Published online Apr 21, 2012. doi: 10.3748/wjg.v18.i15.1732
Revised: September 20, 2011
Accepted: September 27, 2011
Published online: April 21, 2012
AIM: To investigate the influence of macrophages on hepatocyte phenotype and function.
METHODS: Macrophages were differentiated from THP-1 monocytes via phorbol myristate acetate stimulation and the effects of monocyte or macrophage-conditioned medium on HepG2 mRNA and protein expression determined. The in vivo relevance of these findings was confirmed using liver biopsies from 147 patients with hepatitis C virus (HCV) infection.
RESULTS: Conditioned media from macrophages, but not monocytes, induced a transient morphological change in hepatocytes associated with upregulation of vimentin (7.8 ± 2.5-fold, P = 0.045) and transforming growth factor (TGF)-β1 (2.6 ± 0.2-fold, P < 0.001) and downregulation of epithelial cadherin (1.7 ± 0.02-fold, P = 0.017) mRNA expression. Microarray analysis revealed significant upregulation of lipocalin-2 (17-fold, P < 0.001) and pathways associated with inflammation, and substantial downregulation of pathways related to hepatocyte function. In patients with chronic HCV, real-time polymerase chain reaction and immunohistochemistry confirmed an increase in lipocalin-2 mRNA (F0 1.0 ± 0.3, F1 2.2 ± 0.2, F2 3.0 ± 9.3, F3/4 4.0 ± 0.8, P = 0.003) and protein expression (F1 1.0 ± 0.5, F2 1.3 ± 0.4, F3/4 3.6 ± 0.4, P = 0.014) with increasing liver injury. High performance liquid chromatography-tandem mass spectrometry analysis identified elevated levels of matrix metalloproteinase (MMP)-9 in macrophage-conditioned medium, and a chemical inhibitor of MMP-9 attenuated the change in morphology and mRNA expression of TGF-β1 (2.9 ± 0.2 vs 1.04 ± 0.1, P < 0.001) in macrophage-conditioned media treated HepG2 cells. In patients with chronic HCV infection, hepatic mRNA expression of CD163 (F0 1.0 ± 0.2, F1/2 2.8 ± 0.3, F3/4 5.3 ± 1.0, P = 0.001) and MMP-9 (F0 1.0 ± 0.4, F1/2 2.8 ± 0.3, F3/4 4.1 ± 0.8, P = 0.011) was significantly associated with increasing stage of fibrosis.
CONCLUSION: Secreted macrophage products alter the phenotype and function of hepatocytes, with increased expression of inflammatory mediators, suggesting that hepatocytes actively participate in liver injury.
- Citation: Melino M, Gadd VL, Walker GV, Skoien R, Barrie HD, Jothimani D, Horsfall L, Jones A, Sweet MJ, Thomas GP, Clouston AD, Jonsson JR, Powell EE. Macrophage secretory products induce an inflammatory phenotype in hepatocytes. World J Gastroenterol 2012; 18(15): 1732-1744
- URL: https://www.wjgnet.com/1007-9327/full/v18/i15/1732.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i15.1732
Most of the morbidity/mortality from chronic liver diseases occurs in subjects with advanced fibrosis or cirrhosis, who are at risk of developing complications of end-stage liver disease including hepatocellular cancer. Activated liver macrophages have a key role in the progression of liver injury and repair, and knowledge about their interaction with hepatocytes and other cells in the liver microenvironment may provide new targets for antifibrotic therapy. In experimental models of liver disease, an increase in the number of macrophages correlates with the extent of injury, and damage can be attenuated by depletion of these cells[1-6]. Similarly, in diseased human liver, particularly chronic viral hepatitis, there is an increase in the density and size of macrophages[7,8]. This is seen largely at inflammatory sites with prominent mononuclear cell infiltration or within inflamed portal tracts. These tissue-resident macrophages appear to be derived from circulating monocytes even in steady state conditions, although there is a marked increase in recruitment of these cells to the liver following inflammatory insults[9]. A recent study has demonstrated that monocytes increase in the circulation as well as in the liver of patients during progression of chronic liver disease, and that they are activated and release high amounts of proinflammatory cytokines and reactive oxygen species[10].
Secreted products from activated macrophages contribute to stellate cell activation and fibrosis. However, relatively little is known about the influence of macrophages on hepatic epithelial cell phenotype and function. Early studies have shown that secretions from activated macrophages influence hepatocyte DNA synthesis and cytochrome P-450 metabolism[11-13]. In a more recent study using a human cell co-culture model, macrophages triggered secretion of proinflammatory cytokines from bile duct epithelial cells, as well as apoptosis[14]. The authors speculated that this cellular interaction provided a mechanism to amplify chronic inflammation and bile duct destruction in vanishing bile duct syndromes[14]. The role of activated macrophages in modulating the hepatocyte inflammatory response to injury has not been determined. Accumulating data also suggest that during inflammatory liver injury, some hepatocytes and cholangiocytes may lose epithelial markers and acquire partial mesenchymal characteristics, although the direct role of macrophages and the contribution of this process to fibrogenesis have not been determined[15,16].
The aim of this study was to investigate the influence of macrophages on hepatocyte phenotype and function, and in particular, to determine whether macrophage-secreted products induce a proinflammatory response in hepatocytes. In order to address this, the effect of monocyte and macrophage-conditioned medium on cell morphology and gene expression was examined in two hepatocyte cell lines, along with the macrophage-secreted products that modulated the hepatocyte phenotype. We also evaluated human liver samples from patients with chronic hepatitis C virus (HCV) infection. Our data indicate that secreted products from activated macrophages induce an inflammatory phenotype in hepatocytes, which may have implications for persistent inflammation and fibrogenesis.
The human hepatoma-derived cell lines, HepG2 and Huh7, and the human acute monocytic leukemia cell line, THP-1, were purchased from American Type Culture Collection (ATCC) (Manassas, VA, United States). Cells were maintained at 37 °C and 5% CO2. Unless otherwise indicated, cells were cultured in “complete medium” comprising Dulbecco’s Modified Eagle’s Medium (Invitrogen, Carlsbad, CA, United States) supplemented with 10% foetal bovine serum (Invitrogen), 100 U/mL penicillin and 100 μg/mL streptomycin (Invitrogen), 2 mmol GlutaMAX™ (Invitrogen) and 20 μmol MEM non-essential amino acids (Invitrogen).
Macrophages were generated as described previously[17,18]. Briefly, THP-1 monocytes were seeded in six-well plates (Nunc, Roskilde, Denmark) at a density of 4 × 106 cells per well in 3 mL of complete medium and incubated for 2 h. Cells were treated with phorbol myristate acetate (PMA) (200 nmol; Sigma-Aldrich, St. Louis, MO, United States) for 24 h, washed three times with 1 × phosphate buffered saline (PBS) and cultured for 42 h in 2 mL of fresh complete medium. The resulting macrophage-conditioned media (MφCM) was collected, clarified by centrifugation at 400 ×g and stored at -20 °C until use. Conditioned media from THP-1 monocytes (MonoCM) was harvested in a similar fashion. For some experiments, MφCM was generated in the presence of matrix metalloproteinase (MMP)-9 Inhibitor I, (100 μmol, Cat# 444278, Calbiochem, Merck Pty Ltd, Kilsyth, Victoria, Australia).
HepG2 and Huh7 cells were seeded in six-well plates at a density of 2 × 105 cells per well in 3 mL of complete medium. After 24 h, cells were washed and cultured with 50% MφCM or 50% MonoCM in complete media for 24 h unless indicated otherwise. Cell morphology was observed by phase contrast microscopy using the Nikon Eclipse TS100. Culture in CM did not influence cell viability as determined by trypan blue exclusion.
Total RNA was extracted from untreated, monocyte CM and MφCM-treated HepG2 cells using TRI Reagent® (Sigma-Aldrich). RNA quality was assessed with an Agilent 2100 BioAnalyser (Agilent Technologies, Santa Clara, CA, United States) and only samples with a RNA integrity number above 8.0 were included. cRNA was generated from 500 ng total RNA using the Illumina TotalPrep cRNA Amplification Kit (Applied Biosciences, Carlsbad, CA, United States) and hybridised to Human HT-12 V3 Expression BeadChips (Illumina, San Diego, CA, United States). Array data were processed using the Illumina GenomeStudio software, transformed by variance stabilization transformation[19] and normalized by robust spline normalization[20] with Lumi[21]. Differential gene expression patterns between MφCM-treated and untreated HepG2 cells were identified using class comparison corrected for multiple testing with BRB ArrayTools (National Cancer Institute, Bethesda, MD, United States)[22]. Gene Ontology (GO), KEGG and BioCarta analyses were applied to the differentially expressed gene sets to identify altered pathways and cell functions. Least-squares (LS)/Kolmogorov-Smirnov (KS) permutation tests, Efron-Tibshirani’s gene set analysis maxmean test and Goeman’s global test were used to identify relevant pathways and a significance threshold of 0.005 was applied.
The study involved 147 consecutive patients with chronic hepatitis C, who had undergone a liver biopsy at the Princess Alexandra Hospital, Brisbane, Australia. Informed consent was obtained from each patient and the protocol was approved by the University of Queensland and Princess Alexandra Hospital Research Ethics Committees. Diagnosis of chronic HCV infection was based on standard serological assays and abnormal serum aminotransferase levels for at least 6 mo. All patients were positive for HCV antibody by the third-generation enzyme-linked immunosorbent assay (Abbott Laboratories, North Chicago, IL, United States), with infection confirmed by detection of circulating HCV RNA by polymerase chain reaction (PCR) using the Amplicor HCV assay (Roche, New Jersey, United States). Viral genotyping was performed using the Inno-Lipa HCV II assay (Innogenetics, Zwijnaarde, Belgium). Patients with other forms of chronic liver disease or antibodies to human immunodeficiency virus were not considered for the analysis. Patients with comorbidity such as acute coronary artery disease, unstable angina, congestive heart failure, significant renal impairment, acute and chronic infectious diseases, autoimmune and rheumatic diseases, cancer, endocrine diseases or inflammatory bowel diseases were not included in the study. Demographic and clinical details for the patients are presented in Table 1.
| No. of patients | n | 147 |
| Age (yr) | mean ± SEM | 42.3 ± 0.9 |
| Sex | M/F | 98/49 |
| Viral genotype | 1/ 2/ 3 | 80/6/61 |
| Stage of fibrosis | 0/1/2/3-4 | 13/65/44/25 |
| Grade of steatosis | 0/1/2-3 | 73/44/29 |
| Necroinflammatory score | 1-3/4-5/6-8 | 56/61/30 |
| BMI (kg/m2) | mean ± SEM | 26.2 ± 0.4 |
| BMI | Lean, overweight, obese1 | 66/51/24 |
| Alcohol - current (g/d) | Median (range) | 1 (0-120) |
| Alcohol - past (g/d) | Median (range) | 30 (0-500) |
| Creatinine (μmol/L) | mean ± SEM | 77.8 ± 1.5 |
| Platelets (× 109/L) | mean ± SEM | 213 ± 6 |
| Red blood cells (× 1012/L) | mean ± SEM | 4.8 ± 0.3 |
| Total WBC (× 109/L) | mean ± SEM | 7.2 ± 0.2 |
| Neutrophils (× 109/L) | mean ± SEM | 4.0 ± 0.1 |
| Lymphocytes (× 109/L) | mean ± SEM | 2.3 ± 0.06 |
| Monocytes (× 109/L) | mean ± SEM | 0.6 ± 0.02 |
A fragment of liver tissue (2-3 mm) was immediately frozen in liquid nitrogen at the time of biopsy and stored at -80 °C until extraction of RNA was performed. The remaining core was fixed in buffered formalin and embedded in paraffin. The degree of inflammation was graded according to the method of Ishak[23], and fibrosis was staged (0-4) according to the method of Scheuer[24]. Steatosis was graded as follows: 0 (< 5% hepatocytes affected); 1 (5%-33% of hepatocytes affected); 2 (34%-66% of hepatocytes affected); or 3 (> 66% of hepatocytes affected).
Total RNA was extracted from liver biopsy tissue, Huh7, HepG2, Mφ and THP-1 cells using TRI Reagent® (Sigma-Aldrich) according to the manufacturer’s instructions and reverse-transcribed to cDNA by SuperScript® III Reverse Transcriptase (Invitrogen). Liver tissue RNA quality was assessed using an Agilent 2100 Bioanalyser (Agilent Technologies) and the RNA 6000 Nano LabChip according to the manufacturer’s instructions. The median RNA Integrity Number (RIN) of RNA extracted from the liver biopsy samples was 8.0 (range: 6.0-9.2). Semi-quantitative real-time PCR (qPCR) for genes of interest was performed using Platinum® SYBR® Green qPCR SuperMix (Invitrogen) and analysed with MxPro QPCR software for MxPro 3000P QPCR systems (Stratagene, La Jolla, CA, United States) as previously described[25]. The expression of the housekeeping genes glyceraldehyde-3-phosphate dehydrogenase, human acidic ribosomal protein and 18S ribosomal RNA was determined using a multiplex real-time PCR protocol as previously described[25]. The relative mRNA expression of genes of interest was normalized to the geometric mean of the expression of the three housekeeping genes. Primers for SYBR® Green assays were custom made by Geneworks (Thebarton, SA, Australia). Custom made primer-probe sets were purchased from Sigma-Aldrich (Castle Hill, NSW, Australia). Specific primer and probe sequences are shown in Table 2.
| E-cadherin | for | 5’-ATTGCAAATTCCTGCCATTC-3’ |
| rev | 5’-GCTGGCTCAAGTCAAAGTCC-3’ | |
| Vimentin | for | 5’-GTTTCCAAGCCTGACCTCAC-3’ |
| rev | 5’-GCTTCAACGGCAAAGTTCTC-3 | |
| TGF-β1 | for | 5’-AAGTGGACATCAACGGGTTC-3’ |
| rev | 5’-TGCGGAAGTCAATGTACAGC-3 | |
| MMP-9 | for | 5’-TTCGACGTGAAGGCGCAGATGG-3’ |
| rev | 5’-AACTCACGCGCCAGTAGAAGCG-3’ | |
| CD163 | for | 5’-CCAACAAGATGCTGGAGTGAC-3’ |
| rev | 5’-TGACAGCACTTCCACATTCAAG-3’ | |
| Lipocalin-2 | for | 5’-TCACCCTCTACGGGAGAACCAAGG-3’ |
| rev | 5’-TGTGCACTCAGCCGTCGATACAC-3’ | |
| GAPDH | for | 5’-TGCACCACCAACTGCTTAGC-3’ |
| rev | 5’-GGCATGGACTGTGGTCATGAG-3’ | |
| probe | 5’-CCTGGCCAAGGTCATCCATGACAACTT-3’ | |
| HuPO | for | 5’-GCTTCCTGGAGGGTGTCC3-3’ |
| rev | 5’-GGACTCGTTTGTACCCGTTG-3’ | |
| probe | 5’-TGCCAGTGTCTGTCTGCAGATTGG-3’ | |
| 18s | for | 5’-GCCCGAAGCGTTTACTTTGA-3’ |
| rev | 5’-TCCATTATTCCTAGCTGCGGTATC-3’ | |
| probe | 5’-AAAGCAGGCCCGAGCCGCC-3’ |
To visualise epithelial cadherin (E-cadherin) and vimentin protein expression, HepG2 and Huh7 cells were cultured, as described above, on sterile glass cover slips (Deckglaser, Freiburg, Germany). After treatment, cells were fixed with 4% paraformaldehyde (Thermo Fisher Scientific, Waltham, MA, United States), washed with 1% glycine in 1 × PBS and permeabilized for 5 min with 0.2% Triton X-100 (Sigma-Aldrich). Non-specific binding was blocked with 10% heat-inactivated goat serum (Sigma-Aldrich) for 30 min. Cells were incubated with primary antibodies (1/50 dilution in 1% heat-inactivated goat serum) against E-cadherin (mouse monoclonal, ab1416; Abcam, Cambridge, MA, United States) or vimentin (mouse monoclonal, ab8069; Abcam) for 1 h at room temperature. After washing, goat anti-mouse IgG conjugated with Alexa Fluor 488 (Invitrogen; 1/200 dilution) was added for 1 h at room temperature. Cell nuclei were stained with 4’,6-diamidino-2-phenylindole (Invitrogen) and observed using the Zeiss LSM 510 Meta confocal microscope (Carl Zeiss, North Ryde, NSW, Australia).
Immunofluorescence was performed on sections of formalin-fixed, paraffin-embedded liver from patients with chronic HCV using monoclonal antibodies to MMP-9 (diluted 1/400, ab76003; Abcam) and CD163 (diluted 1/300, NB110-59935; Novus Biologicals, Littleton, CO, United States). Antigen retrieval was performed with 10 mmol Tris/1 mmol EDTA, pH 9.0 solution and sections were blocked with 20% heat-inactivated goat serum (Sigma-Aldrich) to prevent non-specific binding. Positively stained macrophages were observed using the Zeiss LSM 510 Meta confocal microscope.
Immunohistochemistry was also performed on nine biopsy specimens using an antibody directed against lipocalin-2 (LCN2) (diluted 1/35, ab23477; Abcam). Immunoreactivity was revealed using the Novolink Polymer detection system (Leica Microsystems Pty Ltd, North Ryde, NSW, Australia) according to the manufacturer’s instructions, and sections were photographed using the NanoZoomer (Olympus, Centre Valley, PA, United States). The sections were assessed for intensity of staining (weak, 1; moderate, 2; heavy, 3) and multiplied by the proportion of cells stained (0%-24%, 1; 25%-49%, 2; 50%-74%, 3; 75%-100%, 4) to give a value between 1 and 12.
CM from Mφ and THP-1 cells were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) on 4%-20% polyacrylamide gels (Invitrogen) and stained with SimplyBlue™ Safestain (Invitrogen). Bands differentially displayed in MφCM (n = 10) were precisely excised, de-stained, in-gel-trypsin-digested and analyzed by high performance liquid chromatography-tandem mass spectrometry (HPLC/MS/MS) and database searching[26,27]. One hundred and thirty-six proteins were identified. Following pruning to eliminate proteins of incorrect molecular weight or proteins not known to be secreted, database and literature searching identified probable candidates, of which MMP-9 was further investigated.
MφCM (PMA-differentiated THP-1 cells) and MonoCM (THP-1 cells) was subjected to SDS-PAGE on 4%-20% polyacrylamide gels (Invitrogen) and transferred to Immobilon P membrane (Millipore) using standard methods. Blots were incubated overnight at 4 °C with monoclonal anti-MMP-9 (diluted 1/5000, ab76003; Abcam) followed by Envision horseradish-peroxidase-linked anti-mouse polymer (Dako Australia, Botany, NSW, Australia; dilution 1/30). Blots were washed as previously described[28]. Proteins were detected with the enhanced chemiluminescence (ECL+) system (GE Healthcare Bio-Sciences, Rydalmere, Sydney, NSW, Australia).
Zymography was performed on CM from Mφs (PMA-differentiated THP-1 cells) and monocytes (THP-1 cells) using Novex® 10% Zymogram (Gelatin) Gel (Invitrogen) according to the manufacturer’s instructions. Gels were stained with SimplyBlue™ Safestain (Invitrogen) to visualize bands of protease activity.
Continuous normally distributed variables were represented graphically as mean ± SEM. Grade of steatosis, grade of hepatic inflammation, stage of fibrosis, alcohol consumption and RIN were summarized as median and range. For normally distributed variables, analysis of variance (ANOVA) or Student’s t test was performed to compare the means between groups. To determine differences between groups not normally distributed, medians were compared using the Kruskal-Wallis or Mann-Whitney U test.
Multivariate analysis was performed, including terms for age at biopsy, sex, viral genotype, stage of fibrosis, body mass index (BMI), alcohol consumption, total inflammatory score and grade of steatosis. Independent effects of normally distributed variables were assessed by ANOVA using general linear models. A backward elimination approach was used to remove non-significant variables and determine the most parsimonious model. All analyses were performed using SPSS version 17.0 (SPSS Inc, Chicago, IL, United States) and P < 0.05 was considered significant.
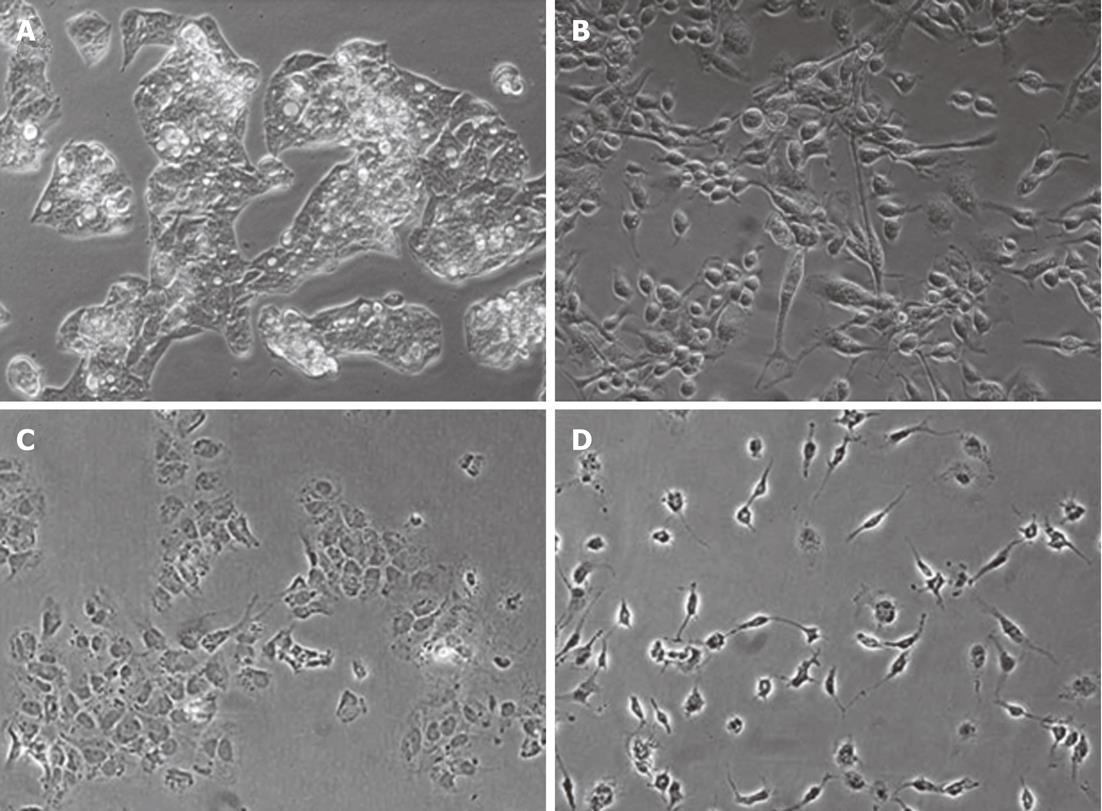
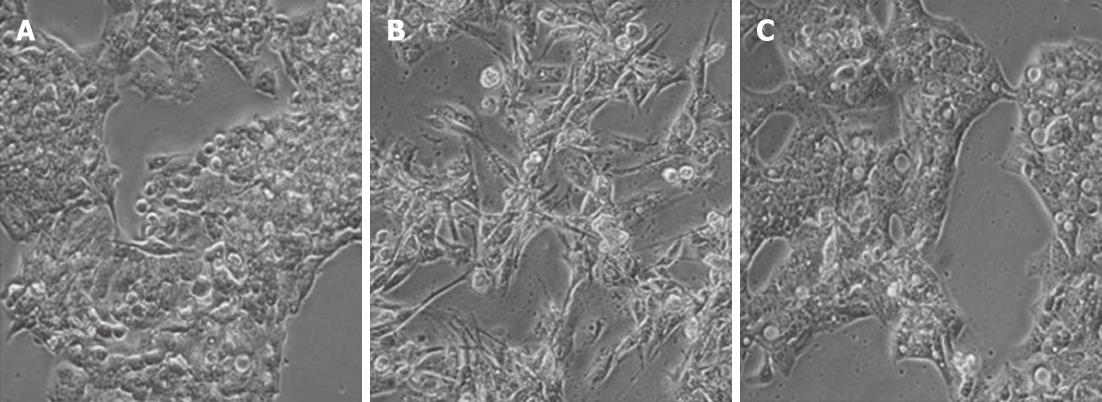
We began by assessing the effects of macrophage culture supernatants on hepatocyte morphology and gene expression. HepG2 and Huh7 cells typically form epithelial clusters with well-developed cell junctions (Figure 1A-C). Upon treatment with MφCM, these hepatocytes displayed an observable change (Figure 1B-D), acquiring an elongated spindle shape with loss of cell-cell contact. To determine if the morphological change in hepatocytes was permanent, after the initial MφCM treatment, HepG2 and Huh7 cells were washed and cultured in fresh growth medium for a further 2 d. Following release from treatment with MφCM, cells that were initially elongated and spindle-shaped after 24 h of treatment, formed epithelial clusters similar to untreated hepatocytes (Figure 2).
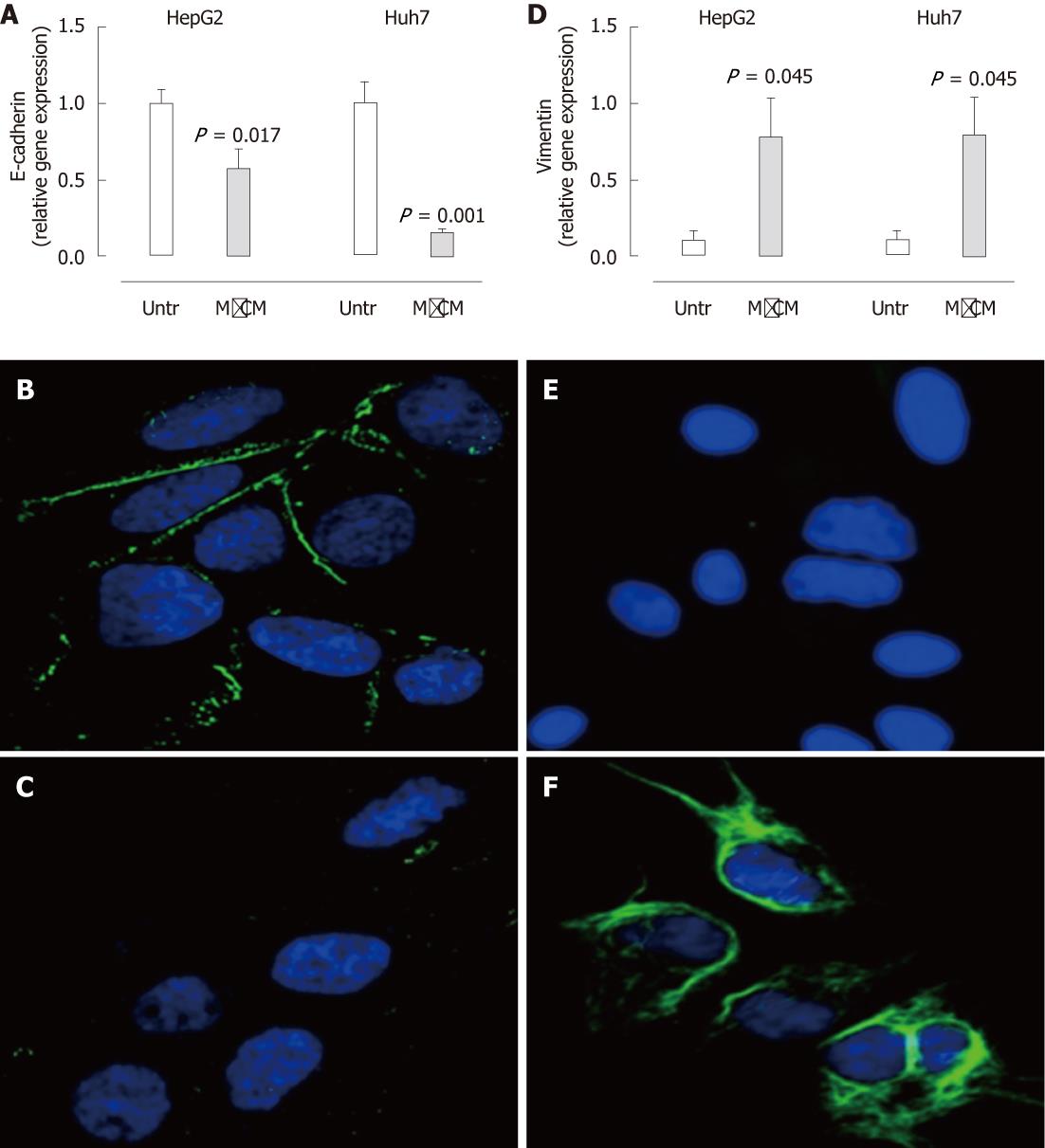
In association with the observable morphological change, HepG2 and Huh7 cells treated with MφCM demonstrated a substantial reduction in mRNA and protein expression of the epithelial marker E-cadherin (Figure 3A-C). mRNA and protein levels of vimentin, an intermediate filament and a marker of mesenchymal cells, were substantially enhanced in MφCM-treated hepatocytes (Figure 3D-F). mRNA expression of the pro-fibrogenic cytokine transforming growth factor-β1 (TGF-β1) was also increased (HepG2, 2.9-fold induction, P < 0.001; Huh7, 3.2-fold induction, P = 0.002), whereas collagen and α-smooth muscle actin (SMA) levels were not significantly altered (not shown).
In contrast to MφCM, no morphological changes or alteration in the levels of epithelial and mesenchymal markers were observed in hepatocytes treated with THP-1 monocyte CM (data not shown).
Our initial analysis of expression of epithelial and mesenchymal marker genes was next extended more globally through expression profiling. The microarray contained 48 000 probes; 34 693 of which were found to be expressed in at least one sample. Using this probe set, unsupervised hierarchical clustering showed good delineation between untreated and MφCM-treated HepG2 cells. MonoCM-treated HepG2 cells did not cluster differently from untreated cells and were thus not included for further analysis.
GO, KEGG and BioCarta analyses revealed significant upregulation of genes associated with inflammatory pathways; the majority of which included responses to cytokines and reactive oxygen species in addition to antigen processing and presentation (Table 3). Pathway analysis also identified a number of significantly downregulated hepatocyte functional processes including fatty acid and cholesterol metabolism and bile acid biosynthesis (Table 3).
| ID | Biological process (no. genes) | Differential expression | P valuea | |
| Gene ontology | 0034097 | Response to cytokine stimuli (80) | ↑ | 0.00125 |
| 0034341 | Response to interferon-γ (11) | ↑ | 0.00165 | |
| 0000302 | Response to reactive oxygen species (73) | ↑ | 0.00123 | |
| 0031663 | LPS-mediated signalling pathway (12) | ↑ | 0.00144 | |
| 0019882 | Antigen processing and presentation (58) | ↑ | 0.00172 | |
| 0032393 | MHC class I receptor activity (15) | ↑ | 0.00160 | |
| 0019217 | Regulation of fatty acid metabolic process (48) | ↓ | 0.00127 | |
| KEGG | hsa00071 | Fatty acid metabolism (51) | ↓ | 0.00001 |
| hsa00120 | Bile acid biosynthesis (41) | ↓ | 0.00001 | |
| BioCarta | h_tnfr2Pathway | TNFR2 signaling pathway (17) | ↑ | 0.00130 |
| h_ctlPathway | CTL mediated immune response against target cells (15) | ↑ | 0.00157 | |
| h_nkcellsPathway | Ras-independent pathway in NK-cell-mediated cytotoxicity (22) | ↑ | 0.00179 | |
| h_cd40Pathway | CD40L signaling pathway (15) | ↑ | 0.00221 | |
| h_fxrPathway | FXR and LXR regulation of cholesterol metabolism (7) | ↓ | 0.00135 |
To identify genes regulated by MφCM in HepG2 cells, we carried out a two-sample t test (with random variance model) corrected for multiple comparisons. 2665 genes were identified as significantly differentially expressed between untreated and MφCM-treated HepG2 cells with a P value ≤ 0.005 (corrected for false discovery rate). A significant > 4-fold change in expression was seen for 21 genes (17 upregulated and four downregulated) (Table 4). The most differentially expressed gene was LCN2 with an ~18-fold change in expression.
| Gene | Fold change | Differential expression | P value | |
| Lipocalin-2 | Lipocalin-2 | 17.8 | ↑ | < 1E-07 |
| TIMP1 | Metallopeptidase inhibitor 1 | 11.8 | ↑ | < 1E-07 |
| UBD | Ubiquitin D | 10.9 | ↑ | < 1E-07 |
| SERPINA3 | Serpin peptidase inhibitor clade A member 3 | 7.5 | ↑ | < 1E-07 |
| IGFBP1 | Insulin-like growth factor binding protein 1 | 7.5 | ↑ | < 1E-07 |
| S100A3 | S100 calcium binding protein A3 | 6.6 | ↑ | < 1E-07 |
| RASD1 | RAS dexamethasone-induced 1 | 6.4 | ↑ | < 1E-07 |
| CEBPD | CCAAT/enhancer binding protein delta | 6.2 | ↑ | < 1E-07 |
| SERPINE1 | Serpin peptidase inhibitor clade E member 1 | 5.9 | ↑ | 9.00E-07 |
| NDRG1 | N-myc downstream regulated gene 1 | 4.8 | ↑ | < 1E-07 |
| EMP3 | Epithelial membrane protein 3 | 4.6 | ↑ | < 1E-07 |
| DUSP5 | Dual specificity phosphatase 5 | 4.6 | ↑ | < 1E-07 |
| SOD2 | Superoxide dismutase 2, mitochondrial | 4.4 | ↑ | < 1E-07 |
| F2RL1 | Coagulation factor II receptor-like 1 | 4.4 | ↑ | < 1E-07 |
| CCL20 | Chemokine ligand 20 | 4.2 | ↑ | < 1E-07 |
| SDC4 | Syndecan 4 | 4.1 | ↑ | 6.00E-07 |
| TGM2 | Transglutaminase 2 | 4.1 | ↑ | < 1E-07 |
| NR1H4 | Nuclear receptor subfamily 1, group H, member 4 | 4.2 | ↓ | < 1E-07 |
| LIME1 | Lck interacting transmembrane adaptor 1 | 4.2 | ↓ | 1.00E-07 |
| DDC | Dopa decarboxylase | 4.3 | ↓ | < 1E-07 |
| ANKRD38 | Ankyrin repeat domain 38 | 5.1 | ↓ | < 1E-07 |
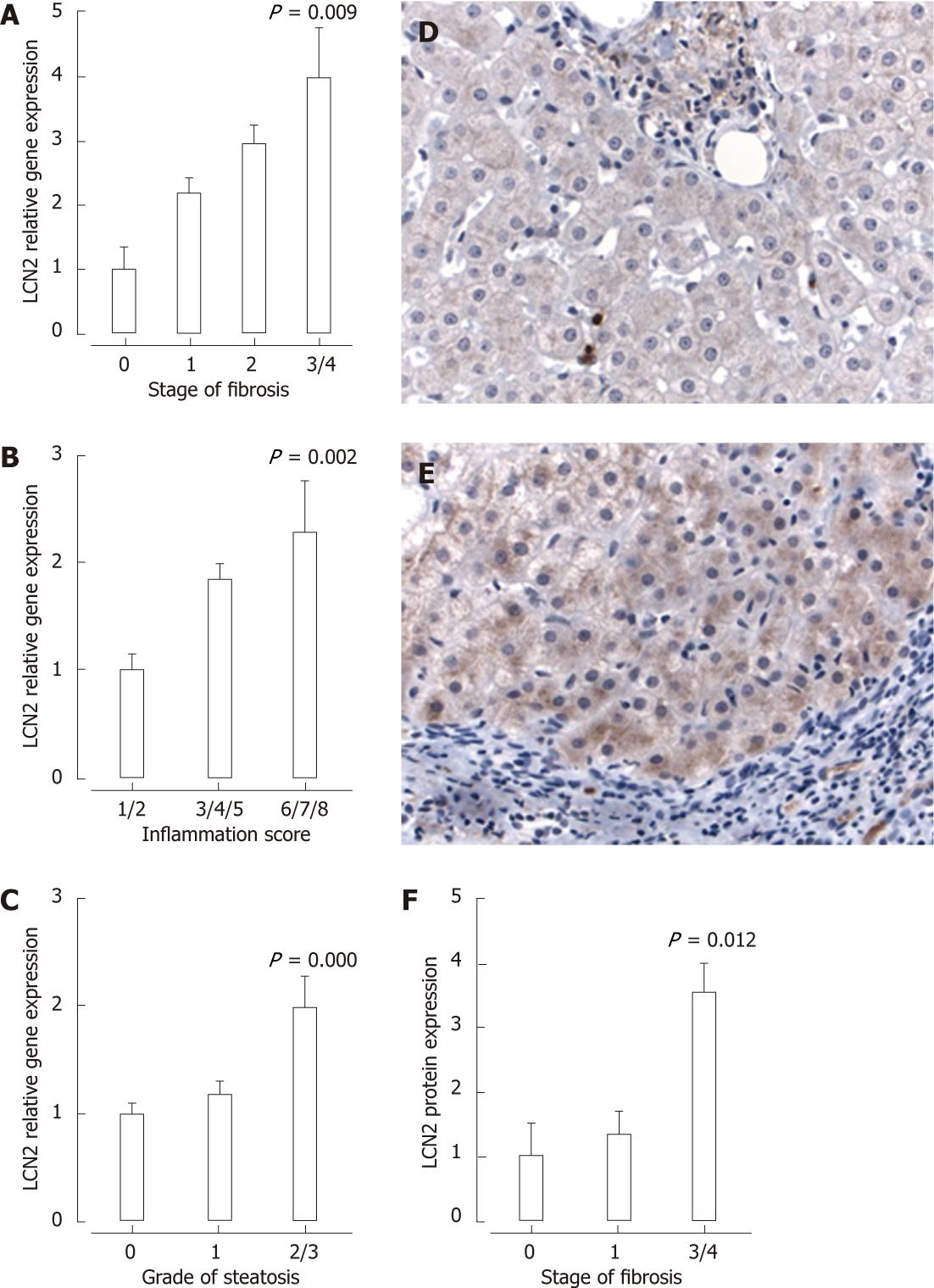
To validate observations from the in vitro studies, we examined the expression of LCN2 in liver biopsies from patients with chronic HCV. A significant increase in hepatic mRNA levels of LCN2 was seen with increasing stage of fibrosis (Figure 4A), grade of inflammation (Figure 4B) and grade of steatosis (Figure 4C). Following multivariate analysis (correcting for BMI, age, sex, alcohol consumption, viral genotype, grade of inflammation and presence of stainable iron), stage of fibrosis (OR: 1.6, 95% CI: 1.1-2.2) and grade of steatosis (OR: 1.7, 95% CI: 1.1-2.5) remained independently associated with hepatic mRNA expression of LCN2.
Immunohistochemistry was performed on a subset of liver biopsies selected due to their varying levels of LCN2 mRNA expression. Although negligible LCN2 staining was observed in patients with minimal fibrosis, enhanced protein expression was evident with increasing stage of fibrosis, predominantly within the hepatocyte cytoplasm, sparse sinusoidal neutrophils and some portal mononuclear cells (Figure 4D and E). To quantify LCN2 expression, tissue sections were scored for intensity of staining (1, weak; 2, moderate; 3, heavy) and multiplied by the proportion of cells stained (1, 1%-24%; 2, 25%-49%; 3, 50%-74%; 4, 75%-100%). In support of the mRNA data, enhanced LCN2 protein expression was observed in patients with stage 3-4 fibrosis (Figure 4F).
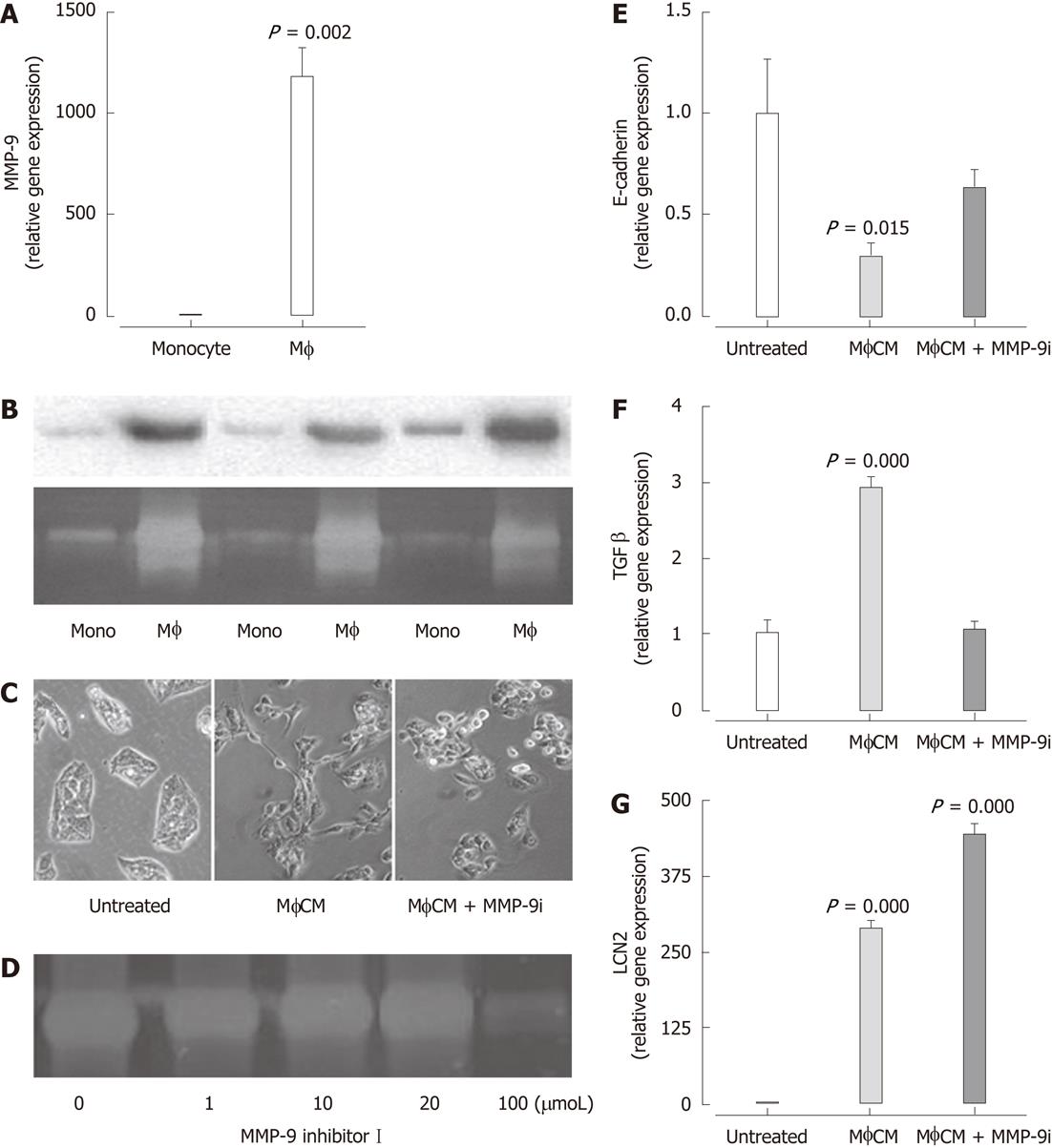
To address mechanisms by which macrophages may regulate hepatocyte function, we surveyed proteins present in MφCM (generated in serum-free medium). HPLC/MS/MS analysis identified the presence of MMP-9 in MφCM but not MonoCM. qPCR (Figure 5A), Western blotting and zymography (Figure 5B) confirmed the significantly enhanced levels of MMP-9 expression in MφCM. Generation of MφCM in the presence of MMP-9 Inhibitor I (100 μmol) prevented the MφCM-induced morphological change in HepG2 cells (Figure 5C). The efficacy of MMP-9 inhibitor I (100 μmol) was confirmed by zymography (Figure 5D). This reduction in MMP-9 activity significantly attenuated the downregulation of E-cadherin (Figure 5E) and the upregulation of TGF-β1 (Figure 5F). In contrast, inducible LCN2 expression was not attenuated by the MMP-9 inhibitor (Figure 5G).
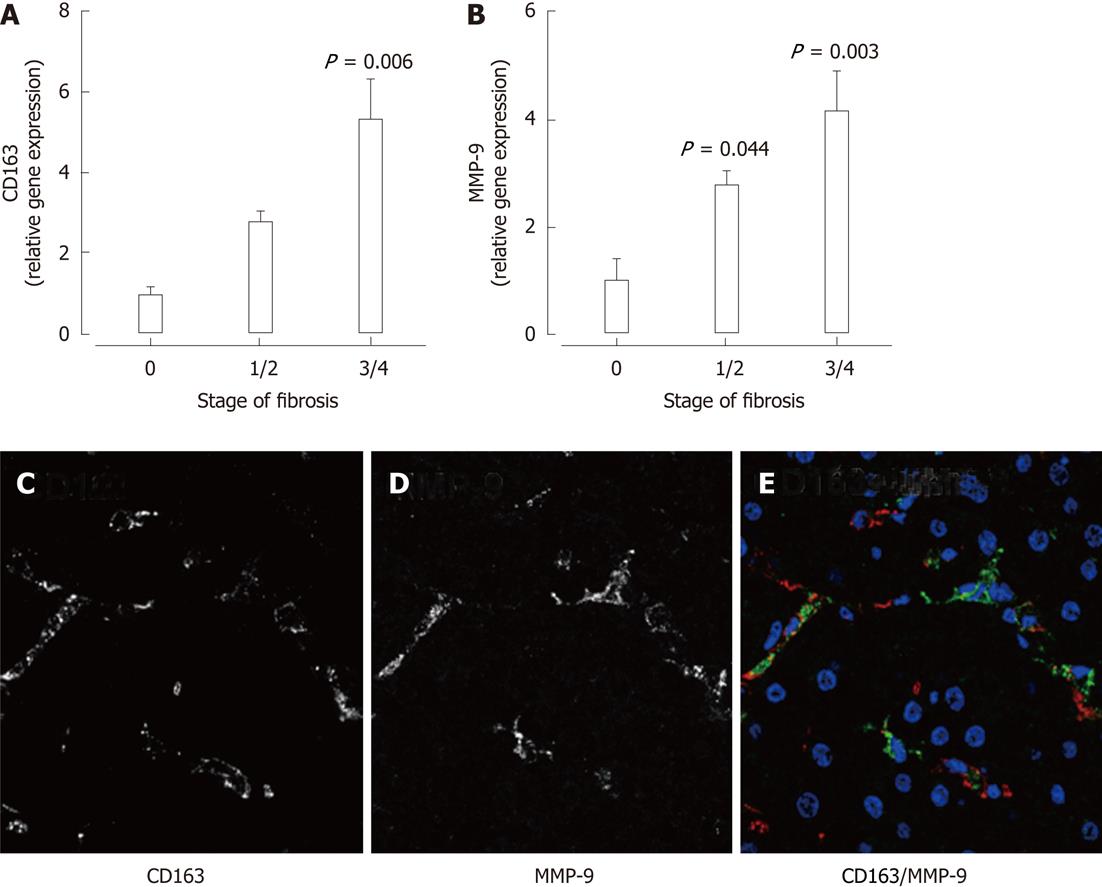
To validate observations from the in vitro studies, we examined expression of MMP-9 and the macrophage marker CD163 by qPCR and immunofluorescence in liver biopsies from patients with chronic HCV. Hepatic mRNA expression of the macrophage marker CD163 (Figure 6A) as well as MMP-9 (Figure 6B) was significantly associated with increasing stage of fibrosis. Immunofluorescence in liver biopsies from patients with chronic HCV demonstrated MMP-9 expression in CD163+ macrophages (Figure 6C-E). Thus, macrophage-expressed MMP-9 may contribute to hepatocyte dysfunction during chronic liver disease, but macrophages are also likely to regulate hepatocyte function independently of this pathway, as demonstrated by the failure of the MMP-9 inhibitor to antagonize inducible LCN2 expression.
Macrophages are a prominent feature of chronic inflammatory liver diseases and have a pivotal role in hepatic stellate cell activation and fibrogenesis[29]. This study was undertaken to determine whether macrophages also have a proinflammatory or profibrogenic effect on other cell populations within the hepatic microenvironment, specifically hepatocytes. Indeed, macrophage-secreted products induced a morphological change in hepatocytes accompanied by an altered gene expression program associated with the production of inflammatory mediators and fibrogenic agonists. This hepatocyte phenotypic change was transient, at least in vitro, with prompt reversal following removal of MφCM.
Relatively little is known about the contribution of liver epithelial cells to the local inflammatory response to injury. Studies performed more than a decade ago showed that human hepatocytes secrete a narrow repertoire of inflammatory cytokines and chemokines in response to stimulation with interleukin (IL)-1β or tumor necrosis factor (TNF)-α[30], and primary hepatocytes isolated from rat liver produce increased IL-8 in the presence of conditioned medium from lipopolysaccharide-stimulated Kupffer cells[31]. In a more recent study, exposure of hepatocytes to bile acids led to increased production of inflammatory mediators, including cytokines, chemokines, adhesion molecules and other proteins that may modulate immune cell accumulation and function[32]. Analogous to hepatocytes, cholangiocytes alter their phenotype in response to co-culture with macrophages, with increased secretion of cytokines involved in inflammation as well as apoptosis[14]. These studies suggest that liver epithelial cells are not simply targets of injury, but actively participate in propagating liver injury by amplifying the inflammatory response.
In the current study, hepatocytes treated with MφCM showed downregulation of genes associated with hepatic metabolism and biosynthetic functions, such as bile acid biosynthesis, fatty acid and cholesterol metabolism. In contrast, genes associated with a number of inflammatory pathways, including the CD40L, interferon-γ and TNF receptor 2 signaling pathways were significantly upregulated. Our data therefore suggest that macrophages both perturb normal homeostatic hepatocyte functions and promote a proinflammatory phenotype within these cells.
MφCM was a potent inducer of LCN2. LCN2 is a small glycoprotein that is secreted by macrophages and antagonizes the actions of bacterial siderophores. It therefore plays a crucial role in innate immunity by limiting iron availability to bacterial pathogens[33]. It also has direct effects on inflammatory cells[34], and can facilitate mucosal regeneration[35]. Some studies have also linked LCN2 expression to chronic disease. During chronic kidney disease progression, epidermal growth factor (EGF) receptor activation leads to LCN2 expression, which subsequently mediates the mitogenic effect of EGF[36]. Circulating LCN2 levels are elevated in obese subjects[37] and patients with non-alcoholic fatty liver disease[38], and mRNA levels of the gene are markedly upregulated in the liver of db/db obese/diabetic mice compared with their lean littermates[37]. A recent study using chronic liver injury models in the rat showed increased expression of LCN2, via an IL-1β-nuclear factor-κB-dependent pathway, in hepatocytes and proliferative bile duct epithelia[39]. The authors suggested that measurement of this protein may have diagnostic value as a biomarker of inflammatory liver damage. In keeping with this view, we demonstrated for the first time that LCN2 was expressed in liver biopsy samples from subjects with chronic HCV infection, and that the level of expression was significantly associated with the extent of liver injury. Furthermore, immunohistochemistry demonstrated LCN2 protein expression in hepatocytes, neutrophils and other inflammatory cells, and immunoreactivity in hepatocytes was particularly prominent in subjects with severe fibrosis. The pathophysiological function of LCN2 in liver disease remains unclear. It is not known at present whether LCN2 has an anti-inflammatory, protective role or whether it contributes to injury. Nevertheless, it appears to be a very good surrogate marker of a proinflammatory state.
Together with the induction of a proinflammatory profile, hepatocytes exposed in vitro to MφCM underwent a reversible change in cell shape. This was accompanied by a decrease in the epithelial marker E-cadherin, and increases in the expression of the mesenchymal marker vimentin, and the profibrogenic cytokine TGF-β1. These epithelial cells did not acquire α-SMA or type 1 collagen expression however, consistent with the concept that the observed morphological change is a response to injury, rather than a permanent transition into a mesenchymal cell[40]. Recent studies in human chronic liver disease have shown dual expression of epithelial and mesenchymal markers in bile duct cells and in some hepatocytes adjacent to portal tracts[16]. In addition, strong expression of TGF-β1 mRNA was seen in epithelial cells comprising the ductular reaction at the interface with parenchyma. In a carbon-tetrachloride-induced liver fibrosis model, abundant TGF-β1 protein expression was seen, not only in inflammatory cells and myofibroblasts, but also in cells with the morphology of hepatocytes immediately adjacent to the scars[41]. Similarly, in liver biopsies from patients with chronic hepatitis, TGF-β1 was detected in the cytoplasm of hepatocytes at the portal tract interface and in close proximity to areas of fibrosis[42]. Our study confirms expression of another secreted protein, LCN2, in hepatocytes adjacent to inflammatory/fibrotic areas in patients with chronic liver disease. Hence, accumulating data suggest that, in response to injury or inflammatory stimuli, hepatocytes are not simply bystanders but may directly contribute to the inflammatory/fibrogenic milieu. Importantly, our in vitro results provide support for the plasticity of this process, with reversal of the phenotypic change following withdrawal of the inflammatory stimulus. This is an important finding from a therapeutic perspective because it implies that, in a chronic setting, it may be possible to reverse the propagation of inflammation.
The inflammatory cell or signal driving the hepatocyte inflammatory response in vivo remains unclear. In the current study, MMP-9 was differentially expressed in MφCM vs MonoCM, and its inhibition prevented the morphological change in hepatocytes and the increase in TGF-β1 production (although chemical inhibitors of MMPs may also inhibit other unknown proteins and so may not be as specific as claimed). In contrast, MMP-9 inhibition led to a paradoxical increase in hepatocyte LCN2 mRNA levels. LCN2 has been shown to co-localize with MMP-9 in chronic vascular disease[43] and in the urine of patients with cirrhosis[44], where it may modulate proteolytic activity by binding to and preventing the degradation of MMP-9[45]. Further studies are required to determine the additional factors responsible for inducing the phenotypic change in hepatocytes.
In conclusion, this study provides evidence that macrophage-secreted products can induce transient phenotypic changes in hepatocytes that may contribute to chronic inflammation and fibrogenesis. Importantly, the data suggest that hepatocytes contribute to LCN2 production during inflammatory liver injury with recruitment and activation of liver macrophage populations. In the future, strategies aimed at blocking macrophage-mediated hepatocyte phenotypic changes could be considered as potential therapeutic approaches for diseases in which liver inflammation contributes to pathology.
We thank Professor Herbert Tilg for his insightful comments on the manuscript.
Activated liver macrophages have a key role in the progression of chronic liver injury and repair, and knowledge about their interaction with hepatocytes and other cells in the liver microenvironment may provide new targets for antifibrotic therapy.
Macrophages are a prominent feature of chronic inflammatory liver diseases. Secreted products from activated macrophages contribute to stellate cell activation and fibrosis. Macrophages also contribute to secretion of proinflammatory cytokines from bile duct epithelial cells, as well as apoptosis. Currently, there is limited knowledge about the effects of macrophage secreted products on hepatic epithelial cell function.
The current study found that macrophage-secreted products induced an altered gene expression program in hepatocytes associated with the production of inflammatory mediators and fibrogenic agonists. Macrophage-conditioned medium is a potent inducer of hepatocyte lipocalin-2 (LCN2) expression. In patients with chronic hepatitis C, fibrosis, inflammation and steatosis are associated with increased expression of LCN2.
Strategies aimed at blocking macrophage-mediated hepatocyte phenotypic changes could be considered as potential therapeutic approaches for diseases in which liver inflammation contributes to pathology.
LCN2 is a small glycoprotein that has been implicated in the innate immune response to bacterial infection, obesity and in the regulation of chronic inflammatory diseases. However, its pathophysiological functions in liver disease remain unclear.
This is a well written and implemented research paper that explores the relationship between Kupffer cells and hepatocytes in human liver disease, using a model of cell lines (the effects of THP-1 conditioned medium, with or without differentiation to macrophages, on HepG2 or Huh7 cells). The importance of the findings with regard to LCN2 was confirmed using samples from human tissues. This is an emerging area of research that has not been studied in great detail. The study is well implemented and the findings are refreshingly not over-interpreted. The only change we would suggest is a comment or two on the fact that chemical inhibitors of matrix metalloproteinases may also inhibit other unknown proteins (so called specific inhibitors tend to not always be as specific as claimed), so the findings with regard to the inhibitors need to be interpreted with caution.
| 1. | Muriel P, Escobar Y. Kupffer cells are responsible for liver cirrhosis induced by carbon tetrachloride. J Appl Toxicol. 2003;23:103-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 68] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 2. | Edwards MJ, Keller BJ, Kauffman FC, Thurman RG. The involvement of Kupffer cells in carbon tetrachloride toxicity. Toxicol Appl Pharmacol. 1993;119:275-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 176] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 3. | Andres D, Sanchez-Reus I, Bautista M, Cascales M. Depletion of Kupffer cell function by gadolinium chloride attenuates thioacetamide-induced hepatotoxicity - Expression of metallothionein and HSP70. Biochem Pharmacol. 2003;66:917-926. [RCA] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 59] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 4. | Thompson WD, Jack AS, Patrick RS. The possible role of macrophages in transient hepatic fibrogenesis induced by acute carbon tetrachloride injury. J Pathol. 1980;130:65-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 27] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 5. | Alric L, Orfila C, Carrere N, Beraud M, Carrera G, Lepert JC, Duffaut M, Pipy B, Vinel JP. Reactive oxygen intermediates and eicosanoid production by kupffer cells and infiltrated macrophages in acute and chronic liver injury induced in rats by CCl4. Inflamm Res. 2000;49:700-707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 40] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 6. | Rivera CA, Bradford BU, Hunt KJ, Adachi Y, Schrum LW, Koop DR, Burchardt ER, Rippe RA, Thurman RG. Attenuation of CCl(4)-induced hepatic fibrosis by GdCl(3) treatment or dietary glycine. Am J Physiol Gastrointest Liver Physiol. 2001;281:G200-G207. [PubMed] |
| 7. | Tomita M, Yamamoto K, Kobashi H, Ohmoto M, Tsuji T. Immunohistochemical phenotyping of liver macrophages in normal and diseased human liver. Hepatology. 1994;20:317-325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 44] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 8. | McGuinness PH, Painter D, Davies S, McCaughan GW. Increases in intrahepatic CD68 positive cells, MAC387 positive cells, and proinflammatory cytokines (particularly interleukin 18) in chronic hepatitis C infection. Gut. 2000;46:260-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 145] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 9. | Klein I, Cornejo JC, Polakos NK, John B, Wuensch SA, Topham DJ, Pierce RH, Crispe IN. Kupffer cell heterogeneity: functional properties of bone marrow derived and sessile hepatic macrophages. Blood. 2007;110:4077-4085. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 232] [Cited by in RCA: 260] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 10. | Zimmermann HW, Seidler S, Nattermann J, Gassler N, Hellerbrand C, Zernecke A, Tischendorf JJ, Luedde T, Weiskirchen R, Trautwein C. Functional contribution of elevated circulating and hepatic non-classical CD14CD16 monocytes to inflammation and human liver fibrosis. PLoS One. 2010;5:e11049. [PubMed] |
| 11. | Billiar TR, Curran RD. Kupffer cell and hepatocyte interactions: a brief overview. JPEN J Parenter Enteral Nutr. 1990;14:175S-180S. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 26] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 12. | West MA, Billiar TR, Curran RD, Hyland BJ, Simmons RL. Evidence that rat Kupffer cells stimulate and inhibit hepatocyte protein synthesis in vitro by different mechanisms. Gastroenterology. 1989;96:1572-1582. [PubMed] |
| 13. | Peterson TC, Renton KW. Kupffer cell factor mediated depression of hepatic parenchymal-cell cytochrome-P-450. Biochem Pharmacol. 1986;35:1491-1497. [RCA] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 49] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 14. | Alabraba EB, Lai V, Boon L, Wigmore SJ, Adams DH, Afford SC. Coculture of human liver macrophages and cholangiocytes leads to CD40-dependent apoptosis and cytokine secretion. Hepatology. 2008;47:552-562. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 43] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 15. | Choi SS, Diehl AM. Epithelial-to-mesenchymal transitions in the liver. Hepatology. 2009;50:2007-2013. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 216] [Cited by in RCA: 260] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 16. | Rygiel KA, Robertson H, Marshall HL, Pekalski M, Zhao L, Booth TA, Jones DE, Burt AD, Kirby JA. Epithelial-mesenchymal transition contributes to portal tract fibrogenesis during human chronic liver disease. Lab Invest. 2008;88:112-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 182] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 17. | Tsuchiya S, Kobayashi Y, Goto Y, Okumura H, Nakae S, Konno T, Tada K. Induction of maturation in cultured human monocytic leukemia cells by a phorbol diester. Cancer Res. 1982;42:1530-1536. [PubMed] |
| 18. | Okamura A, Rakugi H, Ohishi M, Yanagitani Y, Takiuchi S, Moriguchi K, Fennessy PA, Higaki J, Ogihara T. Upregulation of renin-angiotensin system during differentiation of monocytes to macrophages. J Hypertens. 1999;17:537-545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 175] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 19. | Lin SM, Du P, Huber W, Kibbe WA. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res. 2008;36:e11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 342] [Cited by in RCA: 414] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 20. | Workman C, Jensen LJ, Jarmer H, Berka R, Gautier L, Nielser HB, Saxild HH, Nielsen C, Brunak S, Knudsen S. A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biol. 2002;3:research0048. [PubMed] |
| 21. | Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547-1548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1676] [Cited by in RCA: 1733] [Article Influence: 96.3] [Reference Citation Analysis (0)] |
| 22. | Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-ArrayTools. Cancer Inform. 2007;3:11-17. [PubMed] |
| 23. | Ishak K, Baptista A, Bianchi L, Callea F, Degroote J, Gudat F, Denk H, Desmet V, Korb G, Macsween RNM. Histological grading and staging of chronic hepatitis. J Hepatol. 1995;22:696-699. [RCA] [DOI] [Full Text] [Cited by in Crossref: 3521] [Cited by in RCA: 3835] [Article Influence: 123.7] [Reference Citation Analysis (2)] |
| 24. | Scheuer PJ. Classification of chronic viral-hepatitis-a need for reassessment. J Hepatol. 1991;13:372-374. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1130] [Cited by in RCA: 1217] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 25. | Jonsson JR, Barrie HD, O'Rourke P, Clouston AD, Powell EE. Obesity and steatosis influence serum and hepatic inflammatory markers in chronic hepatitis C. Hepatology. 2008;48:80-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 56] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 26. | Riding GA, Hill JR, Jones A, Holland MK, Josh PF, Lehnert SA. Differential proteomic analysis of bovine conceptus fluid proteins in pregnancies generated by assisted reproductive technologies. Proteomics. 2008;8:2967-2982. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 27. | Smout MJ, Laha T, Mulvenna J, Sripa B, Suttiprapa S, Jones A, Brindley PJ, Loukas A. A granulin-like growth factor secreted by the carcinogenic liver fluke, Opisthorchis viverrini, promotes proliferation of host cells. PLoS Pathog. 2009;5:e1000611. [PubMed] |
| 28. | Wu M, Stockley PG, Martin WJ. An improved western blotting technique effectively reduces background. Electrophoresis. 2002;23:2373-2376. [PubMed] |
| 29. | Seki E, De Minicis S, Osterreicher CH, Kluwe J, Osawa Y, Brenner DA, Schwabe RF. TLR4 enhances TGF-beta signaling and hepatic fibrosis. Nat Med. 2007;13:1324-1332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1361] [Cited by in RCA: 1602] [Article Influence: 84.3] [Reference Citation Analysis (1)] |
| 30. | Rowell DL, Eckmann L, Dwinell MB, Carpenter SP, Raucy JL, Yang SK, Kagnoff MF. Human hepatocytes express an array of proinflammatory cytokines after agonist stimulation or bacterial invasion. Am J Physiol. 1997;273:G322-G332. [PubMed] |
| 31. | Mawet E, Shiratori Y, Hikiba Y, Takada H, Yoshida H, Okano K, Komatsu Y, Matsumura M, Niwa Y, Omata M. Cytokine-induced neutrophil chemoattractant release from hepatocytes is modulated by Kupffer cells. Hepatology. 1996;23:353-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 51] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 32. | Allen K, Jaeschke H, Copple BL. Bile acids induce inflammatory genes in hepatocytes: a novel mechanism of inflammation during obstructive cholestasis. Am J Pathol. 2011;178:175-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 314] [Cited by in RCA: 420] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 33. | Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, Akira S, Aderem A. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432:917-921. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1240] [Cited by in RCA: 1423] [Article Influence: 64.7] [Reference Citation Analysis (5)] |
| 34. | Nielsen BS, Borregaard N, Bundgaard JR, Timshel S, Sehested M, Kjeldsen L. Induction of NGAL synthesis in epithelial cells of human colorectal neoplasia and inflammatory bowel diseases. Gut. 1996;38:414-420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 336] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 35. | Playford RJ, Belo A, Poulsom R, Fitzgerald AJ, Harris K, Pawluczyk I, Ryon J, Darby T, Nilsen-Hamilton M, Ghosh S. Effects of mouse and human lipocalin homologues 24p3/lcn2 and neutrophil gelatinase-associated lipocalin on gastrointestinal mucosal integrity and repair. Gastroenterology. 2006;131:809-817. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 86] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 36. | Viau A, El Karoui K, Laouari D, Burtin M, Nguyen C, Mori K, Pillebout E, Berger T, Mak TW, Knebelmann B. Lipocalin 2 is essential for chronic kidney disease progression in mice and humans. J Clin Invest. 2010;120:4065-4076. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 285] [Cited by in RCA: 313] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 37. | Wang Y, Lam KS, Kraegen EW, Sweeney G, Zhang J, Tso AW, Chow WS, Wat NM, Xu JY, Hoo RL. Lipocalin-2 is an inflammatory marker closely associated with obesity, insulin resistance, and hyperglycemia in humans. Clin Chem. 2007;53:34-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 369] [Cited by in RCA: 449] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 38. | Milner KL, van der Poorten D, Xu A, Bugianesi E, Kench JG, Lam KS, Chisholm DJ, George J. Adipocyte fatty acid binding protein levels relate to inflammation and fibrosis in nonalcoholic fatty liver disease. Hepatology. 2009;49:1926-1934. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 135] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 39. | Borkham-Kamphorst E, Drews F, Weiskirchen R. Induction of lipocalin-2 expression in acute and chronic experimental liver injury moderated by pro-inflammatory cytokines interleukin-1β through nuclear factor-κB activation. Liver Int. 2011;31:656-665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 111] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 40. | Kisseleva T, Brenner DA. Is it the end of the line for the EMT? Hepatology. 2011;53:1433-1435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 24] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 41. | Duffield JS, Forbes SJ, Constandinou CM, Clay S, Partolina M, Vuthoori S, Wu S, Lang R, Iredale JP. Selective depletion of macrophages reveals distinct, opposing roles during liver injury and repair. J Clin Invest. 2005;115:56-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1126] [Cited by in RCA: 1211] [Article Influence: 57.7] [Reference Citation Analysis (0)] |
| 42. | Turato C, Calabrese F, Biasiolo A, Quarta S, Ruvoletto M, Tono N, Paccagnella D, Fassina G, Merkel C, Harrison TJ. SERPINB3 modulates TGF-beta expression in chronic liver disease. Lab Invest. 2010;90:1016-1023. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 43] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 43. | Hemdahl AL, Gabrielsen A, Zhu C, Eriksson P, Hedin U, Kastrup J, Thorén P, Hansson GK. Expression of neutrophil gelatinase-associated lipocalin in atherosclerosis and myocardial infarction. Arterioscler Thromb Vasc Biol. 2006;26:136-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 259] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 44. | Kim JW, Lee SH, Jeong SH, Kim H, Ahn KS, Cho JY, Yoon YS, Han HS. Increased urinary lipocalin-2 reflects matrix metalloproteinase-9 activity in chronic hepatitis C with hepatic fibrosis. Tohoku J Exp Med. 2010;222:319-327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 45. | Yan L, Borregaard N, Kjeldsen L, Moses MA. The high molecular weight urinary matrix metalloproteinase (MMP) activity is a complex of gelatinase B/MMP-9 and neutrophil gelatinase-associated lipocalin (NGAL). Modulation of MMP-9 activity by NGAL. J Biol Chem. 2001;276:37258-37265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 504] [Cited by in RCA: 547] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
Peer reviewers: Fabrizio Montecucco, MD, Assistant, Division of Cardiology, Department of Internal Medicine, University of Geneva, Avenue de la Roseraie 64, 1211 Geneva, Switzerland; Wendy M Mars, PhD, Department of Pathology, University of Pittsburgh, S-411B South Biomedical Science Tower Pittsburgh, PA 15261, United States
S- Editor Zhang DN L- Editor Kerr C E- Editor Xiong L