Published online Sep 14, 2012. doi: 10.3748/wjg.v18.i34.4704
Revised: June 11, 2012
Accepted: June 28, 2012
Published online: September 14, 2012
AIM: To investigate the roles of the ribonucleotide reductase M2 (RRM2) subunit in colorectal cancer (CRC) and ultraviolet (UV)-induced DNA damage repair.
METHODS: Immunohistochemical staining of tissue microarray was performed to detect the expression of RRM2. Seven CRC cell lines were cultured and three human colon cancer cell lines, i.e., HCT116, SW480 and SW620, were used. Reverse transcription polymerase chain reaction and Western blotting were performed to determine the mRNA and protein expression levels of RRM2, respectively. Cell proliferation assay, cell cycle analysis were performed. Cell apoptosis was evaluated by double staining with fluorescein isothiocyanate-conjugated Annexin V and propidium iodide (PI) using Annexin V/PI apoptosis kit. The motility and invasion of CRC cells were assessed by the Transwell chamber assay. Cells were irradiated with a 254 nm UV-C lamp to detect the UV sensitivity after RRM2 depletion.
RESULTS: Immunohistochemical staining revealed elevated RRM2 levels in CRC tissues. RRM2 overexpression was positively correlated with invasion depth (P < 0.05), poorly differentiated type (P = 0.0051), and tumor node metastasis stage (P = 0.0015). The expression of RRM2 in HCT116 cells was downregulated after transfection, and HCT116 cell proliferation was obviously suppressed compared to control groups (P < 0.05). In the invasion test, the number of cells that passed through the chambers in the RRM2-siRNA group was 81 ± 3, which was lower than that in the negative control (289 ± 7) and blank control groups (301 ± 7.2). These differences were statistically significant (P < 0.01). Our data suggest that RRM2 overexpression may be associated with CRC progression. RRM2 silencing by siRNA may inhibit the hyperplasia and invasiveness of CRC cells, suggesting that RRM2 may play an important role in the infiltration and metastasis of CRC, which is a potential therapeutic strategy in CRC. In addition, RRM2 depletion increased UV sensitivity.
CONCLUSION: These findings suggest that RRM2 may be a facilitating factor in colorectal tumorigenesis and UV-induced DNA damage repair.
- Citation: Lu AG, Feng H, Wang PXZ, Han DP, Chen XH, Zheng MH. Emerging roles of the ribonucleotide reductase M2 in colorectal cancer and ultraviolet-induced DNA damage repair. World J Gastroenterol 2012; 18(34): 4704-4713
- URL: https://www.wjgnet.com/1007-9327/full/v18/i34/4704.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i34.4704
Colorectal cancer (CRC) is the leading cause of cancer-related death worldwide. In addition, the incidence and mortality rates of CRC are on the rise[1]. CRC develops through a specific series of events, from the transformation of normal colonic epithelium to an adenomatous intermediate, and then ultimately adenocarcinoma[2]; however, the molecular mechanism underlying CRC metastasis is still not fully understood. With evolving insights into the biology of colorectal neoplasia and advances in assay technology, molecular tests are now being developed as accurate, user friendly, safe, and affordable screening tools[3]. For the accurate diagnosis and adequate treatment of CRC, the identification and understanding of the molecules responsible for cancer progression are critical.
Given that all cancer cells rely on changes in metabolism to support their growth and survival, targeting metabolism has the potential to affect cancers arising from many different tissues. Ribonucleotide reductase (RR) is one of these target metabolic enzymes for cancer therapy. RR transforms RNA building blocks to DNA building blocks by catalyzing the substitution of the 2′OH-group of a ribonucleotide with a hydrogen by a mechanism involving protein radicals[4]. In humans, bacteria, and yeast, RR is a highly regulated enzyme in the deoxyribonucleotide synthesis pathway[5]. Human RR is an enzymatic complex that consists of two nonidentical subunits, M1 and M2. M2 has two alternative subunits; namely, ribonucleotide reductase M2 (RRM2), which is involved in DNA replication (the focus of this paper), and p53R2, which is induced by p53 and is involved in supplying dNTPs for mitochondrial DNA replication and repair[6]. In addition, RRM2 is the limiting factor of RR enzymatic complex catalytic activity, which is specifically expressed in the S phase[7,8]. Therefore, RR plays an important role in the regulation of cell proliferation[9-11]. In vitro studies have shown that aggressive tumor proliferation results in overamplification of RR[12,13]. In some types of cancer, elevated RRM2 levels correlate with chemoresistance[14], cellular invasiveness[15], and poor patient outcome[16], which suggests that RRM2 may contribute to malignant progression, and thus be a potential therapeutic target. Recent findings have established that p53R2 suppresses the invasiveness of cancer cells, and its expression is associated with a better survival prognosis for CRC patients[17]; however, the function of RRM2 in CRC is unclear. Here, we demonstrate that RRM2 may play an important role in the development of CRC and may contribute to the response to ultraviolet (UV) irradiation.
This study consisted of a tissue microarray (TMA, Shanghai Outdo Biotech, National Engineering Center for Biochip at Shanghai, China) containing 56 CRC tissues and 56 adjacent normal mucosa, which were laparoscopically resected at Shanghai Minimal Invasive Surgery Centre (Shanghai Jiao Tong University School of Medicine) between 2007 and 2009. In addition, another 78 specimens of CRC tissues were laparoscopic resected between 2011 and 2012. None of the patients received preoperative treatment such as radiation or chemotherapy. All research protocols in the present study were approved by the Institutional Review Board. Staging of the tumors was performed according to the tumor node metastasis classification updated by the American Joint Commission on Cancer and the World Health Organization[18]. Pathological diagnoses were verified by two different pathologists. TMA blocks (5 μmol/L) were sectioned. Preparation of sections from paraffin blocks was performed as previously described[19]. Immunohistochemical and immunocytochemical analysis of RRM2 was performed with an anti-RRM2 antibody [R2 (E-16): SC-10846] (1:200, Santa Cruz Biotechnology, Santa Cruz, CA, United States). Relative RRM2 expression and subcellular location were classified into 5 groups based on the intensity of RRM2 staining and its distribution between the cytoplasm and nucleus.
Seven human CRC cell lines were purchased from American Type Culture Collection (Manassas, VA, United States). The CRC cell lines, HCT116 and HT-29, were cultured in McCoy’s 5A medium supplemented with 10% fetal bovine serum (FBS), penicillin-streptomycin, L-glutamine, and nonessential amino acids. SW1116, SW620, SW480, LoVo and Colo205 cells were maintained in the same medium, except that McCoy’s 5A medium was substituted with RPMI 1640 medium (Sigma Aldrich, St Louis, MO, United States). Cells were maintained at 37 °C in a 5% CO2 incubator, and were grown according to standard protocols.
Total RNA was extracted with TRIzol (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions. Reverse transcription was performed with oligo dT primers. Real-time polymerase chain reaction (PCR) was carried out in an Applied Biosystems 7500 System with Power SYBR Green PCR Master Mix (2 ×, Applied Biosystems, Warrington, United Kingdom). Relative levels of gene expression were determined with glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as the control.
Transfection of cells with siRNAs targeting RRM2 was carried out by mixing the siRNAs with diluted Lipofectamine™ 2000 at a final siRNA concentration of 20 nmol/L. The target sequences for siRNAs were as follows: 5'-AAACCCGAGGAGAGAUAUUTT-3', 5′-GGAGCGAUUUAGCCAAGAATT-3′ and 5′-GCACUCUAAUGAAGCAAUATT-3′. siRNAs targeting RRM2 and a negative control siRNA (5'-UUCUCCGAACGUGUCACGUTT-3') were purchased from Genepharma (Shanghai, China). Each experiment was performed in triplicate.
The following sets of oligonucleotides were used to knockdown RRM2 expression; shRRM2-sense: CACCGGAGCGATTTAGCCAAGAAGTCACCACTTCTTGGCTAAATCGCTCC and shRRM2-antisense: AAAAGGAGCGATTTAGCCAAGAAGTGGTGACTTCTTGGCTAAATCGCTCC. Each set of oligonucleotides were annealed and cloned into pGPH1/Neo (Genepharma, Shanghai, China).
An anti-RRM2 antibody (0.2 μg/mL) or goat polyclonal anti-GAPDH antibody (Santa Cruz Biotechnology; 0.2 μg/mL) was used as the primary antibody. Seven human CRC cell lines were resuspended in ice-cold NP-40 lysis buffer, and cell extracts from the colorectal mucosa were prepared by scraping mucosal epithelial cells into ice-cold NP-40 lysis buffer (50 mmol/L Tris-HCl, pH 7.5, 150 mmol/L NaCl, 1 mmol/L EGTA, and 1% NP-40) containing protease inhibitors. The isolated protein was quantified by a commercially available modified Bradford assay (Bio-Rad Laboratories, Hercules, CA, United States).
Cells were seeded at 2.5 × 103 cells per well in 96-well plates and cultured for 120 h. Cells were counted using the CASY TT Cell Counter (Roche Diagnostics, Indianapolis, United States). Cell viability was determined by adding 10 μL of Cell Counting Kit-8 (Dojindo Molecular Technologies, Kumamoto, Japan) to each well at 37 °C in a 5% CO2 incubator. Each experiment was performed three times. 96-well plates were read using an Epoch Microplate Spectrophotometer (BioTek, Winooski, United States) with Gen5™ Microplate Data Analysis Software included.
Motility and invasion capabilities in vitro were measured in a transwell chamber assay (Becton Dickinson Labware, Bedford, MA, United States). HCT116 cells (2.5 × 105) were seeded in McCoy’s’ 5A medium without FBS, and then plated on each 8.0 μmol/L pore size membrane insert in 24-well plates. McCoys’ 5A medium plus 20% FBS was plated in the bottom wells as chemoattractants. After 24 h, cells on the top side of the inserts were removed with a cotton swab. Cells that had migrated to the underside of the inserts were stained with 0.05% crystal violet for 1 h before rinsing with phosphate buffered saline (PBS) for 20 min. Cells on each insert were counted at a 20 × magnification in 10 random fields of view. The invasion assay was done in the same way except that the 8.0-μmol/L pore size membrane insert was coated with Matrigel that was diluted in McCoys’ 5A medium (1:8 dilution). The results were expressed as mean ± SD.
HCT116 cells after different treatments were rinsed twice with saline PBS. Cells were dissociated, followed by the addition of 5 μL Annexin V-FITC. Cells were then gently vortexed and incubated for 15 min at 4-8 °C in the dark. Propidium iodide (PI, 10 μL) was added to the tube for another 5 min at 4-8 °C in the dark. The labeled cells were analyzed using the BD FACS Vantage System (Becton, Dickinson and Company, Franklin Lakes, NJ, United States) in accordance with the manufacturer’s protocol. Gating was implemented on the basis of the staining profiles of the negative control.
Cells were irradiated with a 254 nm UV-C lamp (UVP Inc., Upland, CA), and doses were measured with a UVX radiometer (Upland).
All continuous values were expressed as mean ± SD and all experiments were repeated three times. The results were subjected to a nonparametric Mann-Whitney U test. A paired Student’s test was also used to analyze the intragroup differences. All statistical analyses were done using Stat View 5.0 for Windows (SAS Institute Inc., Cary, NC, United States). Fisher’s exact test and Cochram-Artimage test were applied to analyze the relationship between RRM2 immunoreactivity and clinicopathological features. Student’s t-test was also used to test differences in cell viability assays. A P value less than 0.05 was considered statistically significant.
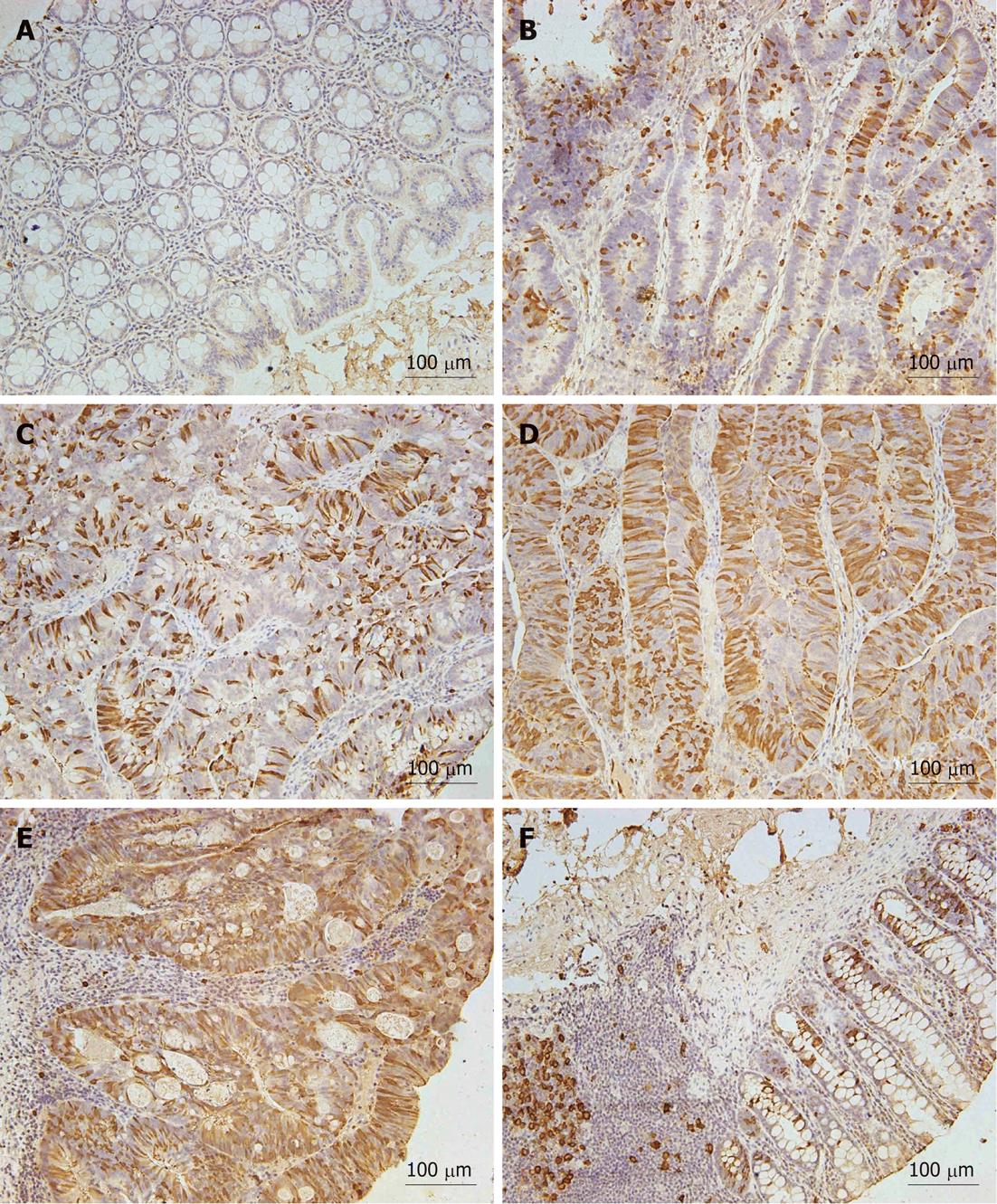
To examine RRM2 protein expression in normal mucosa and CRC tissues, we performed hRRM2 immunohistochemistry on the TMA. The normal human colon exhibits occasional RRM2+ cells located at the base of the crypt (six cases). In comparison, 38 of 56 tumor samples exhibited a substantial increase in overall RRM2 expression and in the number of RRM2+ cells. In the majority of these tumors, RRM2 was present in the cytoplasm (Figure 1). We set the cutoff point of RRM2 overexpression as 10%, because this is obviously higher than the positivity in normal colorectal mucosa, and because it was easy to determine positive or negative expression with this simple cutoff value. Among the 56 CRC tissues included in the TMA, RRM2 overexpression (≥ 10% cancer cells stained) was observed in 38 cases (67.9%). The relationship between RRM2 expression and clinicopathologic characteristics in CRC patients is summarized in Table 1. RRM2 overexpression was significantly positively correlated with invasion depth (P = 0.0015) and differentiation (P = 0.0051), although the frequency of RRM2 overexpression was lower in T4 tumors (20%) compared to T3 tumors (75%). However, no significant differences were observed with respect to gender, age, tumor size, or lymph node metastasis. To further elucidate the clinicopathologic significance of RRM2 overexpression in CRC, especially the correlation with invasion depth and poor differentiation, we examined RRM2 expression in another 78 CRC tissues. The results are summarized in Table 2 and RRM2 immunostaining was categorized further as score 0-3[20]. These data are in accordance with previous studies showing correlation between RRM2 overexpression, invasion depth (P < 0.0001) and differentiation (P < 0.0001).
| Clinical or pathologic feature | Total (n) | RRM2+ | P value |
| All cases | 56 | 38 (67.9) | |
| Gender | 0.0908 | ||
| Male | 33 | 20 (60.6) | |
| Female | 23 | 18 (78.3) | |
| Age (yr) | 0.2094 | ||
| ≤ 64 | 29 | 19 (65.5) | |
| ≥ 65 | 27 | 19 (70.4) | |
| Tumor stage (invasion depth) | 0.0015 | ||
| Tis/T1 | 3 | 0 | |
| T2 | 16 | 6 (37.5) | |
| T3 | 32 | 24 (75.0) | |
| T4 | 5 | 1 (20.0) | |
| Tumor size (cm2, average = 17.3) | 0.0677 | ||
| ≤ 17 | 31 | 20 | |
| > 17 | 25 | 11 | |
| Lymph node metastasis (N) | 0.2281 | ||
| Absent | 25 | 14 (56.0) | |
| 1-3 | 21 | 16 (76.2) | |
| ≥ 4 | 10 | 8 (80.0) | |
| Classification (differentiation) | 0.0051 | ||
| High-grade | 5 | 1 (20.0) | |
| Mid-grade | 45 | 35 (77.8) | |
| Low-grade | 6 | 2 (33.3) |
| Pathologic feature | Total (n) | RRM2- | RRM2+ | DF | P value | ||
| Score 0 | Score 1 | Score 2 | Score 3 | ||||
| Tumor stage (invasion depth) | 1 | < 0.0001 | |||||
| Tis/T1 | 17 | 12 | 4 | 1 | 0 | ||
| T2 | 26 | 1 | 6 | 16 | 3 | ||
| T3 | 21 | 0 | 2 | 8 | 11 | ||
| T4 | 14 | 0 | 2 | 5 | 7 | ||
| Classification (differentiation) | 1 | < 0.0001 | |||||
| High | 20 | 9 | 9 | 2 | 0 | ||
| Mid | 39 | 4 | 3 | 20 | 12 | ||
| Low | 19 | 0 | 2 | 8 | 9 | ||
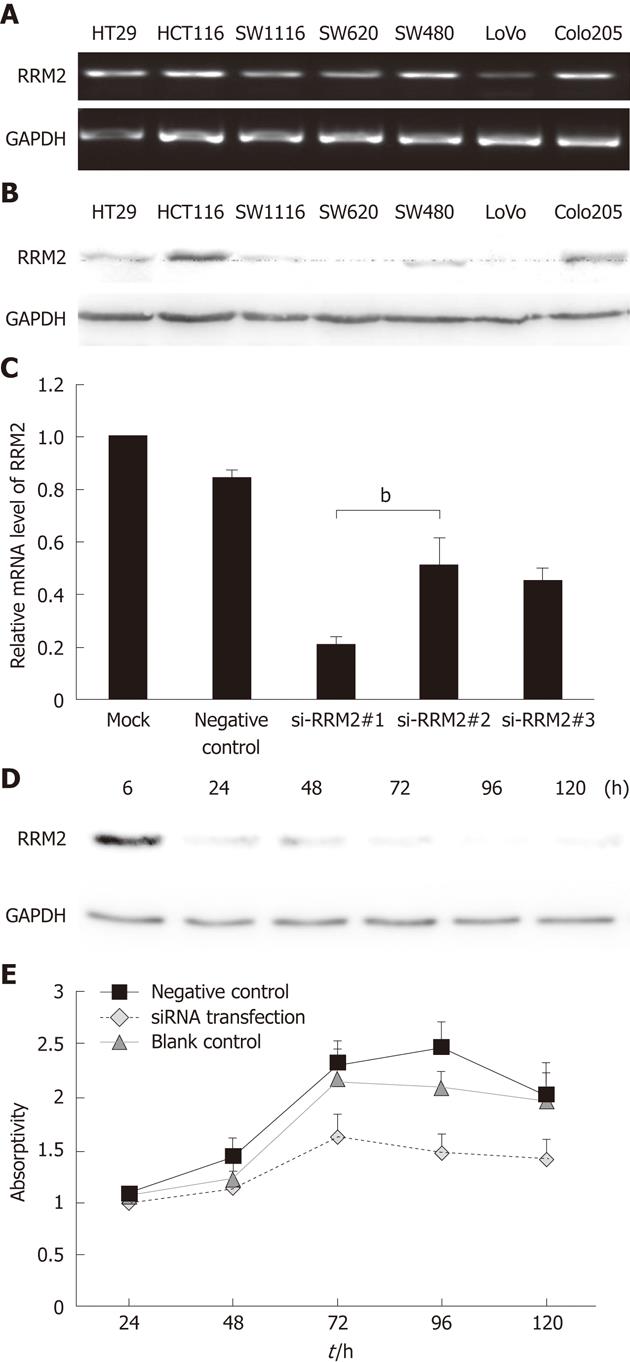
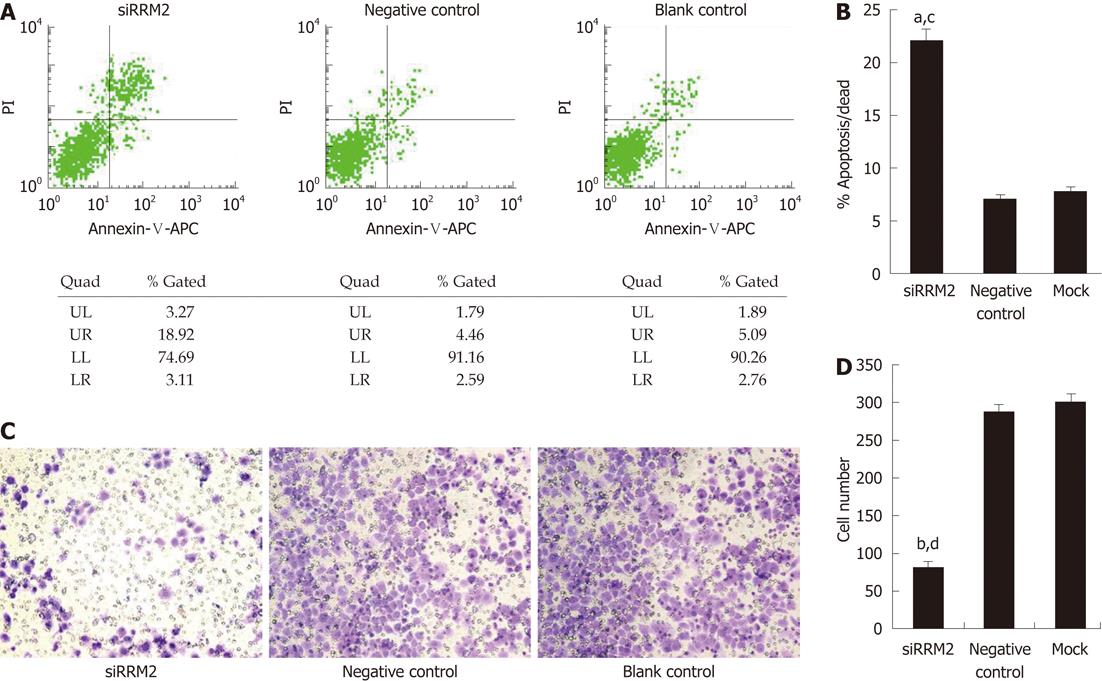
We next sought to determine whether RRM2 overexpression is functionally important for the in vitro proliferation of colonic cancer cell lines. RRM2 had the highest expression levels in HCT116 cells (Figure 2A and B). Then, HCT116 cells were transfected with three different siRNA sequences or a scrambled siRNA control. Quantitative PCR analysis of the HCT116 cell line demonstrated that the expression of RRM2 mRNA was reduced by three siRNA oligos segments to varying degrees (Figure 2C). We used the most efficient one to down-regulate RRM2 and evaluated cell growth; efficient knockdown of RRM2 protein levels after transfection was observed (Figure 2D). Analysis of cell growth in the presence or absence of the RRM2 siRNAs revealed that blocking RRM2 protein synthesis significantly inhibited CRC cell growth (Figure 2E). We used Annexin V/PI staining to identify the dead cells and then stratified them into categories of necrosis, early apoptosis, and both later apoptosis and necrosis. In each flow cytometry graph, later apoptotic or necrotic cells are shown in the upper-right quadrant. The total death rate 48 h after siRNA treatment showed a 2-3 fold increase. Flow cytometric analysis showed evidence of apoptosis when RRM2 functions were abrogated (Figure 3A and B). In the invasion assay, the number of cells that crossed through the chambers in the RRM2-siRNA group was 81 ± 3, which was lower than that in the negative control (289 ± 7) and empty control groups (301 ± 7.2). These differences were statistically significant (P < 0.01) (Figure 3C and D).
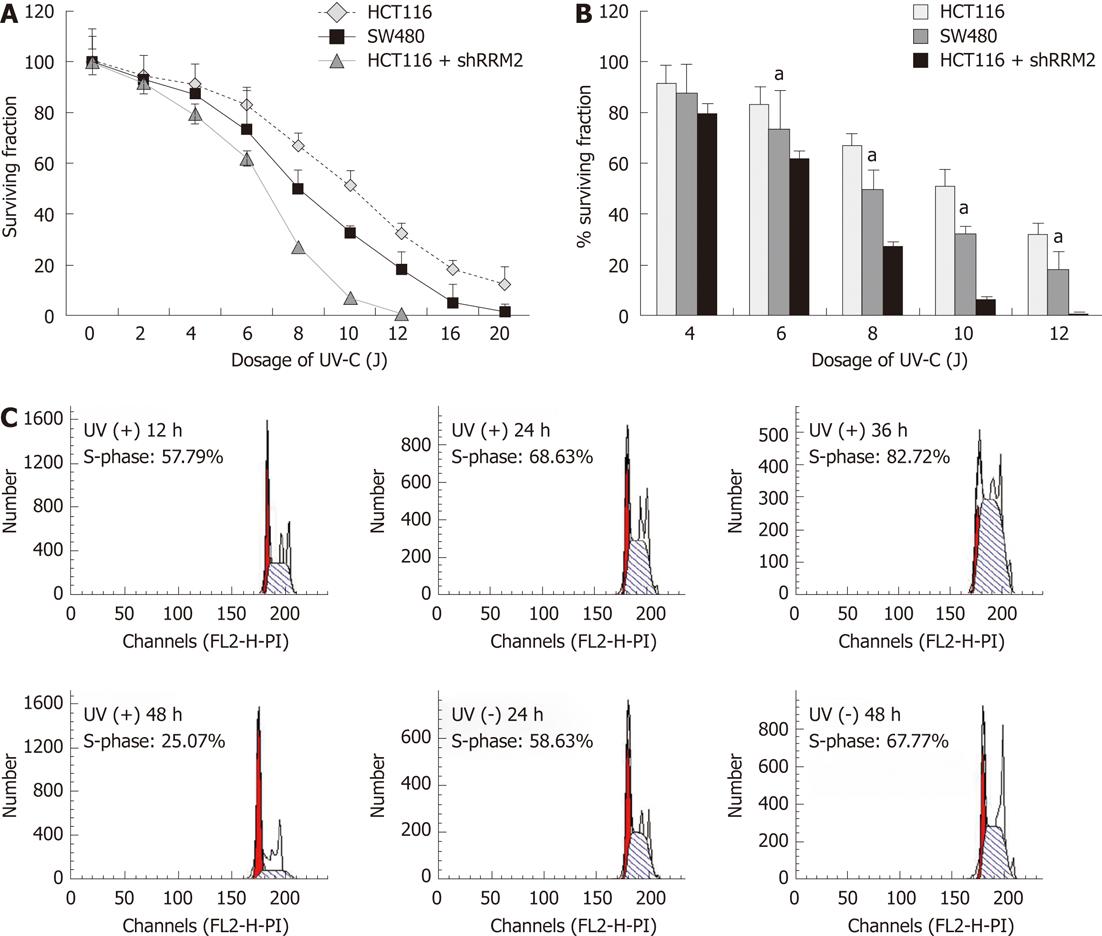
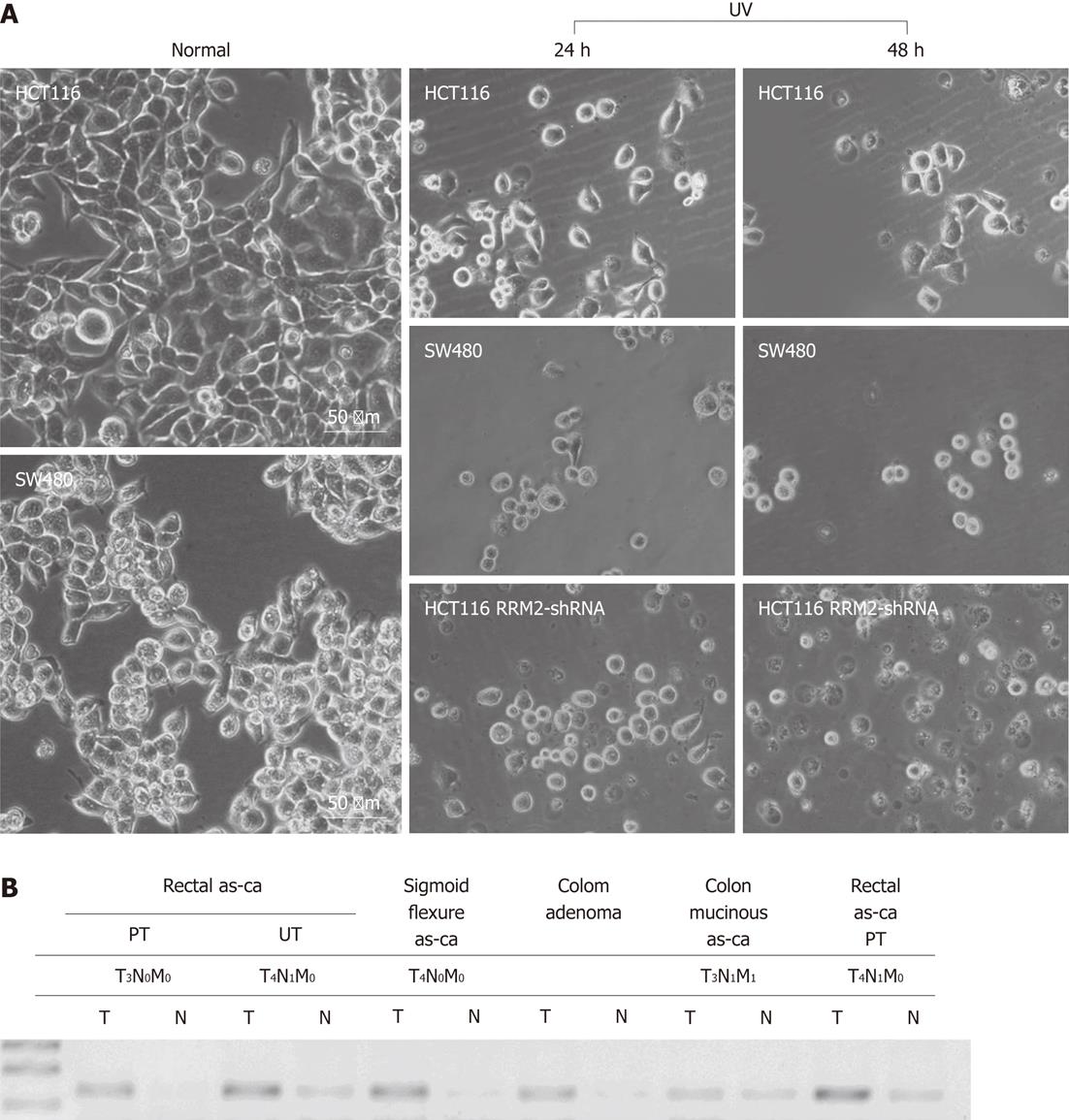
Four human CRC cell lines with low RRM2 expression, namely, SW1116 (p53 mutant-type), SW620 (mutated p53), HT29 (mutated p53), SW480 (p53 mutant-type), and LoVo (p53 wild-type), all proved to be more sensitive to UV irradiation in colony formation assays than HCT116 cells (p53 wild-type). HCT116 cells transfected with shRRM2 revealed significant RRM2 depletion, and its colony-forming ability under UV radiation was suppressed. At a dosage of 10 J/m2, the colony-forming ability of the HCT116 cells transfected with RRM2 antisense was inhibited by 93.13%, whereas wild-type HCT116 cells maintained 51.28% viability, and SW620 cells (RRM2-/-) maintained 32.40% viability (Figure 4A and B). In addition, the cell cycle distribution of the CRC cells did not significantly change at the various time points in the UV (-) panel. After a sublethal dose of UV radiation, an increased number of cells in the S-phase could be seen at 24-36 h (Figure 4C). Cellular morphology was observed by light microscopy. HCT116 cells with high levels of RRM2 expression, and SW480 cells, which do not express RRM2, were cultured under normal conditions or exposed to UV radiation for 48 h. Under these conditions, SW480 cells showed UV hypersensitivity, whereas the UV sensitivity of HCT116 cells with suppressed RRM2 levels increased and cells went through necrosis. Thus, endogenous RRM2 functions to suppress hypersensitivity to UV radiation. Figure 5A gives a visual depiction of the morphological changes that occurred.
The importance of altered metabolism in the context of tumorigenesis has received renewed attention as metabolic changes entwined with the molecular hallmarks of cancer have been elucidated. Previous studies have shown that alterations in RR levels may significantly influence the biological properties of cells, including tumor promotion and tumor progression, suggesting that RR may be implicated in tumorigenesis. Thus, we were interested in determining the effect of RRM2 on tumor cell growth. The discovery of p53R2, an analog of RRM2 in mammalian cells, provided an interesting link between RR and cancer because it is a downstream target for the p53 tumor suppressor. Previous studies have shown that p53R2 is negatively related to tumor invasion and lymph node involvement. In addition, investigations have revealed that p53R2 correlates with a better survival of CRC with advanced grade III and grade IV tumors, rather than earlier grade I and grade II tumors[18]. The genes for p53R2 and RRM2 are located on separate chromosomes. p53R2 is 80%-90% identical to RRM2, but lacks its 33 amino-terminal residues, including a KEN box required for degradation during mitosis. p53R2 can substitute for RRM2 and form a highly active RR, which is believed to be active in DNA repair after DNA damage[4]. However, the current knowledge about RRM2 function in human CRCs is still preliminary[20,21]. Therefore, determining if RRM2 is essential for CRC progression may provide promising therapeutic opportunities.
Our findings showed that RRM2 expression is substantially upregulated in CRC using microarray analysis and RT-PCR. Moreover, RRM2 overexpression was significantly associated with invasion depth and differentiation, and clinical tissue specimens also showed that the expression levels of RRM2 may be associated with tumor stage (Figure 5B). These data suggest that RRM2 overexpression could be associated with CRC progression. Several previous reports demonstrated the frequent overexpression of the RRM2 protein and its possible role in other cancers[20-23]. Recent observations showed that top 400 gene lists is produced by microarrays for each of the 13 cancer types and RRM2 appeared in nine lists of significantly altered genes in cancers[23], the results point to the value of RRM2 in normal to cancer transformation. RRM2 is upregulated in esophageal/gastric cancers and melanomas compared to pancreatic cancer[24]. Other groups have also suggested that RRM2 may be a novel diagnostic marker and potential therapeutic target in bladder and gastric cancers. Furthermore, the studies suggested that RRM2 overexpression is associated with the male sex, the presence of muscularis propria invasion, the presence of Epstein-Barr virus, and expression of survivin, but not with age, histology, tumor size, lymph node metastasis, or expression of the phosphatase and tensin homolog, the phosphorylated signal transducer, and p53. Suppression of RRM2 synthesis inhibited the growth of gastric cancer cell lines in vitro[20]. In addition, RRM2 is one of the putative tumor markers that is strongly expressed in the very early stages of premalignancy and preneoplasia of breast carcinomas[25], and hepatocellular carcinoma cells are highly dependent on these genes for proliferation[26]. Analyses also revealed that hRRM2 positively correlates with poor tumor differentiation of non-small cell lung cancer[27]. It may be that the altered expression of metabolic enzymes or changes in the regulation of metabolic pathways also occur downstream of many oncogenes and tumor suppressor genes, and cancers with specific genetic lesions are dependent upon some of these metabolic changes. As was previously shown, RRM2 is upregulated in several cancers, and abrogating its expression reduces the proliferation of cancer cells, and triggered the development of RRM2-targeted siRNA-containing nanoparticles.
We also demonstrated that siRNA-mediated suppression of RRM2 synthesis inhibited the growth of the CRC cell lines investigated. In addition, abrogating its expression induced cell apoptosis. Therefore, it is likely that RRM2 overexpression is essential for the survival of CRC cells. Our finding is consistent with recent observations demonstrating that cells with altered RRM2 expression exhibit significantly reduced subcutaneous tumor latency and increased tumor growth rates in syngeneic mice, and show markedly elevated metastatic potential in lung metastasis assays[28]. Their studies demonstrate that RRM2 can participate in other critical cellular functions in addition to ribonucleotide reduction. For example, RRM2 overexpression may increase pancreatic cancer cell invasion in an NF-κB-dependent manner, and RNA interference mediated silencing of RRM2 expression attenuated cell invasion and matrix metallopeptidase 9 activity[16]. Another laboratory also reported that RRM2 overexpression plays an important role in tumor invasiveness[29,30]. Our results are consistent with these previous reports, and suggest that targeting RRM2 has the potential for the treatment of CRC.
RRM2 is known to be involved in chemoresistance. The suppression of RRM2 can sensitize colon cancer cells to chemotherapeutic agents and significantly inhibit the proliferation of melanoma cell lines with different oncogenic mutations with synergistic enhancement in combination with temozolomide[31]. The relationship between RRM2 expression and chemotherapeutic effects in clinical settings has also been investigated. For instance, GTI-2040, an antisense oligonucleotide targeting RRM2, has been tested for the treatment of advanced metastatic solid tumors in clinical settings[32]. Radiation is also a key component of carcinoma therapy. Unfortunately, it results in significant side effect in some patients. Thus, agents that radiosensitize CRC cells would be very useful. Here we show that RRM2 depletion decreased the surviving fraction of tumor cells in response to radiation and increased the sensitizer enhancement ratios. Recent observations suggest that targeting polo-like kinase 1 (PLK1) with small molecule inhibitors, in combination with radiation therapy, is a novel strategy in the treatment of some cancers[33]. In addition, it has been hypothesized that there are some links between RRM2 and PLK1, because PLK1 was one of the most drastically downregulated genes following RRM2 silencing[34]. Our data suggest RRM2 depletion increased UV sensitivity and make an argument for further exploring the role of RRM2 inhibition in CRC. It will be important to elucidate in detail the specific mechanisms by which RRM2 mediates tumor cell radiosensitivity.
In conclusion, we performed both functional and correlative studies on RRM2 in CRCs and provide strong evidence that RRM2 overexpression may be associated with CRC progression; thus suppressing its function may be a potential therapeutic strategy in CRC.
Previous studies have shown that alterations in ribonucleotide reductase (RR) levels may significantly influence the biological properties of cells, including tumor promotion and tumor progression, suggesting that RR may be implicated in tumorigenesis. Thus, the authors were interested in determining the effect of ribonucleotide reductase M2 (RRM2) on colorectal cancer (CRC) cells.
Recently, metabolic genes have received increasing and specific attention due to their potential role in carcinogenesis. Overexpression of RRM2 has been revealed in breast cancer, renal cell carcinoma, prostate cancer and gastric cancer to elevated cellular proliferation and invasiveness.
Few studies have described the correlation between RRM2 and the development of CRC. And the possible mechanism by which RRM2 mediates CRC progression is unclear. It is well established that in some types of cancer, elevated RRM2 levels correlate with chemoresistance, but whether it may contribute to the response to ultraviolet (UV) irradiation is unclear. The results of this study suggest that RRM2 may be a facilitating factor in colorectal tumorigenesis and UV-induced DNA damage repair.
The data suggests that RRM2 subunit overexpression is associated with the CRC progression. RRM2 silencing may inhibit the hyperplasia and invasiveness of CRC cells. RRM2 may play an important role in the infiltration and metastasis of CRC and that suppression of its function could be a potential therapeutic strategy in CRC.
RR is an essential enzyme required for the de novo conversion of ribonucleoside diphosphates to deoxyribonucleoside diphosphates, and it plays an important role in DNA synthesis and repair. RR consists of two subunits, RRM1 and RRM2/p53R2.
This is an interesting study investigating the importance of the hRRM2 in CRC and UV-induced DNA damage repair.
| 1. | Center MM, Jemal A, Ward E. International trends in colorectal cancer incidence rates. Cancer Epidemiol Biomarkers Prev. 2009;18:1688-1694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 532] [Cited by in RCA: 568] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 2. | Pino MS, Chung DC. The chromosomal instability pathway in colon cancer. Gastroenterology. 2010;138:2059-2072. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 576] [Cited by in RCA: 626] [Article Influence: 39.1] [Reference Citation Analysis (1)] |
| 3. | Ahlquist DA. Molecular detection of colorectal neoplasia. Gastroenterology. 2010;138:2127-2139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 99] [Article Influence: 6.2] [Reference Citation Analysis (2)] |
| 4. | Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681-706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 821] [Cited by in RCA: 884] [Article Influence: 44.2] [Reference Citation Analysis (0)] |
| 5. | Jordan A, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 1998;67:71-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 588] [Cited by in RCA: 568] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 6. | Thelander L. Ribonucleotide reductase and mitochondrial DNA synthesis. Nat Genet. 2007;39:703-704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 55] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 7. | Chang CH, Cheng YC. Substrate specificity of human ribonucleotide reductase from Molt-4F cells. Cancer Res. 1979;39:5081-5086. [PubMed] |
| 8. | Björklund S, Skog S, Tribukait B, Thelander L. S-phase-specific expression of mammalian ribonucleotide reductase R1 and R2 subunit mRNAs. Biochemistry. 1990;29:5452-5458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 142] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 9. | Cory JG, Sato A. Regulation of ribonucleotide reductase activity in mammalian cells. Mol Cell Biochem. 1983;53-54:257-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 31] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 10. | Thelander L, Berg P. Isolation and characterization of expressible cDNA clones encoding the M1 and M2 subunits of mouse ribonucleotide reductase. Mol Cell Biol. 1986;6:3433-3442. [PubMed] |
| 11. | Zhou B, Mo X, Liu X, Qiu W, Yen Y. Human ribonucleotide reductase M2 subunit gene amplification and transcriptional regulation in a homogeneous staining chromosome region responsible for the mechanism of drug resistance. Cytogenet Cell Genet. 2001;95:34-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 12. | Srinivasan PR, Tonin PN, Wensing EJ, Lewis WH. The gene for ornithine decarboxylase is co-amplified in hydroxyurea-resistant hamster cells. J Biol Chem. 1987;262:12871-12878. [PubMed] |
| 13. | Hurta RA, Samuel SK, Greenberg AH, Wright JA. Early induction of ribonucleotide reductase gene expression by transforming growth factor beta 1 in malignant H-ras transformed cell lines. J Biol Chem. 1991;266:24097-24100. [PubMed] |
| 14. | Chiaramonte RM, Rich MA, Brock WA. Prolapsing congenital polyp of posterior urethra and urethral duplication associated with imperforate anus. Urology. 1992;40:522-524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 182] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 15. | Duxbury MS, Whang EE. RRM2 induces NF-kappaB-dependent MMP-9 activation and enhances cellular invasiveness. Biochem Biophys Res Commun. 2007;354:190-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 78] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 16. | Souglakos J, Boukovinas I, Taron M, Mendez P, Mavroudis D, Tripaki M, Hatzidaki D, Koutsopoulos A, Stathopoulos E, Georgoulias V. Ribonucleotide reductase subunits M1 and M2 mRNA expression levels and clinical outcome of lung adenocarcinoma patients treated with docetaxel/gemcitabine. Br J Cancer. 2008;98:1710-1715. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 90] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 17. | Liu X, Lai L, Wang X, Xue L, Leora S, Wu J, Hu S, Zhang K, Kuo ML, Zhou L. Ribonucleotide reductase small subunit M2B prognoses better survival in colorectal cancer. Cancer Res. 2011;71:3202-3213. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 52] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 18. | Hu H, Krasinskas A, Willis J. Perspectives on current tumor-node-metastasis (TNM) staging of cancers of the colon and rectum. Semin Oncol. 2011;38:500-510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 46] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 19. | Qian-Lin Z, Ting-Feng W, Qi-Feng C, Min-Hua Z, Ai-Guo L. Inhibition of cytosolic chaperonin CCTζ-1 expression depletes proliferation of colorectal carcinoma in vitro. J Surg Oncol. 2010;102:419-423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 27] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 20. | Morikawa T, Hino R, Uozaki H, Maeda D, Ushiku T, Shinozaki A, Sakatani T, Fukayama M. Expression of ribonucleotide reductase M2 subunit in gastric cancer and effects of RRM2 inhibition in vitro. Hum Pathol. 2010;41:1742-1748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 65] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 21. | Yun HJ, Cho YH, Moon Y, Park YW, Yoon HK, Kim YJ, Cho SH, Lee YI, Kang BS, Kim WJ. Transcriptional targeting of gene expression in breast cancer by the promoters of protein regulator of cytokinesis 1 and ribonuclease reductase 2. Exp Mol Med. 2008;40:345-353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 22. | Morikawa T, Maeda D, Kume H, Homma Y, Fukayama M. Ribonucleotide reductase M2 subunit is a novel diagnostic marker and a potential therapeutic target in bladder cancer. Histopathology. 2010;57:885-892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 78] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 23. | Dawany NB, Dampier WN, Tozeren A. Large-scale integration of microarray data reveals genes and pathways common to multiple cancer types. Int J Cancer. 2011;128:2881-2891. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 24. | Kolesar J, Huang W, Eickhoff J, Hahn K, Alberti D, Attia S, Schelman W, Holen K, Traynor A, Ivy P. Evaluation of mRNA by Q-RTPCR and protein expression by AQUA of the M2 subunit of ribonucleotide reductase (RRM2) in human tumors. Cancer Chemother Pharmacol. 2009;64:79-86. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 25. | Kretschmer C, Sterner-Kock A, Siedentopf F, Schoenegg W, Schlag PM, Kemmner W. Identification of early molecular markers for breast cancer. Mol Cancer. 2011;10:15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 158] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 26. | Satow R, Shitashige M, Kanai Y, Takeshita F, Ojima H, Jigami T, Honda K, Kosuge T, Ochiya T, Hirohashi S. Combined functional genome survey of therapeutic targets for hepatocellular carcinoma. Clin Cancer Res. 2010;16:2518-2528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 136] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 27. | Hsu NY, Wu JY, Liu X, Yen Y, Chen CY, Chou MC, Lin CH, Lee H, Cheng YW. Expression status of ribonucleotide reductase small subunits hRRM2/p53R2 as prognostic biomarkers in stage I and II non-small cell lung cancer. Anticancer Res. 2011;31:3475-3481. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 139] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 28. | Fan H, Villegas C, Wright JA. Ribonucleotide reductase R2 component is a novel malignancy determinant that cooperates with activated oncogenes to determine transformation and malignant potential. Proc Natl Acad Sci USA. 1996;93:14036-14040. [PubMed] |
| 29. | Zhou BS, Tsai P, Ker R, Tsai J, Ho R, Yu J, Shih J, Yen Y. Overexpression of transfected human ribonucleotide reductase M2 subunit in human cancer cells enhances their invasive potential. Clin Exp Metastasis. 1998;16:43-49. [PubMed] |
| 30. | Zhou BS, Hsu NY, Pan BC, Doroshow JH, Yen Y. Overexpression of ribonucleotide reductase in transfected human KB cells increases their resistance to hydroxyurea: M2 but not M1 is sufficient to increase resistance to hydroxyurea in transfected cells. Cancer Res. 1995;55:1328-1333. [PubMed] |
| 31. | Zuckerman JE, Hsueh T, Koya RC, Davis ME, Ribas A. siRNA knockdown of ribonucleotide reductase inhibits melanoma cell line proliferation alone or synergistically with temozolomide. J Invest Dermatol. 2011;131:453-460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 37] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 32. | Shibata SI, Doroshow JH, Frankel P, Synold TW, Yen Y, Gandara DR, Lenz HJ, Chow WA, Leong LA, Lim D. Phase I trial of GTI-2040, oxaliplatin, and capecitabine in the treatment of advanced metastatic solid tumors: a California Cancer Consortium Study. Cancer Chemother Pharmacol. 2009;64:1149-1155. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 33. | Harris PS, Venkataraman S, Alimova I, Birks DK, Donson AM, Knipstein J, Dubuc A, Taylor MD, Handler MH, Foreman NK. Polo-like kinase 1 (PLK1) inhibition suppresses cell growth and enhances radiation sensitivity in medulloblastoma cells. BMC Cancer. 2012;12:80. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 66] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 34. | Grade M, Hummon AB, Camps J, Emons G, Spitzner M, Gaedcke J, Hoermann P, Ebner R, Becker H, Difilippantonio MJ. A genomic strategy for the functional validation of colorectal cancer genes identifies potential therapeutic targets. Int J Cancer. 2011;128:1069-1079. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
Peer reviewers: Peter L Lakatos, MD, PhD, Assistant Professor, 1st Department of Medicine, Semmelweis University, Koranyi S 2A, Budapest H1083, Hungary; Vasiliy I Reshetnyak, MD, PhD, Professor, Scientist Secretary of the Scientific Research Institute of General Reanimatology, 25-2, Petrovka str., 107031 Moscow, Russia; Yeun-Jun Chung, MD, PhD, Professor, Director, Department of Microbiology, Integrated Research Center for Genome Polymorphism, The Catholic University Medical College, 505 Banpo-dong, Socho-gu, Seoul 137-701, South Korea
S- Editor Lv S L- Editor A E- Editor Zheng XM
.