Published online Aug 14, 2012. doi: 10.3748/wjg.v18.i30.4051
Revised: March 31, 2012
Accepted: April 9, 2012
Published online: August 14, 2012
AIM: To investigate the association between human papilloma virus (HPV) infection and colorectal cancer.
METHODS: Sixty-nine patients with pathologically confirmed primary colorectal cancer including 6 stage I, 24 stage II, 21 stage III, and 18 stage IV patients were enrolled in this study to investigate whether HPV 16 could be involved in colorectal tumorigenesis. Nested-polymerase chain reaction (nested-PCR) was used to detect HPV16 DNA in colorectal tumor tissues and further confirmed by in situ hybridization (ISH). In addition, immunohistochemistry analysis was performed to examine the E6 oncoprotein in colorectal tumors. To verify whether E6 could inactivate the p53 transcriptional function, the levels of p21 and Mdm2 mRNA expression were evaluated by real-time reverse transcription (RT)-PCR.
RESULTS: Of the 69 colorectal tumors, HPV16 DNA was detected in 11 (16%) by nested-PCR, and HPV16 DNA was present in 8 of the 11 (73%) tumors which was confirmed by ISH. The presence of HPV16 DNA in colorectal tumors was not associated with patients’ clinical parameters including age, gender, smoking status, tumor site; however, HPV16 infection was more common in stage I patients than in late-stages patients (II, III and IV). We next asked whether HPV16 infection could be linked with colorectal cancer development. Immunohistochemical data indicated that 8 of the 11 HPV16 DNA-positive tumors had E6 oncoprotein expression. Moreover, we also observed that the adjacent normal tissues including endothelial cells, lymphocytes, fibroblasts, and gland cells in E6-positive tumors had E6 oncoprotein expression. In addition, 3 of the 4 (75%) E6-positive tumors carrying p53 wild-type had negative immunostaining, but one tumor had less p53 immunostaining. We further examined whether E6-positive and/or p53 mutated tumors reduce p53 transcriptional activity. Real-time RT-PCR analysis indicated that p21 and mdm2 mRNA expression levels in E6/p53-wildtype tumors were significantly lower than in their adjacent normal tissues; as expected, E6-positive/p53-mutated tumors had lower p21 and mdm2 mRNA expression levels compared with their adjacent normal tissues. These results clearly indicate that the E6 oncoprotein expressed in p53 wildtype tumors may reduce p21 and mdm2 expression via p53 inactivation.
CONCLUSION: These results suggest that HPV16 infection may be involved in a subset of colorectal cancer, and we suggest that the transmission of HPV to the colon and rectum might occur through peripheral blood lymphocytes.
- Citation: Chen TH, Huang CC, Yeh KT, Chang SH, Chang SW, Sung WW, Cheng YW, Lee H. Human papilloma virus 16 E6 oncoprotein associated with p53 inactivation in colorectal cancer. World J Gastroenterol 2012; 18(30): 4051-4058
- URL: https://www.wjgnet.com/1007-9327/full/v18/i30/4051.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i30.4051
A high risk of human papilloma virus (HPV) 16/18 infection has been documented to be involved in the development of cervical and anal genital cancers[1]. Among non-genital cancers, the association of HPV16/18 infection and oropharyngeal cancer was recently evidenced[2-4]. However, other non-genital cancers, including lung, breast, and colorectal cancers have not yet been identified[5-12]. This is due to a lack of clarity as to how HPV transmits to internal organs even though blood circulation has been suggested as a possible route of infection[13,14].
In the past two decades, a large body of research has demonstrated an association between HPV infection and colorectal cancer[5,6,15-17]. Studies have shown HPV16 to be the major HPV subtype in colorectal tumors[16,17]. However, inconsistent conclusions have been drawn on this issue because only HPV DNA detection is used to associate HPV and colorectal cancer; there is no evidence to demonstrate that HPV is involved in colorectal cancer development. It is well known that HPV DNA integration into the host chromosome plays a crucial role in HPV-associated tumorigenesis[18]. When HPV DNA was spliced at E2, E6 and E7 oncoproteins were expressed which inactivated the p53 and Rb pathways[18]. Therefore, in the present study, 69 tumors resected from colorectal cancer patients were enrolled to determine the presence of HPV16 DNA by nested polymerase chain reaction (PCR) and in situ hybridization (ISH), and expressions of E6 and p53 proteins were evaluated by immunohistochemistry (IHC) in colorectal tumor paraffin serial sections. We explored whether HPV16 DNA could exist and express E6 oncoprotein to inactivate the p53 pathway in colorectal tumors. We next asked whether HPV16 DNA and E6 oncoprotein could be expressed in adjacent normal tissue cells, such as endothelial cells and lymphocytes, to understand whether colorectal tumors infected with HPV16 could be spread through blood circulation.
We collected tumor specimens from 69 patients with colorectal cancer. All of these patients, including 34 females and 35 males who were admitted to the Department of Surgery at Chung Shan Medical University Hospital, Taichung, Taiwan between 2000 and 2005, were asked to submit a written informed consent approved by the Institutional Review Board. A series of examinations for pathological stages were conducted for each case by board-certified pathologists based on the criteria in the 7th edition of the American Joint Committee on Cancer. We collected information pertaining to personal characteristics from hospital reports. Smokers were defined as those who were active smokers or previous smokers and nonsmokers were those who had never smoked.
Formalin-fixed and paraffin-embedded specimens were sectioned at a thickness of 3 μm. All sections were then deparaffinized in xylene, rehydrated through serial dilutions of alcohol, and washed in phosphate-buffered saline (pH 7.2), the buffer that was used for all subsequent washes. For HPV16 E6 and p53 detection, sections were heated in a microwave oven twice for 5 min in citrate buffer (pH 6.0), and then incubated with a monoclonal anti-human p53 antibody (DAKO, DO7, Denmark; at a dilution of 1:250) for 60 min at 25 °C or with monoclonal anti-HPV16 (Santa Cruz, CA, United States) for 90 min at 25 °C. The conventional streptavidin peroxidase method (DAKO, LSAB Kit K675, Copenhagen, Denmark) was performed to develop signals and the cells were counter-stained with hematoxylin. Negative controls were obtained by leaving out the primary antibody. The intensities of signals were evaluated independently by three observers. The results were evaluated independently by three observers and scored for the percentage of positive nuclei: score 0, no positive staining; score +, from 1% to 10%; score ++, from 1% to 50%; score +++, more than 50% positive cells. Positive control slides for p53 protein detection were purchased from DAKO (Denmark) and the cervical cancer tumor tissues with HPV16 were used as a positive control for HPV16 E6. The antibody dilution buffer replaced the antibodies to serve as a negative control.
Mutations in exons 5-8 of the p53 gene were determined by direct sequencing of PCR products amplified from the DNA of tumor cells isolated by microdissection of the colorectal tumor tissues. DNA lysis buffer was used to lyse cells and then the solution was subjected to proteinase K digestion and phenol-chloroform extraction. Finally, the DNA was precipitated by ethanol. Target sequences were amplified in a 50 μL reaction mixture containing 20 pmol of each primer, 2.5 units of Taq polymerase (TAKARA Shuzo, Shiga, Japan), 0.5 mmol/L dNTPs, 5 μL PCR reaction buffer, and 1 μL genomic DNA as the template. Genomic DNA sequences extracted from the frozen sections were not adequate for an amplification of long fragment DNA sequences, and therefore, PCR products ranging from 200 to 400 bp were amplified for p53 mutation analysis. Primers for β-actin, which act as an internal control, were included in each amplification reaction. The primers used in the reactions were E5S (5’TGCCCTGACTTTCAACTCTG3’) and E5AS (5’GCTGCTCACCATCGCTATC3’) for exon 5, E6S (5’CTGATTCCTCACTGATTGCT3’) and E6AS (5’AGTTGCAAACCAGACCTCAGG3’) for exon 6, E7S (5’CCTGTGTTATCTCCTAGGTTG3’) and E7AS (5’GCACAGCAGGCCAGTGTGCA3’) for exon 7, and E8S (5’GACCTGATTTCCTTACTGCC3’) and E8AS (5’TCTCCTCCACCGCTTCTTGT3’) for exon 8. An initial cycle was performed for 5 min at 94 °C; followed by 35 cycles of 40 s at 94 °C, 40 s at 54 °C, and 1 min at 72 °C. The PCR products were sequenced using an Applied Biosystems 3100 Avant Genetic Analyzer (Applied Biosystems, Foster City, CA, United States), and the same primers were used for both the PCR and for the DNA sequencing. All p53 mutations were confirmed by direct sequencing of both DNA strands.
Genomic DNA was prepared from a tissue section and isolated by conventional phenol-chloroform extraction, ethanol precipitation, and was finally dissolved in 20 μL of sterile distilled water. HPV viral DNA was first amplified with consensus primers MY09 and MY11[19] followed by a second round of amplification with type-specific primers flanking the L1 region to identify the subtype. Ten microliters of the final PCR product were loaded onto a 2% agarose gel, stained with ethidium bromide, and visualized under ultraviolet-visible illumination. Appropriate negative and positive controls were included in each PCR reaction. A part of the β-actin gene in all samples was amplified to exclude false-negative results while DNA preparations from the SiHa cell (containing HPV16) were used as positive controls.
ISH for the detection of HPV type 16 DNA was performed using digenoxenin-labeled (DIG-labeled) oligonucleotide probes and a commercially available hybridization kit (Boehringer Mannheim, Indianapolis, IN). Briefly, the hybridizing probes were prepared by PCR amplification using HPV 16 type-specific primers with DIG-deoxyuridine triphosphate as a substrate according to the manufacturer’s instructions[19]. The deparaffinized and rehydrated 5 μm sections were digested with proteinase K, rinsed with PBS, and dehydrated. The hybridization was performed in a humidified chamber at 48 °C for 16 h followed by a wash with sodium chloride-sodium citrate. Thereafter, the detection reagent (anti-DIG antibody conjugated with peroxidase) was applied to the sections and then the sections were incubated in diaminobenzidine solution to allow the signals to develop. After the signal development, the sections were counterstained with hematoxylin, rinsed briefly in absolute ethanol, mounted, and observed for signals under a microscope.
Total RNA was extracted from the colorectal tumors after homogenization in 1 mL TRIzol reagent, followed by chloroform re-extraction and isopropanol precipitation. Three micrograms of total RNA from the colorectal tumor tissues were reverse transcribed using SuperScript II Reverse Transcriptase (Invitrogen, CA, United States) and oligo d(T)15 primer. Real-time RT-PCR was performed in a final volume of 25 μL containing 1 μL of each cDNA template, 10 pmoles of each primer, and 12.5 μL of a SYBR-Green master mix. The primers were designed using ABI Prism 7000 SDS Software. Quantification was carried out using the comparative threshold cycle (CT) method and water was used as the negative control. An arbitrary threshold was chosen on the basis of the variability of the baseline. CT values were calculated by determining the point at which the fluorescence exceeded the threshold limit. CT was reported as the cycle number at this point. The average of the target gene was normalized to 18S rRNA as the endogenous housekeeping gene.
Statistical analysis was performed using the SPSS statistical software program (Version 11.0 SPSS Inc., Chicago, IL, United States). The χ2 test, Fisher’s exact test (two tailed), and Mann-Whitney test were applied for statistical analysis.
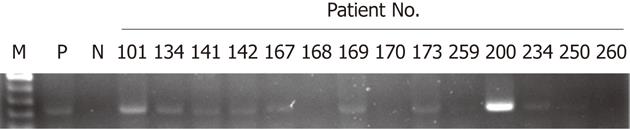
Sixty-nine colorectal tumors were collected to evaluate HPV16 DNA. As shown in Figure 1, the presence of HPV16 DNA in colorectal tumors was determined by nested PCR using MY09/MY11 and specific primers, and 11 of 69 tumors (16%) possessed positive HPV16 DNA signals. The relationships between HPV16 DNA and clinicopathological features were statistically analyzed (Table 1). Our data showed that HPV16 DNA was present more frequently in stageItumors than in late-stage tumors (66.7% for stage I vs 12.5% for stage II, 9.5% for stage III, and 11.1% for stagev IV, P = 0.017). However, the presence of HPV16 DNA in colorectal tumors was not associated with other clinicopathological features including gender, age, smoking status, and tumor site (Table 1). To further confirm the presence of HPV16 DNA, ISH was conducted to determine the presence of HPV16 DNA in 11 colorectal tumor paraffin sections. ISH data indicated that 8 of the 11 tumors (72.7%) had positive HPV16 DNA signals (Table 2 and Figure 2). Collectively, these results clearly indicate that HPV16 DNA exists in a subset of colorectal tumors, at least in this study population.
| Parameters | HPV16 | ||
| Negative | Positive | P value | |
| Gender | 0.782 | ||
| Male (n = 35) | 29 (82.9) | 6 (17.1) | |
| Female (n = 34) | 29 (85.3) | 5 (14.7) | |
| Age (yr) | 0.322 | ||
| < 68 (n = 31) | 28 (90.3) | 3 (9.7) | |
| > 68 (n = 38) | 30 (78.9) | 8 (21.1) | |
| Smoking status | 0.718 | ||
| No (n = 49) | 42 (85.7) | 7 (14.3) | |
| Yes (n = 20) | 16 (80.0) | 4 (20.0) | |
| Stage | 0.017 | ||
| I (n = 6) | 2(33.3) | 4 (66.7) | |
| II (n = 24) | 21 (87.5) | 3 (12.5) | |
| III (n = 21) | 19 (90.5) | 2 (9.5) | |
| IV (n = 18) | 16 (88.9) | 2 (11.1) | |
| Tumor site | 0.554 | ||
| Ascending colon (n = 22) | 19 (86.4) | 3 (13.6) | |
| Transverse colon (n = 5) | 5 (100.0) | 0 (0) | |
| Descending colon (n = 5) | 3 (60.0) | 2 (40.0) | |
| Sigmoid colon (n = 10) | 9 (90.0) | 1 (10.0) | |
| Rectum (n = 27) | 22 (81.5) | 5 (18.5) | |
| PatientNo. | Gender | Age(yr) | Stage | Site | p53 | HPV16 | ||
| Mutation | IHC | ISH | E6 | |||||
| 101 | F | 65 | I | d-colon | No | - | + | + |
| 134 | F | 54 | IIA | Rectum | No | + | + | + |
| 141 | M | 72 | I | Rectum | No | - | + | + |
| 142 | F | 56 | IIA | d-colon | Yes | ++ | + | + |
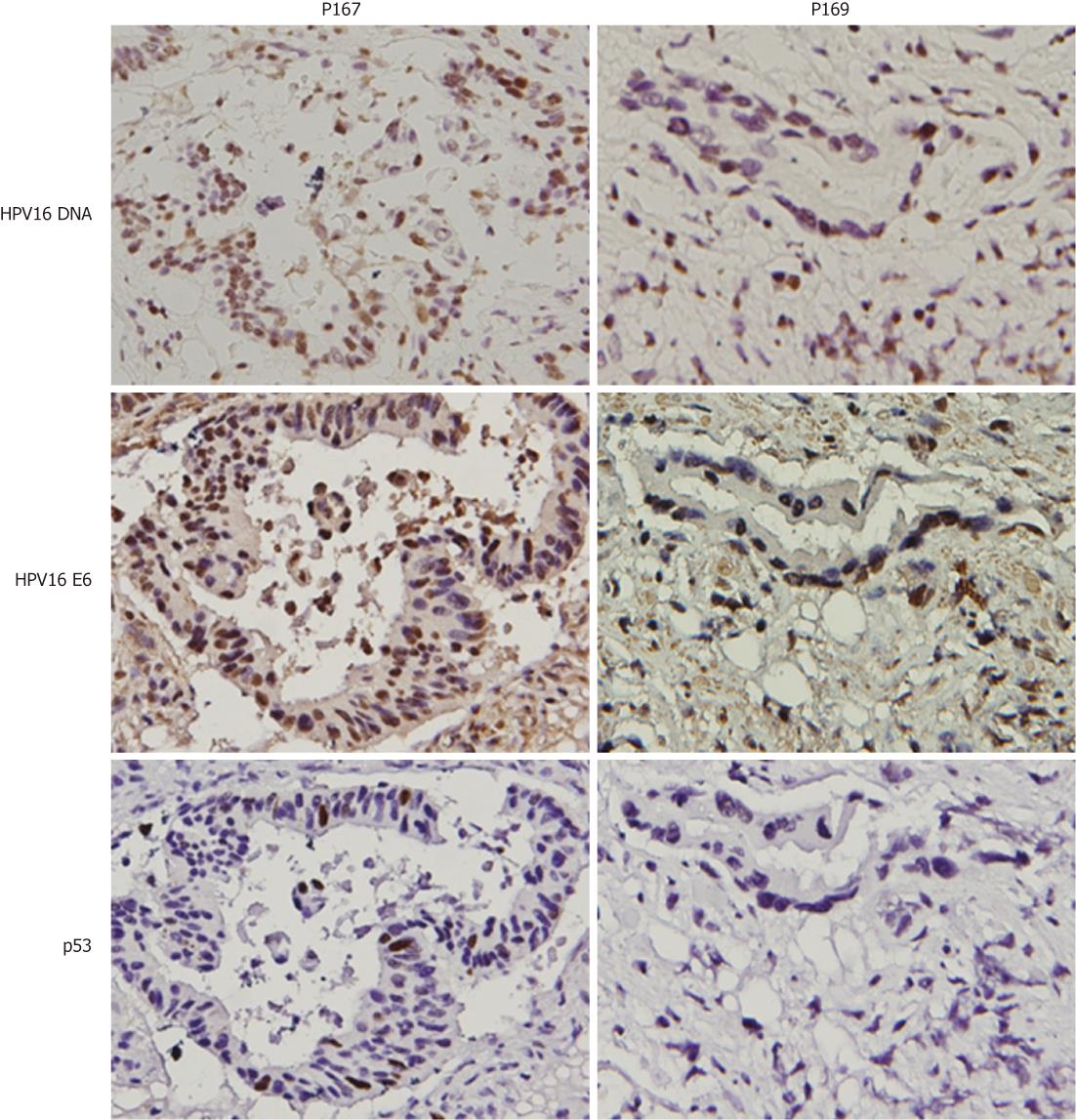
| 167 | M | 70 | IIIB | d-colon | Yes | +++ | + | + |
| 169 | M | 78 | IV | Rectum | No | - | + | + |
| 173 | F | 83 | IIA | s-colon | Yes | + | + | + |
| 200 | M | 74 | IV | a-colon | Yes | + | + | + |
| 226 | F | 78 | I | Rectum | No | - | - | - |
| 234 | F | 76 | I | a-colon | No | + | - | - |
| 259 | F | 79 | IIIB | Rectum | Yes | ++ | - | - |
To explore whether HPV16 infection could be linked with colorectal cancer development, HPV16 E6 oncoprotein was evaluated by IHC in HPV16 DNA-positive colorectal tumors. Our data showed that E6 oncoprotein expression was detected in all HPV16 DNA-positive tumors (Table 2). We next examined whether E6 oncoprotein expressed in four p53 wild-type colorectal tumors (P101, P134, P141 and P169) could degrade p53 protein to produce tumors with p53 negative immunostaining (Figure 2). IHC analysis clearly showed p53 negative immunostaining in three out of four colorectal tumors (P101, P141, and P169), but one tumor (P134) had p53 positive immunostaining (< 10%). The other four E6-positive tumors with p53 mutations (P142, P167, P173 and P200) still had p53 positive immunostaining (Figure 2). To explore whether p53 inactivation could have occurred in p53 wild-type E6-positive tumors, expression levels of the p53-downstream targets Mdm2 and p21 mRNA were decreased compared with adjacent normal tissues. Real-time RT-PCR analysis showed that Mdm2 and p21 mRNA expression levels in these E6-positive tumors with p53 wild-type were significantly lower than in their adjacent normal tissues (P = 0.025 for p21 mRNA; P = 0.032 for Mdm2 mRNA; Table 3). As expected, both gene mRNA expression levels in E6-positive tumors with p53 mutations were lower than in the adjacent normal tissues (P = 0.043 for Mdm2 mRNA); however, p21 mRNA levels were marginally different between tumors and adjacent normal tissues (P = 0.061). These results clearly indicate that E6 oncoprotein is expressed in HPV16 DNA-positive colorectal tumors and may be linked with p53 inactivation in these HPV16 DNA positive tumors, which had the p53 wild-type gene.
| Variable | E6-positive (n = 8) | |||
| p53 wild-type (n = 4) | P | p53-mutation (n = 4) | P | |
| P21 mRNA | 0.025 | 0.061 | ||
| Adjacent normal tissues | 348.46 ± 143.89 | 241.12 ± 84.13 | ||
| Tumor tissues | 49.62 ± 16.58 | 122.96 ± 45.28 | ||
| Mdm2 mRNA | 0.032 | 0.043 | ||
| Adjacent normal tissues | 563.74 ± 252.28 | 541.50 ± 100.56 | ||
| Tumor tissues | 111.60 ± 75.99 | 317.74 ± 137.59 | ||
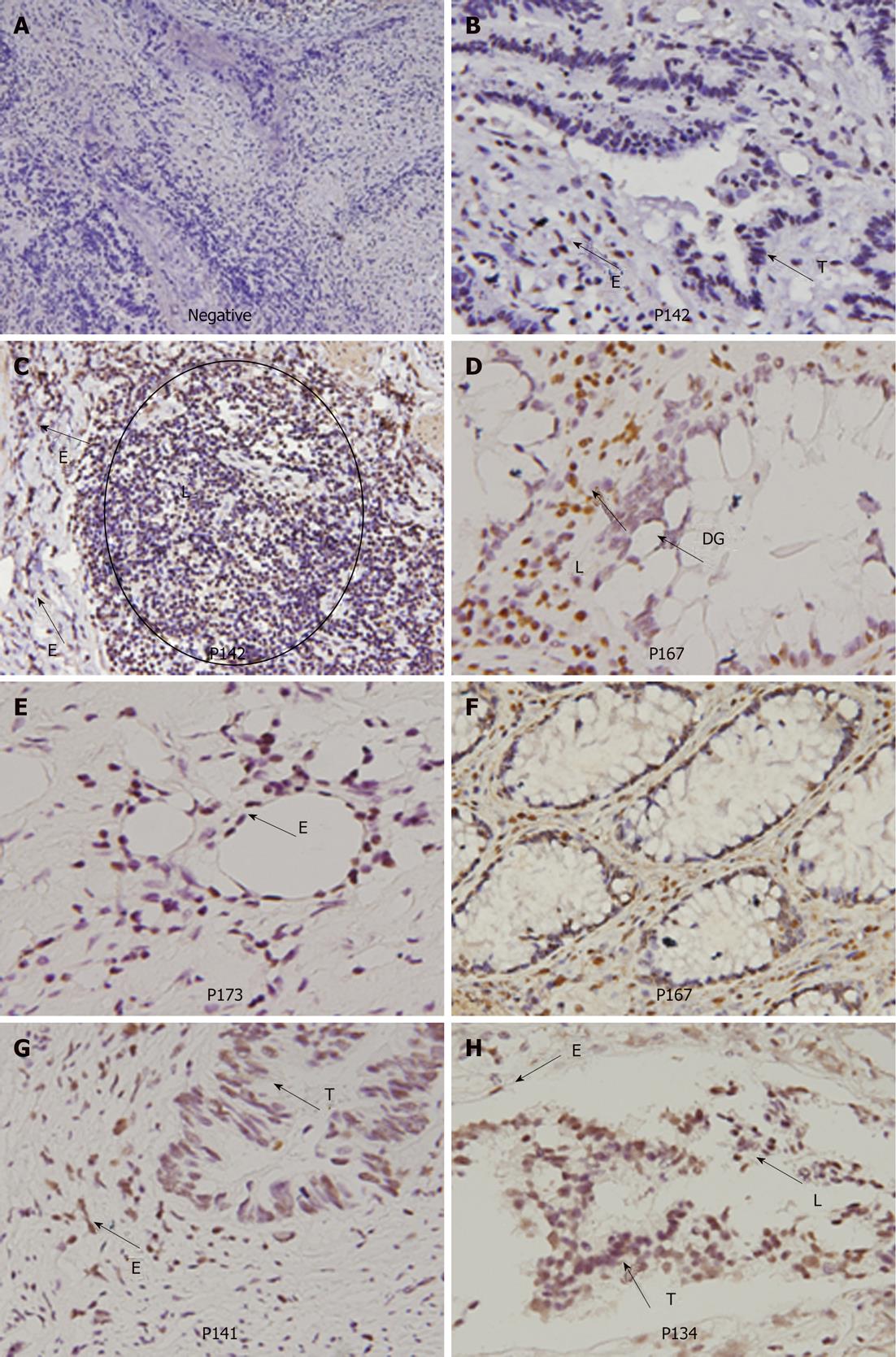
To understand whether HPV-infected colorectal tumors could be mediated through blood circulation, E6 oncoprotein expression in normal parts of the colorectal tumors were examined by IHC (Figure 3). Our data showed that E6 oncoprotein is indeed expressed in endothelial cells of blood vessels and in lymphocytes infiltrating colorectal tumors. In addition, E6 oncoprotein was expressed in normal glands and dysplastic glands in colorectal tumors (Figure 3). These results seem to support the possibility that HPV16 infection in colorectal tumors may occur partially through blood circulation.
The presence of HPV is commonly detected by nested PCR in human tumors including colorectal tumors. In previous studies, only one report has shown the concomitant detection of HPV16 DNA in three colorectal tumors by nested-PCR and in situ PCR[15]. In the present study, nested PCR detected 8 of 11 HPV16-positive colorectal tumors and ISH confirmed this finding as well. Detection of HPV DNA by ISH had markedly lower sensitivity than in situ PCR, suggesting that the HPV16 DNA copy number in the tumors studied herein could be higher than previously reported[19]. In the current study, the association between HPV infection and tumor stage was observed; however, only 6 stage I patients were enrolled in this study. Since the sample size of stage I patients is so small, it might be biased to conclude that HPV infection might play a role in the development of colorectal cancer. In addition, among 4 of the 6 stage I tumors with HPV16 DNA, HPV infection was confirmed by ISH in only 2 tumors. Therefore, the association between HPV infection and tumor stage should be verified by a larger study population.
To explore whether the existence of HPV16 DNA could be involved in colorectal cancer development, the presence of E6 and p53 proteins in tumor tissues and p21 and Mdm2 mRNA expression in tumors and adjacent normal tissues were evaluated in a subset of colorectal tumors (11 of 69, 16%). E6 oncoprotein expression was detected in 8 of the 11 HPV16 DNA-positive tumors. Among these eight E6-positive tumors, p21 and Mdm2 mRNA expression levels were markedly decreased in four E6-positive tumors carrying the wild-type p53 gene; however, no significant decrease in both gene mRNA expression levels was noted in the other four E6-positive tumors carrying the mutated p53 gene. Therefore, these results clearly show that E6 oncoprotein may be involved in this small subset of colorectal cancer development via the inactivated p53 pathway (Table 3).
Sexual activity has been considered to be a major route of transmission for HPV resulting in genital cancers, such as cervical, anal, and oropharyngeal cancers[1,2]. However, there is evidence of HPV infections in infants and female university students who are virgins, revealing that HPV transmission via other routes than sexual intercourse may exist[3,13,14,20-25]. In addition, peripheral blood lymphocytes (PBLs) from healthy donors have been shown to be infected with HPV[13]. Therefore, it has been suggested that HPV infection in internal organ tissues might occur through blood circulation. Our previous lung cancer studies have indicated that HPV16/18 DNA and E6 oncoprotein not only exist in lung tumors but are also expressed in adjacent normal tissues including lymphocytes, endothelial cells, macrophages, and bronchial epithelial cells[26]. We therefore speculated that PBLs may first be infected by HPV to act as a mediator of HPV infection to lung tissues via blood circulation[14,26]. In the present study, IHC analysis clearly indicates that E6 oncoprotein was expressed in tumor-infiltrating lymphocytes of HPV16 DNA-positive colorectal tumors. In addition, E6 oncoprotein expression was also detected in the endothelial cells of the HPV16 DNA-positive colorectal tumors. These results seem to support our previous hypothesis that HPV infection in lung tumor tissues may be mediated through blood circulation and not as a direct contact transmission in cervical and oropharyngeal tumor tissues[14,19,26].
To our knowledge, this is the first report to indicate that HPV16 E6 oncoprotein may downregulate p21 and Mdm2 transcription via inactivation of p53 in the involvement of colorectal cancer development. Similar observations of E6 oncoprotein expression in adjacent normal lung and colorectal tissue cells seem to support the possibility that HPV infection in colorectal tumors might be mediated through blood circulation. Notably, even though our present study provides support for the association between HPV infection and colorectal cancer, the involvement of HPV infection in colorectal cancer development is limited to a small subset of the population.
Human papilloma virus (HPV) DNA integration into the host chromosome plays a crucial role in HPV-associated tumorigenesis. When HPV DNA was spliced at E2, E6 and E7 oncoproteins were expressed to inactivate p53 and Rb pathways. A high risk of HPV16/18 infection has been documented to be involved in the development of cervical and anal genital cancers. Among non-genital cancers, the association of oropharyngeal cancer with HPV16/18 infection was recently evidenced. However, other non-genital cancers, including lung, breast, and colorectal cancers have not yet been identified.
The authors provide the evidence to indicate that HPV16 may be involved in a small subset of colorectal cancer development. Therefore, HPV vaccination not only prevents cervical cancer but also reduces HPV-associated colorectal cancer development.
The association of HPV infection with colorectal cancer has been extensively investigated; however, no strong evidence supports the involvement of HPV in colorectal cancer development. Herein, nested-polymerase chain reaction and in situ hybridization were used to detect the presence of HPV16 DNA in colorectal tumors. Immunohistochemical data further showed that E6 oncoprotein is expressed in HPV16 DNA-positive tumors and E6 expression was negatively correlated with p53 expression. These results suggest that HPV16 might contribute to a small subset of colorectal cancer development.
HPV infection is not only involved in cervical and oropharyngeal cancers but is also linked with internal organ cancers, such as lung and colorectal cancers. Therefore, blood transmission of HPV infection in internal organs might be noted in public.
The authors present an interesting work on the expression of HPV16 E6 oncoprotein in colorectal cancer. The manuscript is generally well written. The topic is very interesting and to some extent provocative. The manuscript is well structured and the cited literature is comprehensive and up-to-date.
| 1. | zur Hausen H. Viruses in human cancers. Eur J Cancer. 1999;35:1878-1885. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 52] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 2. | Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 3. | Veldhuijzen NJ, Snijders PJ, Reiss P, Meijer CJ, van de Wijgert JH. Factors affecting transmission of mucosal human papillomavirus. Lancet Infect Dis. 2010;10:862-874. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 116] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 4. | Syrjänen S. Human papillomavirus (HPV) in head and neck cancer. J Clin Virol. 2005;32 Suppl 1:S59-S66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 287] [Cited by in RCA: 268] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 5. | Cheng JY, Sheu LF, Meng CL, Lee WH, Lin JC. Detection of human papillomavirus DNA in colorectal carcinomas by polymerase chain reaction. Gut. 1995;37:87-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 51] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 6. | Gornick MC, Castellsague X, Sanchez G, Giordano TJ, Vinco M, Greenson JK, Capella G, Raskin L, Rennert G, Gruber SB. Human papillomavirus is not associated with colorectal cancer in a large international study. Cancer Causes Control. 2010;21:737-743. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 49] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 7. | Li N, Bi X, Zhang Y, Zhao P, Zheng T, Dai M. Human papillomavirus infection and sporadic breast carcinoma risk: a meta-analysis. Breast Cancer Res Treat. 2011;126:515-520. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 65] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 8. | Baltzell K, Buehring GC, Krishnamurthy S, Kuerer H, Shen HM, Sison JD. Limited evidence of human papillomavirus in [corrected] breast tissue using molecular in situ methods. Cancer. 2012;118:1212-1220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 9. | Li YJ, Tsai YC, Chen YC, Christiani DC. Human papilloma virus and female lung adenocarcinoma. Semin Oncol. 2009;36:542-552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 10. | Klein F, Amin Kotb WF, Petersen I. Incidence of human papilloma virus in lung cancer. Lung Cancer. 2009;65:13-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 111] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 11. | Uronis HE, Bendell JC. Anal cancer: an overview. Oncologist. 2007;12:524-534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 103] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 12. | Hoots BE, Palefsky JM, Pimenta JM, Smith JS. Human papillomavirus type distribution in anal cancer and anal intraepithelial lesions. Int J Cancer. 2009;124:2375-2383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 305] [Cited by in RCA: 325] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 13. | Chen AC, Keleher A, Kedda MA, Spurdle AB, McMillan NA, Antonsson A. Human papillomavirus DNA detected in peripheral blood samples from healthy Australian male blood donors. J Med Virol. 2009;81:1792-1796. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 56] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 14. | Chiou HL, Wu MF, Liaw YC, Cheng YW, Wong RH, Chen CY, Lee H. The presence of human papillomavirus type 16/18 DNA in blood circulation may act as a risk marker of lung cancer in Taiwan. Cancer. 2003;97:1558-1563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 15. | Bodaghi S, Yamanegi K, Xiao SY, Da Costa M, Palefsky JM, Zheng ZM. Colorectal papillomavirus infection in patients with colorectal cancer. Clin Cancer Res. 2005;11:2862-2867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 109] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 16. | Yavuzer D, Karadayi N, Salepci T, Baloglu H, Dabak R, Bayramicli OU. Investigation of human papillomavirus DNA in colorectal carcinomas and adenomas. Med Oncol. 2011;28:127-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 30] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 17. | Pérez LO, Abba MC, Laguens RM, Golijow CD. Analysis of adenocarcinoma of the colon and rectum: detection of human papillomavirus (HPV) DNA by polymerase chain reaction. Colorectal Dis. 2005;7:492-495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 59] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 18. | zur Hausen H. Papillomaviruses causing cancer: evasion from host-cell control in early events in carcinogenesis. J Natl Cancer Inst. 2000;92:690-698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 897] [Cited by in RCA: 862] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 19. | Cheng YW, Chiou HL, Sheu GT, Hsieh LL, Chen JT, Chen CY, Su JM, Lee H. The association of human papillomavirus 16/18 infection with lung cancer among nonsmoking Taiwanese women. Cancer Res. 2001;61:2799-2803. [PubMed] |
| 20. | Cason J, Mant CA. High-risk mucosal human papillomavirus infections during infancy & amp; childhood. J Clin Virol. 2005;32 Suppl 1:S52-S58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 56] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 21. | Smith EM, Ritchie JM, Yankowitz J, Swarnavel S, Wang D, Haugen TH, Turek LP. Human papillomavirus prevalence and types in newborns and parents: concordance and modes of transmission. Sex Transm Dis. 2004;31:57-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 68] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 22. | Winer RL, Lee SK, Hughes JP, Adam DE, Kiviat NB, Koutsky LA. Genital human papillomavirus infection: incidence and risk factors in a cohort of female university students. Am J Epidemiol. 2003;157:218-226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 675] [Cited by in RCA: 622] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 23. | Bodaghi S, Wood LV, Roby G, Ryder C, Steinberg SM, Zheng ZM. Could human papillomaviruses be spread through blood? J Clin Microbiol. 2005;43:5428-5434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 86] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 24. | Müller M, Gissmann L, Cristiano RJ, Sun XY, Frazer IH, Jenson AB, Alonso A, Zentgraf H, Zhou J. Papillomavirus capsid binding and uptake by cells from different tissues and species. J Virol. 1995;69:948-954. [PubMed] |
| 25. | Mercer J, Helenius A. Virus entry by macropinocytosis. Nat Cell Biol. 2009;11:510-520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 585] [Cited by in RCA: 636] [Article Influence: 37.4] [Reference Citation Analysis (0)] |
| 26. | Cheng YW, Wu MF, Wang J, Yeh KT, Goan YG, Chiou HL, Chen CY, Lee H. Human papillomavirus 16/18 E6 oncoprotein is expressed in lung cancer and related with p53 inactivation. Cancer Res. 2007;67:10686-10693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 99] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
Peer reviewer: Michael Linnebacher, PhD, Department of Molecular Oncology and Immunotherapy, Section of General Surgery, Rostock University Hospital, Schillingallee 35, 18055 Rostock, Germany
S- Editor Lv S L- Editor O’Neill M E- Editor Xiong L