system was high in this study, false-negative PCR results may still occur because of the presence of inherent inhibitors or because of very low numbers of organisms.
Interestingly, when C. jejuni were inoculated into stool specimens, the detection limit by the PCR assay was 103 CFU/g. The PCR detection limit from stool samples was similar to that reported in a previous study, but the detection limit of the direct culture methods mostly depended on the culture methods used, particularly the choice of plating medium, which may influence the efficiency of C. jejuni isolation during direct plating of fecal samples[25,26]. Instant plating of the inoculated samples in this study might increase the laboratory isolation capacity. Meanwhile, the non-uniformity of the clinical stool specimens in terms of physical matter, target organisms, the associated fecal flora and transportation status would result in variability of the culture results. In this study, although we used selective medium under specific microaerobic atmosphere conditions, some samples were contaminated with Proteus bacilli, and the suspected colonies could not be picked because of the rapid spread of the contaminants. Proteus bacilli may be one of the agents that reduced culture sensitivity, and the use of cotton swabs rather than inoculating loops for streaking on plates might have increased this contamination. In this study, we used QIAamp DNA stool mini kits to extract the DNA; the QIAamp kit has the highest sensitivity and requires the least manipulation of template DNA prior to PCR. More research is required to develop an in-house procedure for DNA extraction from different specimens and for optimizing the extraction process to increase the desired yield.
In conclusion, the real-time PCR assay developed in this study is sufficiently sensitive and accurate for C. jejuni detection from stool specimens. Although the PCR results do not permit its use as the standard method of diagnosing C. jejuni infection in the clinical setting, this newly developed method will benefit large-scale epidemiology investigations and pre-screening for bacterial isolation.
Campylobacter jejuni (C. jejuni) is a major food-borne pathogen. In addition to enteritis, C. jejuni infection can also cause Guillain-Barré syndrome, an autoimmune neuropathy in humans. The “gold standard” diagnostic methods for C. jejuni infection include bacterial culture and culture-based biochemical tests. Primary in vitro culture of C. jejuni is difficult because of its special growth requirements, low infectious doses, and potential for entering a viable, but not cultivable, state. In addition to its fastidious culture conditions, current methods for discrimination and differentiation of Campylobacter spp. are complicated and fallible. Specific DNA detection, particularly real-time polymerase chain reaction (PCR), is a promising tool for the rapid, reliable, and more sensitive detection and diagnosis of C. jejuni infection. In this study, the authors developed and validated a real-time PCR assay method for the detection of C. jejuni from stool specimens.
C. jejuni is a fastidious organism and requires microaerophilic conditions for in vitro culture. Nucleic acid-based methods for the detection of C. jejuni infection are research hotspots, particularly PCR methods, which are promising tools for the rapid, reliable, and more sensitive detection of C. jejuni for laboratory-based diagnosis.
In the present study, the authors developed a sensitive and specific real-time PCR assay using a DNA fragment of the hipO gene in C. jejuni, which can directly differentiate C. jejuni from other pathogens and other Campylobacter spp. from stool specimens obtained from diarrheal patients.
The real-time PCR assay developed in this study is sensitive and accurate for the detection of C. jejuni from stool specimens. This method will benefit both large-scale epidemiology investigations and the clinical diagnosis of C. jejuni infection in humans.
This is an excellent and well-written manuscript. The authors are to be congratulated on developing and validating their own in-house PCR-based technique to detect C. jejuni in stool specimens. The hipO gene is specific to C. jejuni; therefore, by definition, this is a highly specific test.
| 1. | Centers for Disease Control and Prevention (CDC). Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food--selected sites, United States, 2003. MMWR Morb Mortal Wkly Rep. 2004;53:338-343. [PubMed] |
| 2. | Allos BM. Campylobacter jejuni Infections: update on emerging issues and trends. Clin Infect Dis. 2001;32:1201-1206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 803] [Cited by in RCA: 792] [Article Influence: 31.7] [Reference Citation Analysis (0)] |
| 3. | Dasti JI, Tareen AM, Lugert R, Zautner AE, Gross U. Campylobacter jejuni: a brief overview on pathogenicity-associated factors and disease-mediating mechanisms. Int J Med Microbiol. 2010;300:205-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 273] [Cited by in RCA: 272] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 4. | Fredricks DN, Relman DA. Application of polymerase chain reaction to the diagnosis of infectious diseases. Clin Infect Dis. 1999;29:475-486; quiz 487-488. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 133] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 5. | Holland JL, Louie L, Simor AE, Louie M. PCR detection of Escherichia coli O157: H7 directly from stools: evaluation of commercial extraction methods for purifying fecal DNA. J Clin Microbiol. 2000;38:4108-4113. [PubMed] |
| 6. | Josefsen MH, Löfström C, Hansen TB, Christensen LS, Olsen JE, Hoorfar J. Rapid quantification of viable Campylobacter bacteria on chicken carcasses, using real-time PCR and propidium monoazide treatment, as a tool for quantitative risk assessment. Appl Environ Microbiol. 2010;76:5097-5104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 131] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 7. | Liu J, Gratz J, Maro A, Kumburu H, Kibiki G, Taniuchi M, Howlader AM, Sobuz SU, Haque R, Talukder KA. Simultaneous detection of six diarrhea-causing bacterial pathogens with an in-house PCR-luminex assay. J Clin Microbiol. 2012;50:98-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 63] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 8. | Wiemer D, Loderstaedt U, von Wulffen H, Priesnitz S, Fischer M, Tannich E, Hagen RM. Real-time multiplex PCR for simultaneous detection of Campylobacter jejuni, Salmonella, Shigella and Yersinia species in fecal samples. Int J Med Microbiol. 2011;301:577-584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 70] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 9. | Zhang M, Li Q, He L, Meng F, Gu Y, Zheng M, Gong Y, Wang P, Ruan F, Zhou L. Association study between an outbreak of Guillain-Barre syndrome in Jilin, China, and preceding Campylobacter jejuni infection. Foodborne Pathog Dis. 2010;7:913-919. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 43] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 10. | Swanson EC, Collins MT. Use of the API 20E system to identify veterinary Enterobacteriaceae. J Clin Microbiol. 1980;12:10-14. [PubMed] |
| 11. | Gómez-Duarte OG, Bai J, Newell E. Detection of Escherichia coli, Salmonella spp., Shigella spp., Yersinia enterocolitica, Vibrio cholerae, and Campylobacter spp. enteropathogens by 3-reaction multiplex polymerase chain reaction. Diagn Microbiol Infect Dis. 2009;63:1-9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 66] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 12. | Bullman S, O’Leary J, Corcoran D, Sleator RD, Lucey B. Molecular-based detection of non-culturable and emerging campylobacteria in patients presenting with gastroenteritis. Epidemiol Infect. 2012;140:684-688. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 53] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 13. | Nachamkin I, Allos BM, Ho T. Campylobacter species and Guillain-Barré syndrome. Clin Microbiol Rev. 1998;11:555-567. [PubMed] |
| 14. | Yuki N, Hartung HP. Guillain-Barré syndrome. N Engl J Med. 2012;366:2294-2304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 786] [Cited by in RCA: 809] [Article Influence: 57.8] [Reference Citation Analysis (0)] |
| 15. | On SL. Identification methods for campylobacters, helicobacters, and related organisms. Clin Microbiol Rev. 1996;9:405-422. [PubMed] |
| 16. | Inglis GD, Kalischuk LD. Direct quantification of Campylobacter jejuni and Campylobacter lanienae in feces of cattle by real-time quantitative PCR. Appl Environ Microbiol. 2004;70:2296-2306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 74] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 17. | Steinhauserova I, Ceskova J, Fojtikova K, Obrovska I. Identification of thermophilic Campylobacter spp. by phenotypic and molecular methods. J Appl Microbiol. 2001;90:470-475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 35] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 18. | Englen MD, Ladely SR, Fedorka-Cray PJ. Isolation of Campylobacter and identification by PCR. Methods Mol Biol. 2003;216:109-121. [PubMed] |
| 19. | Stacy-Phipps S, Mecca JJ, Weiss JB. Multiplex PCR assay and simple preparation method for stool specimens detect enterotoxigenic Escherichia coli DNA during course of infection. J Clin Microbiol. 1995;33:1054-1059. [PubMed] |
| 20. | Inglis GD, Kalischuk LD. Use of PCR for direct detection of Campylobacter species in bovine feces. Appl Environ Microbiol. 2003;69:3435-3447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 115] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 21. | Lund M, Nordentoft S, Pedersen K, Madsen M. Detection of Campylobacter spp. in chicken fecal samples by real-time PCR. J Clin Microbiol. 2004;42:5125-5132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 150] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 22. | Maher M, Finnegan C, Collins E, Ward B, Carroll C, Cormican M. Evaluation of culture methods and a DNA probe-based PCR assay for detection of Campylobacter species in clinical specimens of feces. J Clin Microbiol. 2003;41:2980-2986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 70] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 23. | Bessède E, Delcamp A, Sifré E, Buissonnière A, Mégraud F. New methods for detection of campylobacters in stool samples in comparison to culture. J Clin Microbiol. 2011;49:941-944. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 95] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 24. | Gun-Munro J, Rennie RP, Thornley JH, Richardson HL, Hodge D, Lynch J. Laboratory and clinical evaluation of isolation media for Campylobacter jejuni. J Clin Microbiol. 1987;25:2274-2277. [PubMed] |
| 25. | Potturi-Venkata LP, Backert S, Lastovica AJ, Vieira SL, Norton RA, Miller RS, Pierce S, Oyarzabal OA. Evaluation of different plate media for direct cultivation of Campylobacter species from live broilers. Poult Sci. 2007;86:1304-1311. [PubMed] |
| 26. | Goossens H, De Boeck M, Coignau H, Vlaes L, Van den Borre C, Butzler JP. Modified selective medium for isolation of Campylobacter spp. from feces: comparison with Preston medium, a blood-free medium, and a filtration system. J Clin Microbiol. 1986;24:840-843. [PubMed] |
P- Reviewers Bourke B, Louwen R, Rossignol JF, Salazar-Lindo E S- Editor Wen LL L- Editor Stewart GJ E- Editor Li JY