Published online Jul 28, 2012. doi: 10.3748/wjg.v18.i28.3727
Revised: November 21, 2010
Accepted: May 12, 2012
Published online: July 28, 2012
AIM: To establish methods for quantitative polymerase chain reaction (PCR) for hepcidin using RNAs isolated from paraffin-embedded sections and in situ hybridization of hepatocellular carcinoma (HCC).
METHODS: Total RNA from paraffin-embedded sections was isolated from 68 paraffin-embedded samples of HCC. Samples came from 54 male and 14 female patients with a mean age of 66.8 ± 7.8 years. Quantitative PCR was performed. Immunohistochemistry and in situ hybridization for hepcidin were also performed.
RESULTS: Quantitative PCR for hepcidin using RNAs isolated from paraffin-embedded sections of HCC was performed successfully. The expression level of hepcidin mRNA in cancer tissues was significantly higher than that in non-cancer tissues. A method of in situ hybridization for hepcidin was established successfully, and this demonstrated that hepcidin mRNA was expressed in non-cancerous tissue but absent in cancerous tissue.
CONCLUSION: We have established novel methods for quantitative PCR for hepcidin using RNAs isolated from paraffin-embedded sections and in situ hybridization of HCC.
-
Citation: Sakuraoka Y, Sawada T, Shiraki T, Park K, Sakurai Y, Tomosugi N, Kubota K. Analysis of hepcidin expression:
In situ hybridization and quantitative polymerase chain reaction from paraffin sections. World J Gastroenterol 2012; 18(28): 3727-3731 - URL: https://www.wjgnet.com/1007-9327/full/v18/i28/3727.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i28.3727
Hepcidin is a key regulator of iron metabolism, binding to the iron-exporter ferroportin and triggering internalization and degradation[1]. Because both iron deficiency and overload are toxic for the human body, iron concentration is tightly regulated, and hepcidin plays a major role in this process.
Hepcidin was discovered initially as an anti-microbial peptide[2,3], and a recent study has indicated that hepcidin functions as an acute inflammatory molecule[4]. Furthermore, the roles of hepcidin in other diseases, including cancer, have been extensively studied[5,6]. We have previously reported that the expression of hepcidin mRNA is suppressed in hepatocellular carcinoma (HCC)[7]. In the present study, we isolated RNAs from fresh surgical specimens. There have been no previous reports describing the isolation of RNA from paraffin-embedded sections and quantitative reverse-transcriptional polymerase chain reaction (RT-PCR). It would be valuable if quantitative RT-PCR could be performed using RNAs isolated from paraffin-embedded sections. However, it is well known that hepcidin is a difficult molecule to analyze in this way. Here we report a simple way to isolate hepcidin RNA for RT-PCR from paraffin-embedded sections, and also in situ hybridization for hepcidin.
A total of 68 paraffin-embedded samples of HCC were used in this study. The sections were taken from areas that were clearly in the border zone between cancerous and non-cancerous tissues. The mean age of the patients was 66.8 ± 7.8 years, and there were 54 males and 14 females. Liver cirrhosis was observed in 42 patients and chronic hepatitis was diagnosed in 21; only 5 patients lacked chronic hepatitis or liver cirrhosis. Tumor stage was determined based on the classification of the Liver Cancer Study Group of Japan.
Isolation of total RNA from paraffin-embedded sections was performed using an Agencourt Formapure Kit (Beckman-Coulter, Beverly, MA). Briefly, tissue sections were moistened with 20 μL of lysis buffer, and cancerous and non-cancerous parts were separately cut off with a scalpel and transferred to a 96-well plate (IWAKI, Tokyo, Japan). The removed samples were heated at 72 °C for 60 min, then 20 μL proteinase K was added, followed by incubation at 55 °C for 60 min. After 2 min of cooling, the lysates were mixed with 150 μL of Bind I buffer and 320 μL of Bind II buffer, and incubated at 55 °C for 5 min. The plate containing the digests was placed on a magnetic sheet for 5 min, and then the supernatants were removed. The RNA and bead complexes were washed twice, and the complexes were resuspended with 70% ethanol. The plate was then placed back on the magnetic sheet and the supernatant was again removed. Then, the plate was removed from the magnetic sheet and 100 μL of DNase solution was added. After 15 min of incubation at 37 °C, 550 μL of Wash Buffer was added and the plate was left at room temperature for 5 min. It was then placed again on the magnetic sheet for 10 min, and the supernatant was again removed. The plate was then removed from the magnetic sheet and 750 μL of 70% ethanol was added, followed by replacement on the magnetic sheet. After 5 min, the supernatant was removed, the plate was taken from the magnetic sheet, and the complex was resuspended in 500 μL of 90% isopropanol. The plate was then incubated at 70 °C for 3 min, followed by removal from the magnetic sheet and aspiration of the supernatant. The complex was resuspended in 750 μL of 70% ethanol and the plate was placed back on the magnetic sheet for 5 min, followed by removal of the supernatant. The complex was then air-dried for 10 min and resuspended in 80 μL nuclease-free water. The plate was then incubated at 65 °C for 30 s, placed back on the magnetic sheet for 1 min, and the supernatant containing RNA was obtained.
Reverse transcription reactions were performed using a Rever Tra Ace α-First Strand cDNA Synthesis Kit (Toyobo, Osaka, Japan). Briefly, 1 μg of total RNA, oligo dT-primer, and dNTPs were incubated at 65 °C for 5 min, then 10 μL of a cDNA synthesis mixture was added and the mixture was incubated at 50 °C for 50 min. The reaction was terminated by adding 1 μL of RNaseH and incubating the mixture at 37 °C for 20 min.
Real-time PCR was performed with an ABI Prism 7700 sequence detector (Applied Biosystems, Warrington, United Kingdom). The PCR reaction was carried out in a final volume of 2 μL cDNA, 12.5 μL 2 × SYBR Green (Applied Biosystems), 0.5 μL of 25 nmol/L sense and antisense primers, and H2O up to 25 μL. The PCR conditions consisted of 40 cycles at 95 °C for 30 s and 60 °C for 30 s. The sequences of the primers were as follows: GAPDH: sense-primer 5’-CCACCCAGAAGACTGTGGAT-3’, anti-sense 5’-TTCAGCTCAGGGATGACCTT-3’; β-actin: sense-primer 5’-GTCGTACCACTGGCATTGTG-3’, anti-sense 5’-CCATCTCTTGCTCGAAGTCC-3’; hepcidin 1: sense-primer 5’-CACAACAGACGGGACAACTT-3’, anti-sense 5’-CGCAGCAGAAAATGCAGATG-3’; hepcidin 2: sense-primer 5’-GACCAGTGGCTCTGTTTTCC-3’, anti-sense 5’-CACATCCCACACTTTGATCG-3’. The primer sets for GAPDH, β-actin, and hepcidin 2 were designed using Primer 3 software (ver. 4.0), and that for hepcidin 1 was based on the previous report[8].
The level of expression was calculated using the formula: Relative expression (t) = (Copy number of target molecule/Copy number of GAPDH)[8]. Samples were assayed in triplicate. Means and standard deviations were calculated from the data obtained. The t value was calculated from the mean of three different assays.
The tissues were fixed with Tissue Fixative (Genostaff), and then embedded in paraffin, and sectioned at 6 μm. Tissue sections were deparaffinized with xylene and rehydrated through an ethanol series and Tris-buffered saline (TBS). Antigen retrieval was performed by microwave treatment for 10 min at 500 W in 1 mmol/L ethylenediaminetetracetic acid (EDTA) buffer, pH 9.0. Endogenous peroxidase was blocked with 3% H2O2 in methanol for 15 min, followed by incubation with Protein Block (DAKO). The sections were incubated with anti-hepcidin-25 antibody (HEPC13-S, Alpha Diagnostic International, San Antonio, TX) at 4 °C overnight. After washing with TBS, the sections were treated with a Biotin Blocking System (DAKO) and biotin-conjugated goat anti-rabbit Ig (DAKO) diluted 1:600, for 30 min at room temperature, followed by addition of peroxidase-conjugated streptavidin (Nichirei) for 5 min. Peroxidase activity was visualized using diaminobenzidine. The sections were counterstained with Mayer’s hematoxylin (Muto), dehydrated, and then mounted with Malinol (Muto). Some sections were also stained with hematoxylin-eosin.
The tissue sections were dewaxed with xylene and rehydrated through an ethanol series and phosphate buffered saline (PBS). The sections were then fixed with 4% paraformaldehyde in PBS for 15 min and washed with PBS. The sections were treated with 15 μg/mL proteinase K in PBS for 30 min at 37 °C, washed with PBS, refixed with 4% paraformaldehyde in PBS, washed again with PBS, and placed in 0.2 mol/L HCl for 10 min, followed by acetylation by incubation in 0.1 mol/L triethanolamine-HCl, pH 8.0, and 0.25% acetic anhydride for 10 min. After a wash with PBS, the sections were dehydrated through an ethanol series. Hybridization was performed with digoxigenin-labeled RNA probes at a concentration of 100 ng/mL in Probe Diluent (Genostaff Co., Ltd., Tokyo, Japan) at 60 °C for 16 h. After hybridization, the sections were washed in 5 × HybriWash (Genostaff Co., Ltd.), equal to 5 × SSC, at 60 °C for 20 min and then in 50% formamide, 2 × HybriWash at 60 °C for 20 min, followed by RNase treatment in 50 μg/mL RNaseA in 10 mmol/L Tris-HCl, pH 8.0, 1 mol/L NaCl and 1 mmol/L EDTA. Then the sections were washed twice with 2 × HybriWash at 60 °C for 20 min, twice with 0.2 × HybriWash at 60 °C for 20 min and once with TBS containing 0.1% Tween 20 (TBST). After treatment with 0.5% blocking reagent (Roche) in TBST for 30 min, the sections were incubated with anti-DIG alkaline phosphatase (AP) conjugate (Roche) diluted 1: 1000 with TBST for 2 h. The sections were washed twice with TBST and then incubated in 100 mmol/L NaCl, 50 mmol/L MgCl2, 0.1% Tween 20, and 100 mmol/L Tris-HCl, pH 9.5. Coloring reactions were performed with BM purple AP substrate (Roche) overnight and then washed with PBS. The sections were counterstained with Kemechtrot stain solution (Muto), dehydrated, and then mounted with Malinol (Muto). The DNA fragment used for the probes was a 246-bp fragment corresponding to nucleotide positions 72-317 of human hepcidin (GenBank accession number NM_021175).
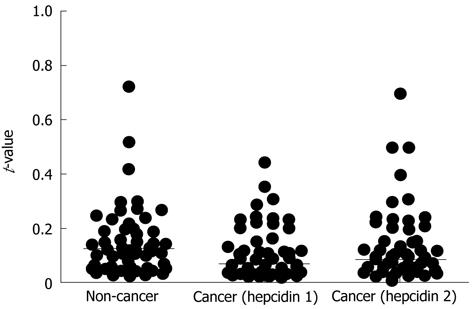
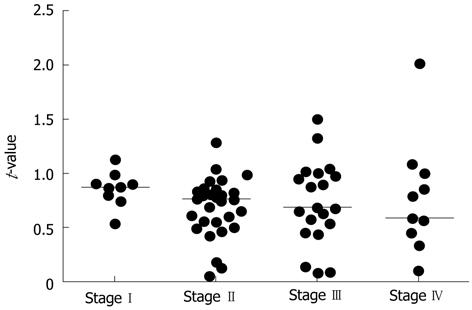
Using the method described in Materials and Methods, quantitative RT-PCR was performed for all samples. The median t-values for non-cancer and cancer tissues with the hepcidin 1 and hepcidin 2 primers were 0.124 (0.021-3.433), 0.067 (0.009-2.063), and 0.0825 (0.007-2.1), respectively. The t-values for cancer tissues were significantly higher than those for non-cancer tissues (P = 0.0057) (Figure 1). The assays were repeated using primers for β-actin, and the results were the same as those shown in Figure 1. Figure 2 shows the ratio of t-values for non-cancer to cancer tissues according to tumor stage. The median ratio of t-values for stages I, II, III and IV were 0.874 (0.537-1.127), 0.754 (0.052-1.286), 0.678 (0.069-1.494), and 0.579 (0.090-2.013), respectively (P = 0.3381; one-way analysis of variance).
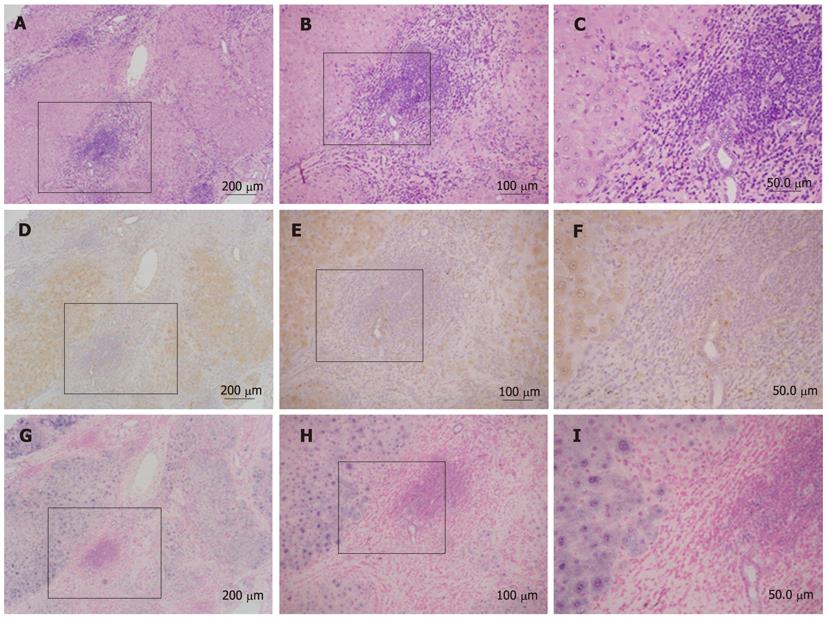
Figure 3 shows a representative result of in situ hybridization for hepcidin. Positive staining for hepcidin was observed in non-cancerous tissues by immunohistochemistry (Figure 3D-F). Accordingly, by in situ hybridization, hepcidin mRNA was expressed in non-cancerous tissue but was absent in cancerous tissue (Figure 3G-I).
As we have reported previously, hepcidin gene expression was decreased in HCC[7]. In the previous study, we extracted RNAs from freshly resected surgical specimens. This time, however, we aimed to investigate hepcidin gene expression using RNAs obtained from paraffin-embedded sections. However, for reasons that are unclear, this was not an easy process. We finally succeeded in isolating hepcidin RNA using the method described here. The expression of hepcidin mRNA was lower in cancerous than in non-cancerous tissues in most of the samples, and although not statistically significant, the ratios of the level of hepcidin mRNA in non-cancer to cancer tissues tended to be lower in poorly differentiated than in well differentiated HCCs.
Visualizing the expression of hepcidin is very important when analyzing which types of cells express or do not express the molecule. For this purpose, in situ hybridization is an ideal approach, but so far no studies have succeeded with in situ hybridization for hepcidin. As shown in Figure 3, hepcidin mRNA was clearly detected in non-cancerous tissue, and not in cancerous tissue. Thus, the present in situ hybridization technique should contribute to investigations of the role of hepcidin in HCC.
Hepcidin is a key regulator of iron metabolism, binding to the iron-exporter ferroportin and triggering internalization and degradation. Hepcidin functions as an acute inflammatory molecule. The authors have previously reported that the expression of hepcidin mRNA is suppressed in hepatocellular carcinoma (HCC). In the present study, the authors isolated RNAs from fresh surgical specimens. There have been no previous reports describing the isolation of RNA from paraffin-embedded sections and quantitative reverse-transcriptional polymerase reaction (RT-PCR). It would be valuable if quantitative RT-PCR could be performed using RNAs isolated from paraffin-embedded sections. However, it is well known that hepcidin is a difficult molecule to analyze in this way. In the present study, the authors devised a simple way to isolate hepcidin RNA for RT-PCR from paraffin-embedded sections, and also performed in situ hybridization for hepcidin.
There have been no previous reports of isolation of RNA from paraffin-embedded sections and quantitative RT-PCR. It would be valuable if quantitative RT-PCR could be performed using RNAs isolated from paraffin-embedded sections.
The authors established novel methods for quantitative PCR for hepcidin using RNAs isolated from paraffin-embedded sections and in situ hybridization of HCC.
The methods described in this paper will help to clarify the role of hepcidin in various disorders.
This technical report relates to the analysis of hepcidin expression in the liver tissues of paraffin sections from patients with hepatocellular carcinoma using in situ hybridization and quantitative polymerase chain reaction. It is a nice technical note and well written, and should be relevant to other investigators in general.
| 1. | Hentze MW, Muckenthaler MU, Galy B, Camaschella C. Two to tango: regulation of Mammalian iron metabolism. Cell. 2010;142:24-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1714] [Cited by in RCA: 1576] [Article Influence: 98.5] [Reference Citation Analysis (0)] |
| 2. | Krause A, Neitz S, Mägert HJ, Schulz A, Forssmann WG, Schulz-Knappe P, Adermann K. LEAP-1, a novel highly disulfide-bonded human peptide, exhibits antimicrobial activity. FEBS Lett. 2000;480:147-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 890] [Cited by in RCA: 898] [Article Influence: 34.5] [Reference Citation Analysis (0)] |
| 3. | Park CH, Valore EV, Waring AJ, Ganz T. Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J Biol Chem. 2001;276:7806-7810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1500] [Cited by in RCA: 1530] [Article Influence: 61.2] [Reference Citation Analysis (0)] |
| 4. | De Domenico I, Zhang TY, Koening CL, Branch RW, London N, Lo E, Daynes RA, Kushner JP, Li D, Ward DM. Hepcidin mediates transcriptional changes that modulate acute cytokine-induced inflammatory responses in mice. J Clin Invest. 2010;120:2395-2405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 151] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 5. | Ward DG, Roberts K, Brookes MJ, Joy H, Martin A, Ismail T, Spychal R, Iqbal T, Tselepis C. Increased hepcidin expression in colorectal carcinogenesis. World J Gastroenterol. 2008;14:1339-1345. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 74] [Cited by in RCA: 81] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 6. | Tseng HH, Chang JG, Hwang YH, Yeh KT, Chen YL, Yu HS. Expression of hepcidin and other iron-regulatory genes in human hepatocellular carcinoma and its clinical implications. J Cancer Res Clin Oncol. 2009;135:1413-1420. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 54] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 7. | Kijima H, Sawada T, Tomosugi N, Kubota K. Expression of hepcidin mRNA is uniformly suppressed in hepatocellular carcinoma. BMC Cancer. 2008;8:167. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 66] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 8. | Okada T, Sawada T, Osawa T, Adachi M, Kubota K. MK615 inhibits pancreatic cancer cell growth by dual inhibition of Aurora A and B kinases. World J Gastroenterol. 2008;14:1378-1382. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 23] [Cited by in RCA: 21] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
Peer reviewers: Matias A Avila, Professor and Senior Staff Scientist, Division of hepatology and gene therapy, University of Navarra, Avda. Pio XII, n55, Pamplona 31008, Spain; Xian-Ming Chen, Associate Professor, MD, Department of Medical Microbiology and Immunology, Creighton University, 2500 California Plaza, Omaha, NE 68178, United States
S- Editor Cheng JX L- Editor O’Neill M E- Editor Zhang DN