Published online Mar 21, 2012. doi: 10.3748/wjg.v18.i11.1216
Revised: May 26, 2011
Accepted: June 3, 2011
Published online: March 21, 2012
AIM: To search for and validate differentially expressed proteins in patients with gastric adenocarcinoma.
METHODS: We used two-dimensional gel electrophoresis and mass spectrometry to search for differentially expressed proteins in patients with gastric adenocarcinoma. A set of proteins was validated with immunoblotting.
RESULTS: We identified 30 different proteins involved in various biological processes: metabolism, development, death, response to stress, cell cycle, cell communication, transport, and cell motility. Eight proteins were chosen for further validation by immunoblotting. Our results show that gastrokine-1, 39S ribosomal protein L12 (mitochondrial precursor), plasma cell-induced resident endoplasmic reticulum protein, and glutathione S-transferase mu 3 were significantly underexpressed in gastric adenocarcinoma relative to adjacent non-tumor tissue samples. On the other hand, septin-2, ubiquitin-conjugating enzyme E2 N, and transaldolase were significantly overexpressed. Translationally controlled tumor protein was shown to be differentially expressed only in patients with cancer of the gastric cardia/esophageal border.
CONCLUSION: This work presents a set of possible diagnostic biomarkers, validated for the first time. It might contribute to the efforts of understanding gastric cancer carcinogenesis.
- Citation: Kočevar N, Odreman F, Vindigni A, Grazio SF, Komel R. Proteomic analysis of gastric cancer and immunoblot validation of potential biomarkers. World J Gastroenterol 2012; 18(11): 1216-1228
- URL: https://www.wjgnet.com/1007-9327/full/v18/i11/1216.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i11.1216
Gastric cancer (GC) is the fourth most common cancer worldwide[1]. However, incidence rates have steadily declined; this is largely determined by trends in the fundus and distal stomach[1]. Gastric cardia cancer, on the other hand, is on the increase, and is usually of the diffuse histological type[2]. However, despite declining incidence rates, GC is still the second leading cause of cancer death and thus remains a major health problem[1]. Radical surgery still offers the only chance of a cure for GC that invades the muscular layer, but only half of patients qualify for this at the time of their diagnosis[2]. The majority of patients are diagnosed at an advanced stage, where a systemic spread of the tumor cells has to be anticipated. The 5-year survival rate is very low and despite consistent improvements the prognosis remains poor[3]. On the other hand, the 5-year survival rate exceeds 90% for patients in an early stage of the disease[4].
A definitive diagnosis of GC requires a gastroscopic or surgical biopsy[5], which means there is a need for a non-invasive test, e.g., the detection of serum/plasma biomarkers. The most widely investigated serum markers include carcinoembryonic antigen, carbohydrate antigen (CA) 19.9, CA 72.4, cytokeratins and the β subunit of human chorionic gonadotropin. However, their low sensitivity and specificity precludes their use in screening and early diagnosis. Therefore, a search for more appropriate biomarkers is necessary.
One possible source of potential new biomarkers is the proteome. As opposed to static genomic data, the proteome is necessary for understanding the dynamic processes in cells[6]. It is also more complex, e.g., due to alternative splicing and post-translational modification, and the protein levels often do not correlate with the mRNA[7]. Proteomics is a rapidly developing field and is now applied to all areas of the life sciences[8]. Gel-based approaches, for example, belong to the most frequently used assays in protein separation and analysis[9]. Two-dimensional gel electrophoresis (2-DE) is still the method of choice for complex protein samples, although, due to its limitations with hydrophobic and large proteins, alternative or complementary methods based on mass spectrometry (MS) are gaining popularity.
2-DE, dating back to the 1970s[10,11], in conjunction with MS, has allowed the identification of differentially expressed proteins in various diseases, including GC[12-17]. However, the process of translating basic proteomic discoveries to the clinic is very time consuming and expensive, and despite all the efforts put into biomarker research, very few diagnostic protein biomarkers have been approved by the United States Food and Drug Administration[8]. The lack of specific and sensitive biomarkers thus creates a persistent need for the expedited development of biomarkers and their use in improving the diagnosis and treatment of cancer. Furthermore, the validity of potential biomarkers, as found by proteome analysis, needs to be investigated and incorporated as the second step of the research, e.g., by immunohistochemical and/or western blot analyses, in order to fulfill the criterion of a systematic investigation of the protein biomarkers in GC.
In this study, we used 2-DE and MS to search for differentially expressed proteins in patients with gastric adenocarcinoma: a total of 30 different protein alterations were found. Gastrokine-1 precursor (GKN1), 39S ribosomal protein L12 (mitochondrial precursor) (MRPL12), plasma cell-induced resident endoplasmic reticulum protein (PACAP), glutathione S-transferase mu 3 (GSTM3), septin-2 (SEPT2), ubiquitin-conjugating enzyme E2 N (UBE2N), transaldolase (TALDO1), and translationally controlled tumor protein (TPT1) were further validated with the immunoblot analysis, making them a set of possible biomarkers for gastric adenocarcinoma.
A total of 32 pairs of gastric adenocarcinoma tissues and adjacent control tissue from 32 patients were obtained from the tissue bank of the Institute of Oncology Ljubljana after the National Medical Ethics Committee’s approval. They were stored at -70 °C until further use. The patients’ and tumors’ parameters are listed in Table 1.
| Characteristics | n (%) |
| Sex | |
| Male | 19 (59.4) |
| Female | 13 (40.6) |
| Lauren’s classification | |
| Intestinal | 20 (62.5) |
| Diffuse | 11 (34.4) |
| Mixed | 1 (3.1) |
| Location | |
| Corpus | 8 (25) |
| Antrum | 7 (21.9) |
| Cardia/gastroesophageal border | 10 (31.2) |
| Several parts | 7 (21.9) |
| Grade | |
| Well-differentiated | 2 (6.2) |
| Moderately differentiated | 12 (37.5) |
| Poorly differentiated | 12 (37.5) |
| Undifferentiated | 6 (18.8) |
| pT | |
| pT1b | 1 (3.1) |
| pT2a | 5 (15.6) |
| pT2b | 15 (46.9) |
| pT3 | 11 (34.4) |
| pN | |
| pN0 | 8 (25) |
| pN1 | 16 (50) |
| pN2 | 6 (18.8) |
| pN3 | 2 (6.2) |
The tissues were ground with a mortar and pestle in the presence of liquid nitrogen and then lysed with 7 mol/L urea, 2 mol/L thiourea, 40 g/L 3-[ (3-cholamidopropyl)-dimethylammonio]-1-propane sulfonate (CHAPS), 20 mmol/L dithiothreitol (DTT) and a protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO, United States). For every 10 mg of tissue, 50 μL lysis buffer was added. After sonication on ice, three times for 10 s, the samples were incubated for 1 h on ice with occasional vortexing and then centrifuged for 1 h at 20 000 ×g at 4 °C. The supernatant was collected and the protein concentration was determined according to a commercial Bradford reagent (Thermo Fisher Scientific, Waltham, MA, United States) with bovine serum albumin as the standard.
2-DE was conducted on 12 pairs of tissues and repeated in triplicate. For the isoelectric focusing, 100 μg protein was diluted in 450 μL rehydration solution (7 mol/L urea, 2 mol/L thiourea, 20 g/L CHAPS, 20 mmol/L DTT, 0.5% immobilized pH gradient (IPG) buffer, pH 4-7), loaded by in-gel rehydration onto IPG strips with an acidic pH range of 4-7 (GE Healthcare, Stockholm, Sweden), and focused in an Ettan IPGphor II isoelectric focusing system (GE Healthcare) to a total of 63.5 kVh. Next, the IPG strips were equilibrated for 15 min in 6 mol/L urea, 30% glycerol, 20 g/L sodium dodecyl sulfate (SDS), 50 mmol/L Tris-HCl, pH 8.8, with 10 g/L DTT, and for another 15 min in the same solution with DTT replaced by 25 g/L iodoacetamide. The strips were then transferred to 12% polyacrylamide gels and the second dimension was carried out in an Ettan DALTsix electrophoresis unit (GE Healthcare) under constant power in a Laemmli buffer system until the bromophenol blue reached the end of the gel.
The gels were silver stained according to Mortz et al[18]. Briefly, they were fixed overnight in 50% methanol, 12% acetic acid, and 0.05% formalin, washed for 2 × 20 min with 35% ethanol and sensitized for 2 min with 0.02% sodium thiosulfate. Next, they were washed for 3 × 5 min with water and stained for 20 min in 0.2% silver nitrate and 0.076% formalin. Then they were washed again for 2 × 1 min with water and developed with 6% sodium carbonate, 0.05% formalin and 0.0004% sodium thiosulfate. The reaction was stopped with 50% methanol and 12% acetic acid and the gels were left in 1% acetic acid until the scanning.
The gels were scanned with an ImageScanner II (GE Healthcare) at 300 dpi and analyzed with an ImageMaster 2D Platinum v7 (GE Healthcare). The spots were detected, matched and quantified by relative volume. The normalization of the spot values (relative spot volumes) was based on the total spot volume. Next, the data were analyzed with Excel. The spots of interest were determined using a pair-by-pair comparison. The spots were considered to be differentially expressed if they matched the following criteria: at least a twofold change in the relative spot volume, the occurrence of this change in at least three patients, and statistical significance.
The enzymatic digestion of the excised spots was performed using a Progest robot (Genomic Solutions, Holliston, MA, United States). Briefly, the protein spots were excised from the gel, placed into 96-well plates and washed with 50 μL acetronitrile (CH3CN) for 3 min and then with 50 μL 25 mmol/L ammonium bicarbonate (NH4HCO3) for 3 min. This washing step was repeated three times to eliminate SDS, Tris and glycine. After the washing procedure, the excised spots were treated with 100 μL 10 mmol/L DTT for the reduction step and the reaction was left to proceed at 57 °C for 1 h. After DTT removal, 100 μL 55 mmol/L iodoacetamide was added for the cysteine carbamidomethylation and the reaction was left to proceed at room temperature (RT) for 1 h. After removal of the supernatant, the washing procedure was repeated three times and the gel slices were dried with a SpeedVac (5 min). Based on the gel-slice volume, 5-10 μL 12 ng/μL Porcine Trypsin (Promega, Madison, W, United States) was added. The enzyme was freshly diluted in 25 mmol/L NH4HCO3 and the digestion was performed overnight at RT. Trypsin was the protease of choice for the MS protein identification because of its reliability and its substrate specificity, yielding peptides with C-terminal basic residues (Arg and Lys), which facilitated positive ionization and subsequent MS detection. Finally, 10 μL of 39% H2O/60% CH3CN/1% HCOOH was added and the samples were left for 3 h at RT to favor the extraction of peptides from the gel.
Trypsin-generated peptide mixtures were analyzed by matrix-assisted laser desorption ionization coupled with a time-of-flight analyzer (MALDI-TOF) (Voyager, Applied Biosystems, Carlsbad, CA, United States). Samples that provided an ambiguous identification with MALDI-TOF were analyzed by tandem MS (MS/MS) using nanoliquid-chromatography electrospray ionization coupled with the quadrupole and time-of-flight analyzer (nanoLC-ESI-Q-TOF) (nanoAcquity, Q-TOF Premier; Waters, Milford, MA, United States).
For MALDI-TOF MS, the peptide extracts (0.5 μL) were mixed with an equal volume of 2,5-dihydroxybenzoic acid (10 mg/mL; Sigma-Aldrich) dissolved in 20% CH3CN. Trypsin-digested ovalbumin was used for external calibration. Crystals were obtained using the dried-droplet method on a 100-spots metallic plate and approximately 500 MALDI mass spectra were averaged per spot to optimize the signal-to-noise ratio. The laser fluency was adjusted to the threshold to achieve the best resolution and mass accuracy. The MS measurements were carried out at a maximum accelerating potential of 20 kV, in the positive reflectron mode. The acquisition range was set to m/z 800-3500 with a low-mass gate at m/z 700. For m/z values of about 1500, the mean mass resolution was 15 000.
Proteins were identified by peptide mass fingerprinting with two search programs: Mascot and ProFound. The following search parameters were applied: SWISS-PROT and NCBI were used as the protein-sequence databases; a mass tolerance of 50 ppm and one missed cleavage were allowed; the alkylation of cysteine by carbamidomethylation was considered complete; while the oxidation of methionine was considered as a possible modification.
For nanoLC-ESI-Q-TOF MS/MS analysis, peptide mixtures were SpeedVac-treated for 10 min to eliminate CH3CN, then 25 mmol/L NH4HCO3 was added before injection into the nanoAcquity/Q-TOF system equipped with a trapping column (Symmetry C18, 180 μm × 20 mm, 5 μm particle size) and an analytical column (BEH130 C18, 75 μm × 100 mm, 1.7-μm particle size) (Waters). The aqueous solvent (buffer A) was 0.1% formic acid and the organic phase (buffer B) was acetonitrile with 0.1% formic acid. A 2%-40% B gradient was set for 25 min. The MS parameters were as follows: positive ion mode; capillary voltage, 3 kV; cone voltage, 40 V; ion-source block temperature, 80 °C; and collision energy ramping from 15 to 40 eV. For the exact mass measurements, the glufibrinopeptide reference (m/z = 785.8426) was continuously supplied during the nanoLC-MS/MS experiments using the lockspray device. The peptide mass measurements were corrected by the PLGS software (ProteinLynx Global Server; Waters) during data processing. Peak lists were generated by PLGS and the processed data were submitted to Mascot searching using the following parameters: data bank, NCBI; peptide tolerance, 15 ppm; fragment tolerance, 0.1 Da; number of missed cleavages, one; variable modifications, oxidation; and fixed modifications, carbamido methylation.
To validate the differential expression from the 2-DE gels, an immunoblot analysis was performed on an expanded number of samples, on 27 pairs. For TPT1, additional four pairs of cardia/gastroesophageal border adenocarcinoma were used. A total of 30 μg protein per sample was loaded on 12% or any kD gels (Bio-Rad, Hercules, CA, United States), transferred onto PDVF membranes (Millipore, Billerica, MA, United States), and blocked in 50 g/L skimmed milk overnight at 4 °C. Primary antibodies were used in the following dilutions: anti-GKN1 at 0.5 μg/mL (WH0056287M1; Sigma–Aldrich), anti-MRPL12 at 1 μg/mL (WH0006182M1; Sigma-Aldrich), anti-PACAP at 1:1000 (ab96308; Abcam, Cambridge, United Kingdom), anti-GSTM3 at 0.75 μg/mL (ab74749; Abcam), anti-SEPT2 at 1 μg/mL (ab88657; Abcam), anti-UBE2N at 0.5 μg/mL (ab25885; Abcam), anti-TALDO1 at 1:1000 (ab67467; Abcam), and anti-TPT1 at 1 μg/mL (WH0007178M1; Sigma-Aldrich). Horseradish-peroxidase-conjugated secondary antibodies were used in the following dilutions: goat anti-mouse at 1:5000 (115-035-062; Jackson ImmunoResearch, Newmarket, Suffolk, United Kingdom) and goat anti-rabbit at 1:10 000 (111-035-003; Jackson ImmunoResearch). The proteins were detected chemiluminescently with an LAS-4000 CCD camera (Fujifilm, Tokyo, Japan) at 10-s intervals. The blots were then quantified with Multi Gauge software (Fujifilm) and the intensity values were exported to Excel. Differential expression was determined after normalization to Ponceau S-stained membranes for loading and transfer differences and statistical significance was assessed.
To assess the statistical significance of differential protein expression (tumor vs non-tumor) in 2-DE as well as in immunoblotting, Wilcoxon signed-rank test was used. The test was double-sided and values with P < 0.05 and a 95% CI were considered to be statistically significant. To assess the correlation of the differential protein expression from immunoblotting with the histopathological parameters, repeated-measures analysis of variance was used. The values with P < 0.05 and a 95% CI were considered to be statistically significant. Bonferroni post-test was used to narrow down where the differences were significant. All analyses were performed using Microsoft Office Excel 2007 (Microsoft Corporation, Redmond, WA, United States) and GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA, United States).
Altogether, we used 32 pairs of human gastric adenocarcinoma and adjacent normal tissue samples from 32 patients. The mean age of the patients was 68.3 ± 10.5 (range 40-84) years. The subject group contained predominantly males (59.4%). According to Lauren’s classification, the majority of tumors were intestinal (62.5%) and the largest part was located in the cardia/gastroesophageal border (31.2%). The tumors were mainly moderately (37.5%) and poorly (37.5%) differentiated. According to the TNM classification, most were pT2b (46.9%) and pN1 (50%). The data on pM were only available for some patients and were thus not included in any further analysis. For more details, refer to Table 1.
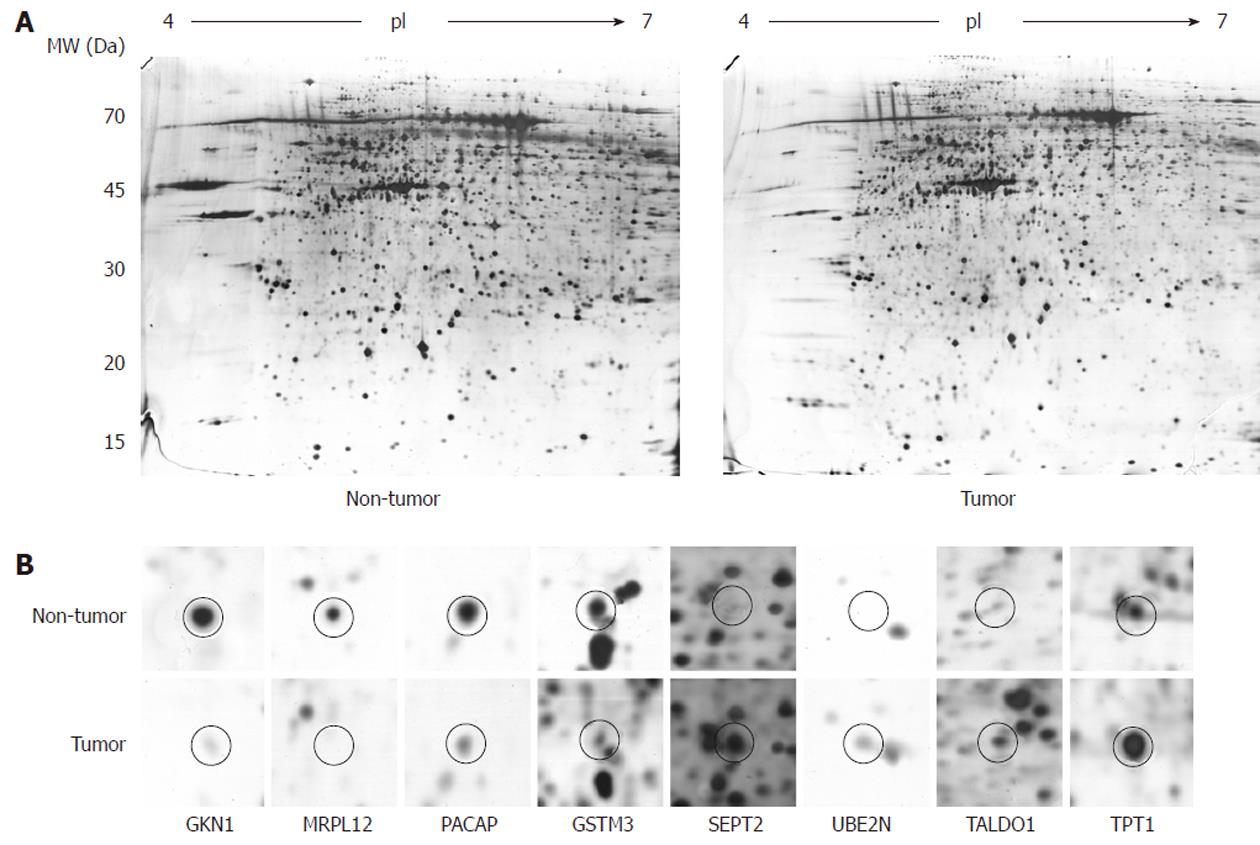
To determine the differentially expressed proteins in the gastric adenocarcinoma patients, we first performed 2-DE on 12 pairs of human gastric tissue samples. For good resolution we used large (24 cm) IPG strips with an acidic pH gradient (4-7). We performed the experiments in triplicate, which produced well-resolved spots and reproducible 2-DE patterns. On average, 1197 ± 150 spots per gel were detected after silver staining. Figure 1 shows a typical image of a tumor- and a non-tumor-sample-derived gel. In the lower part of the figure, one can see differentially expressed spots from the tumor/non-tumor pairs, obtained from a pair-by-pair comparison.
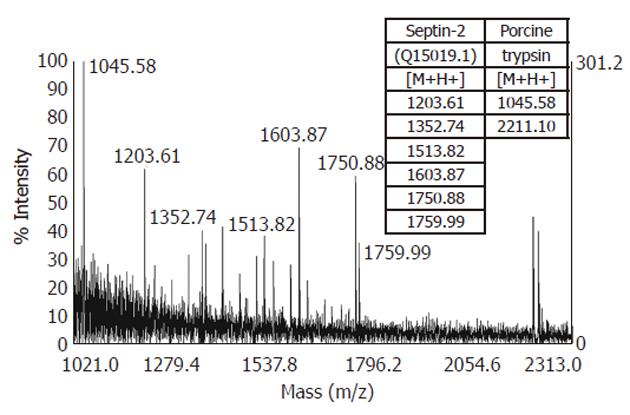
After the proteolytic digestion of 32 excised spots, we identified 30 different proteins by MALDI-TOF MS or nanoLC-ESI-Q-TOF MS/MS. Eleven proteins were found to be underexpressed, while 19 were found to be overexpressed in the gastric adenocarcinoma relative to adjacent non-tumor tissue samples. These results are summarized in Table 2 and an example of the MS identification is shown in Figure 2.
| Protein name | Gene name | Swiss-Prot ID | pI/MW | Frequency1 | Fold2 | P value | Coverage3 | No. of pep.4 |
| Down-regulated | ||||||||
| Gastrokine-1 precursor | GKN1 | Q9NS71 | 5.3/21.0 | 75% (9/12) | 15.17 | 0.000 | 16% | 6 |
| 15-hidroxyprostaglandin dehydrogenase isoform 1 | HPGD | P15428 | 5.6/25.2 | 66% (8/12) | 3.85 | 0.016 | 23% | 5 |
| Enoyl CoA hydratase, mitochondrial precursor | ECHS1 | P30084 | 5.8/27.9 | 66% (8/12) | 3.38 | 0.001 | 49% | 15 |
| 39S ribosomal protein L12, mitochondrial precursor | MRPL12 | P52815 | 5.1/20.6 | 58.3% (7/12) | 10.95 | 0.016 | 16% | 4 |
| Acyl-CoA dehydrogenase | ACADS | P16219 | 6.1/39.9 | 58.3% (7/12) | 4.94 | 0.005 | 12% | 5 |
| NADH dehydrogenase (ubiquinone) flavoprotein 2, mitochondrial precursor | NDUFV2 | P19404 | 5.7/24.2 | 50% (6/12) | 2.26 | 0.027 | 58% | 16 |
| Phenazine biosynthesis-like domain-containing protein isoform a | PBLD | P30039 | 6.1/31.8 | 50% (6/12) | 3.60 | 0.007 | 25% | 5 |
| Plasma cell-induced resident endoplasmic reticulum protein | PACAP | Q8WU39 | 5.2/22.0 | 41.6% (5/12) | 4.41 | 0.042 | 39% | 5 |
| ATP synthase subunit d, mitochondrial | ATP5H | O75947 | 5.2/20.5 | 41.6% (5/12) | 2.40 | 0.012 | 26% | 5 |
| Glutathione S-transferase mu 3 | GSTM3 | P21266 | 5.3/26.3 | 33.3% (4/12) | 3.35 | 0.042 | 29% | 4 |
| Transthyretin | TTR | P02766 | 5.4/16.4 | 25% (3/12) | 2.74 | 0.027 | 53% | 7 |
| Up-regulated | ||||||||
| Septin-2 | SEPT2 | Q15019 | 6.0/41.0 | 75% (9/12) | 4.58 | 0.009 | 34% | 7 |
| Maspin | SERPINB5 | P36952 | 5.6/40.2 | 58.3% (7/12) | 4.99 | 0.007 | 14% | 5 |
| Phosphoglycerate kinase 1 | PGK1 | P00558 | 6.0/40.1 | 58.3% (7/12) | 6.46 | 0.016 | 22% | 6 |
| Pyruvate kinase, muscle | PKM2 | P14619 | 6.0/38.8 | 58.3% (7/12) | 5.25 | 0.021 | 20% | 8 |
| Actin, cytoplasmic 1 | ACTB | P60709 | 4.8/32.2 | 58.3% (7/12) | 7.77 | 0.005 | 44% | 12 |
| Tumor necrosis factor receptor superfamily member 16 precursor | NGFR | P08138 | 5.2/35.5 | 50% (6/12) | 3.94 | 0.002 | 17% | 5 |
| Heat shock 70 kDa protein 1A/1B | HSPA1 | P08107 | 5.1/36.9 | 50% (6/12) | 4.77 | 0.009 | 14% | 8 |
| Heterochromatin-like protein 1 | CBX3 | Q13185 | 5.2/22.0 | 50% (6/12) | 4.12 | 0.027 | 19% | 3 |
| Ubiquitin-conjugating enzyme E2 N | UBE2N | P61088 | 5.7/16.4 | 41.6% (5/12) | 4.94 | 0.001 | 40% | 5 |
| Transaldolase | TALDO1 | P37837 | 5.9/38.5 | 41.6% (5/12) | 3.12 | 0.003 | 18% | 5 |
| Cathepsin D precursor | CTSD | P07339 | 5.3/28.4 | 41.6% (5/12) | 3.49 | 0.034 | 27% | 11 |
| Annexin A4 | ANXA4 | P09525 | 5.6/31.3 | 41.6% (5/12) | 13.31 | 0.027 | 54% | 26 |
| Myotrophin | MTPN | P58546 | 5.1/13.5 | 41.6% (5/12) | 2.45 | 0.000 | 22% | 4 |
| Enolase 1 variant | ENO1 | P06733 | 5.5/42.6 | 41.6% (5/12) | 3.95 | 0.034 | 19% | 7 |
| Nicotinamide N-methyltransferase | NNMT | P40261 | 5.4/28.5 | 41.6% (5/12) | 4.24 | 0.000 | 9% | 3 |
| Eukaryotic translation initiation factor 2 subunit 1 | EIF2S1 | P05198 | 4.9/33.5 | 41.6% (5/12) | 6.61 | 0.042 | 24% | 8 |
| Translationally-controlled tumor protein | TPT1 | P13693 | 4.8/20.7 | 25% (3/12) | 2.50 | 0.002 | 18% | 4 |
| Annexin A5 | ANXA5 | P08758 | 5.0/32.4 | 25% (3/12) | 3.24 | 0.005 | 70% | 25 |
| Nucleoside diphosphate kinase A isoform a | NME1 | P15531 | 5.7/19.0 | 25% (3/12) | 3.11 | 0.016 | 33% | 5 |
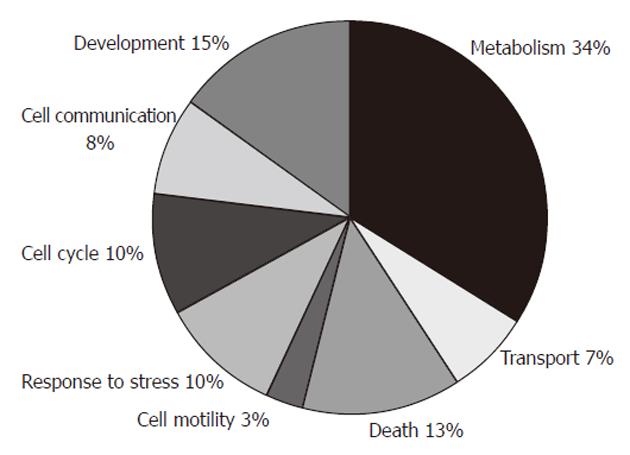
The identified differentially expressed proteins could be classified into eight groups according to the biological process in which they are involved (Figure 3), by information from the GO Classification for Homo sapiens from European Molecular Biology Laboratory - European Bioinformatics Institute (http://www.ebi.ac.uk/integr8/GOAnalysisPage.do?orgProteomeID=25). These groups were: metabolism (HPGD, ECHS1, MRPL12, ACADS, NDUFV2, PBLD, PACAP, ATP5H, GSTM3, PGK1, ACTB, HSPA1, CBX3, UBE2N, TALDO1, MTPN, ENO1, NNMT, EIF2S1, NME1), development (HPGD, NDUFV2, GSTM3, SEPT2, SERPINB5, ACTB, NGFR, MTPN, NME1), death (PACAP, PKM2, NGFR, CTSD, ANXA4, TPT1, ANXA5, NME1), response to stress (ACTB, HSPA1, UBE2N, ENO1, EIF2S1, ANXA5), cell cycle (GKN1, HPGD, SEPT2, NGFR, HSPA1, NME1), cell communication (GSTM3, ACTB, NGFR, ANXA4, ANXA5), transport (ATP5H, TTR, SEPT2, TPT1), and cell motility (SERPINB5, ACTB).
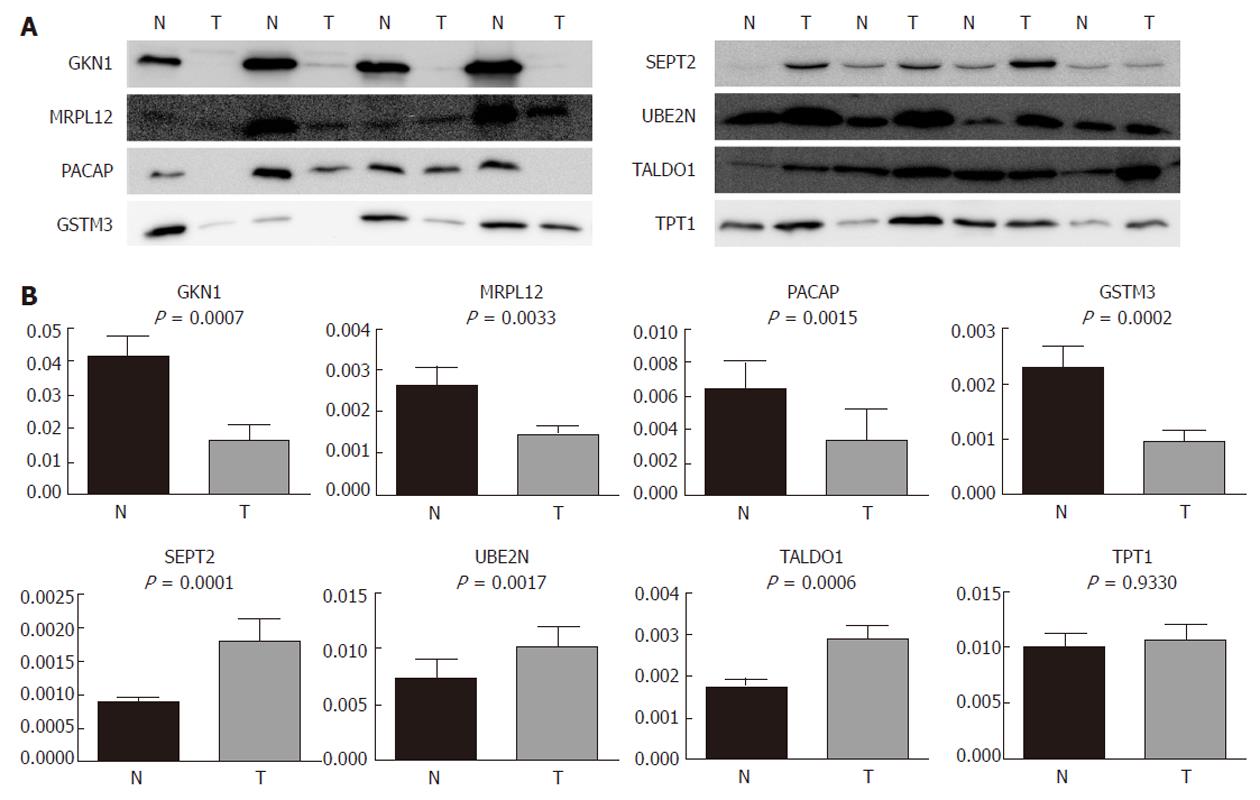
In order to validate the results from the 2-DE and to investigate the possibility of the identified proteins becoming relevant biomarkers, we performed an immunoblot analysis on an expanded group of samples: 11 that were used for 2-DE as well as an additional 16 (n = 27). The selection of proteins subjected to the analysis was based on their putative relevance. GKN1 was the most abundantly underexpressed protein and was already validated at the protein level[19], so it was chosen as a general control in our 2-DE and immunoblot experiments. MRPL12 has, to the best of our knowledge, not yet been found to be associated with gastric cancer, while SEPT2 (as the most abundantly overexpressed), UBE2N, TALDO1, TPT1, PACAP, and GSTM3 were already found to be associated with gastric adenocarcinoma, but were not previously validated at the protein level.
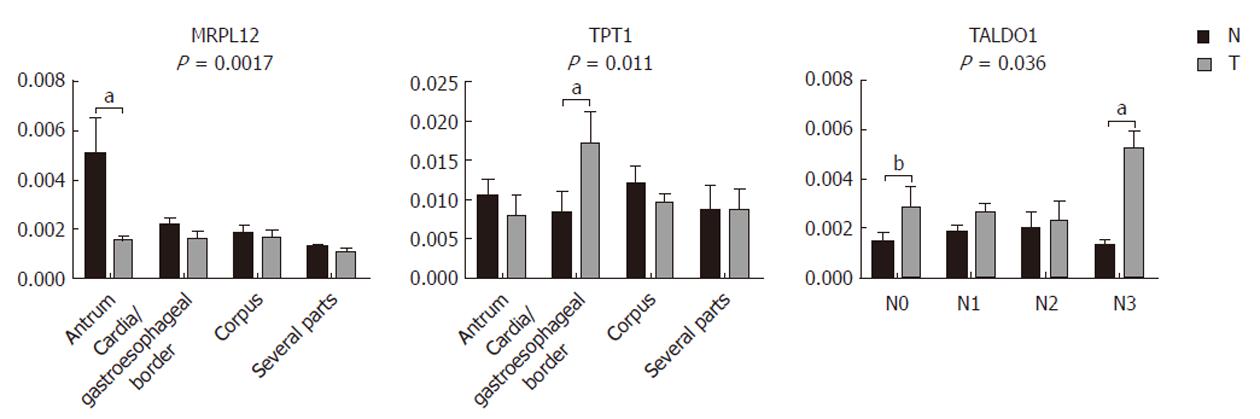
Our results (Figure 4) show that GKN1 (P = 0.0007), MRPL12 (P = 0.0033), PACAP (P = 0.0015), and GSTM3 (P = 0.0002) were significantly underexpressed in gastric adenocarcinoma. SEPT2 (P = 0.0001), UBE2N (P = 0.0017), and TALDO1 (P = 0.0006), on the other hand, were significantly overexpressed. The immunoblot results confirmed our results from the 2-DE analysis. TPT1, on the other hand, did not appear as generally differentially expressed in the tumor samples compared to the non-tumor samples. However, we also checked for a correlation of the differential expression with the histopathological parameters. We observed (Figure 5) significant differences in the MRPL12 expression and tumor location (P = 0.017), as well as in TPT1 expression and tumor location (P = 0.011). TALDO1 expression was observed to correlate with pN status (P = 0.036). A higher rate of MRPL12 overexpression was found in the antrum (P < 0.001) and a higher rate of TPT1 overexpression was found in the cardia/gastroesophageal border (P < 0.01). A higher rate of TALDO1 overexpression was found in pN0 (P < 0.05) and pN3 (P < 0.01) tumors.
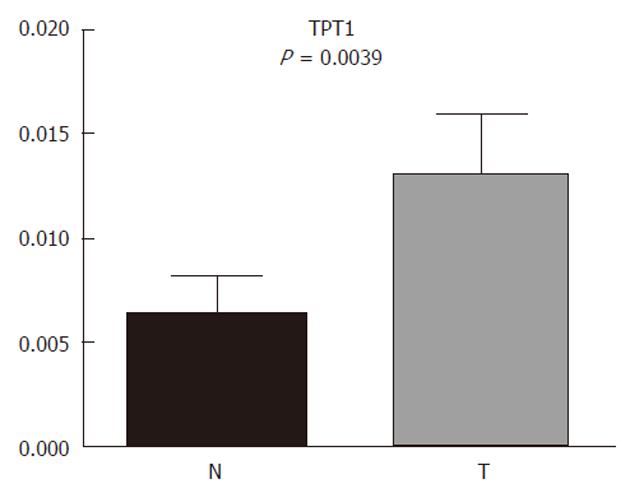
Due to the interesting results of TPT1 and cardia/gastroesophageal border correlation, we used additional tissue samples from four patients with cardia/gastroesophageal border adenocarcinoma to confirm it. The trend remained the same: TPT1 was upregulated. When combining tissues from all 10 patients with cardia/gastroesophageal border adenocarcinoma (six from before and four additional), Wilcoxon signed-rank test confirmed the differential expression (Figure 6).
Using a gel-based proteomic approach, we aimed to find and validate the differentially expressed proteins in a set of gastric adenocarcinoma patients. A total of 30 different proteins were identified in the study. They belonged to different biological processes, including metabolism, development, death, response to stress, cell cycle, cell communication, transport, and cell motility. The largest group, metabolism, contained 20 proteins with at least some role in cell metabolism.
A set of these proteins has already been found in similarly conducted experiments for GC. Among those downregulated in the tumor, enoyl CoA hydratase, mitochondrial precursor[20,21], acyl-CoA dehydrogenase[22], NADH dehydrogenase (ubiquinone) flavoprotein 2, mitochondrial precursor[22], phenazine biosynthesis-like domain-containing protein isoform a[22], and ATP synthase subunit d, mitochondrial[23] have been reported with the same alterations as in our study. In contrast to our observation, ATP synthase subunit d was reported to be upregulated in another study[22]. Among the proteins upregulated in the tumor, phosphoglycerate kinase 1[19], pyruvate kinase isozymes M1/M2[24,25], heat shock 70 kDa protein 1A/1B[24,25], cathepsin D precursor[22,24,26], annexin A4[26], alpha-enolase[24,25], nicotinamide N-methyltransferase[20,27,28], annexin A5[20,29], and actin, cytoplasmic 1 in rat GC metastases[30] have been found elsewhere with the same trend as in our study. However, other studies have reported that pyruvate kinase[22], alpha-enolase[26], and actin[26,31] have the opposite expression patterns compared to our results. This discrepancy could perhaps result from the heterogeneity that is often present in studies of human tumor tissue. In this paper, we present and discuss eight putative biomarkers for gastric carcinogensis that were identified in our study, among which seven were validated for the first time by means of immunoblotting.
GKN1 has already been identified as a downregulated gene in GC[32]. It is suggested to maintain gastric mucosal integrity and mediate repair after injury[33]. The protection of the mucosal barrier is thought to be due to the ability of GKN1 to alter the distribution of specific tight-junction proteins and to stabilize perijunctional actin[34]. It was later demonstrated to bind with F-actin in smooth muscle cells, suggesting a role in cell–cell adhesion and the assembly of actin stress fibers[35]. Recently, GKN1 has been shown to be a modulator of apoptotic signals[36]. It has been confirmed as a secreted protein and as being present in native and metaplastic gastric epithelium, but absent from the gastric carcinoma and the precursor lesion of intestinal metaplasia, making it a possible tumor suppressor in gastric carcinogenesis[37].
GKN1 expression is downregulated in Helicobacter-pylori-positive patients[38]. In another study, a loss of GKN1 occurred, especially in the diffuse-type tumor, but was associated with a significantly worse outcome in the intestinal type[39]. It has been found to be downregulated in GC, using 2-DE[20,23,25,27], and these results have been validated at the protein and mRNA levels[19]. Consistently, we found that GKN1 was underexpressed in tumor tissue, using 2-DE, and we also validated the results by immunoblotting. Our result is in agreement with other reports and, like other research groups, we were also unable to find any correlation with histopathological parameters at the protein level. The overlapping of both steps of the biomarker identification with a number of other studies in this case contributed to the confidence in the approach for the analysis of additional biomarkers found in our proteomic analysis.
MRPL12 is the first cloned and characterized mammalian mitochondrial ribosomal protein encoded by the nucleus[40]. It accumulates in cells at the mRNA and protein levels upon growth-factor stimulation. The enhanced expression later contributes to transcriptional activation[41]. MRPL12 mRNA levels have been detected in different organs, being especially high in the colon and skeletal muscle[41]. Besides being a component of the mitochondrial ribosome (its dimers bind the large ribosomal unit), MRPL12 binds to mitochondrial RNA polymerase and enhances transcription in vitro[42]. It has been speculated that it may either directly couple transcription and translation by binding simultaneously to polymerase and ribosomes, or alone bind to polymerase and activate its transcriptional activity in some way[43]. However, Litonin et al[44] have observed no such stimulation, so further experiments are necessary to clarify the possible role of this protein in transcription.
MRPL12 is differentially expressed, and it has previously been observed by 2-DE to be overexpressed in prostate cancer[45] and in hepatitis B virus (HBV)-associated hepatocellular carcinoma[46]. This is not in accordance with our results because we showed that MRPL12 was downregulated in gastric adenocarcinoma. We also observed it correlation with location: higher rates of underexpression were found in the antrum. In the context of cancer, the knockdown of MRPL12 decreases mitochondrial activity, increases glycolysis and accelerates tumor growth[47]. We speculate that it may also be the case in gastric adenocarcinoma.
PACAP was found as an expressed sequence tag from a microarray analysis where it exhibited downregulation in intestinal-type GC[48]. The protein was found in the endoplasmic reticulum of lymphocytes, where it exhibited upregulation in the course of B-cell differentiation[49,50]. It was found to assist in the oxidative folding of Ig domains; however, both research groups were uncertain as to whether it acted as an oxidoreductase or as a chaperone. In a very recent report[51], PACAP was described as helping to diversify peripheral B-cell functions by regulating Ca2+ stores, antibody secretion and integrin activation.
A patent application[52] has disclosed the use of PACAP as a universal marker of different types of cancer, GC included. However, they claim that increased concentrations of the protein and/or its fragments are associated with cancer, whereas we discovered just the opposite for GC. The same was reported by Huang et al[31] in a 2-DE experiment (although this was without validation), by Hasegawa et al[48] and Katoh et al[53]. However, in the latter two cases, this was at the non-protein level. We validated our results by immunoblotting. Due to its downregulation in the tumor, we strongly support the previous speculation that PACAP might be a candidate tumor-suppressor gene[53].
GSTM3 is a glutathione S-transferase (GST) that belongs to the mu class[54]. It is rather unusual; it is not only about 70% identical in its protein sequence to the other mu-class transferases, but it is also considerably shorter and transcribed in the reverse orientation[55].
Several polymorphisms have been found in GSTs, which can alter the susceptibility to carcinogens and toxins and influence the toxicity and efficacy of drug treatment[54]. They have been studied in relation to several cancers, GC included. For example, Martinez and colleagues have found no association with the GSTM3 genotype and GC risk[56], whereas Tatemichi and co-workers have described a possible association between GSTM3 polymorphisms and Ig titer levels in serum against H. pylori[57].
GSTM3 is found in several normal tissues, including the stomach[58]. A comparison of the differential GSTM3 expression in cancers is made rather difficult by the fact that many studies have focused on the whole GST family or class, but not on individual isoforms. For instance, antral GST enzyme activity has been found to be significantly lower in the stomach of H. pylori-infected patients[59], but the contributions of separate isoforms has not been studied. For GSTM3 specifically, the gene is highly expressed in a subgroup of patients with head and neck squamous cell carcinoma[60]. The protein is upregulated in neuroblastoma[61] and in polycystic ovary syndrome[62]. We found that GSTM3 was downregulated in gastric adenocarcinoma. Our results are in agreement with a study reporting downregulation in the seminomatous germ cell tumor, where the changes were also reflected at the transcriptional level[63]. This reduced expression could be indicative of the decreased detoxification capacity of tumor cells[59].
Septins belong to a family of conserved GTP-binding proteins[64]. They have been implicated in many cellular processes. SEPT2 is thought to be involved in cytokinesis, as well as chromosome congression and segregation[65,66]. Its fibers appear to contact actin bundles and focal adhesion complexes physically, thereby linking it to a functional interaction with actin-based cytoskeletal systems in interphase cells[65]. It has also been found in the microtubule spindle during metaphase and is proposed to form a mitotic scaffold for different effectors to coordinate cytokinesis with chromosome congression and segregation[66]. Several other binding partners and functions have been proposed for SEPT2, such as the DNA damage response, the regulation of the efficiency of vesicular transport, and FCγR-mediated phagocytosis[64]. Recently, it has been reported that SEPT2 is part of a diffusion barrier between the primary cilia and the cell and it is essential for retaining receptor-signaling pathways in primary cilia[67]. Also, in response to physiological and pathological stimuli, SEPT2 redistributes and its interaction with actin increases, which allows for the dynamic modulation of the airway epithelial barrier function[68]. Despite all the progress, the exact molecular mechanisms, cellular, and physiological functions of septins are still poorly understood and interactome studies could help[69].
Septins have been linked to diseases such as neurodegeneration and cancer[70]. It is proposed that altered SEPT2 expression can lead to disordered chromosomal dynamics and underlie the development of the aneuploidy common to cancers[66]. SEPT2, among others, has been found to be a fusion partner of the mixed lineage leukemia (MLL) gene in therapy-related acute myeloid leukemia[71]. Such fusion is associated with downregulation of SEPT2 and MLL in myeloid neoplasia[72]. It has also been shown to be downregulated in glioblastoma[73]. On the other hand, it has been identified as upregulated in hepatoma carcinoma cells, where its phosphorylation on Ser218 by casein kinase 2 has been determined as crucial for hepatoma carcinoma cell proliferation[74]. In a 2-DE experiment, SEPT2 was, in agreement with our results, determined to be upregulated in GC[22]; however, no validation was carried out. The same expression pattern was found in renal cell carcinoma[75] and in late-stage human colon cancer tissue[76], and it is abundantly expressed in several brain tumors and brain-tumor cell lines[77]. Taken as a whole, these results suggest its possible role as an oncogene.
Ubiquitination is a post-translational modification carried out in several steps[78], one of them being conjugation of an activated ubiquitin to an ubiquitin-conjugating enzyme (E2) via a highly conserved catalytic cysteine residue. By directly influencing the type of lysine used to label the substrates, they influence the fate of the substrates. One of the E2s is UBE2N, which acts as part of a complex that enables the formation of the non-canonical Lys63-mediated polyubiquitin chains[79]. As opposed to Lys48-mediated ones, these do not target proteasome degradation but mediate other processes. Among other functions, it has been shown that UBE2N in a complex with Mms2 functions via Lys63-mediated polyubiquitination in DNA repair, whereas in a complex with Uev1A, it functions in activating nuclear factor-κB signaling[78,80]. Both of the partner proteins, however, are dispensable for the RNF8-dependent propagation of DNA damage signals via ubiquitination[81], although there are some questions as to the importance of this activity[82].
UBE2N is differentially expressed between different types of leukemia and lymphoma cell lines[83]. It has significantly lower transcriptional expression levels in non-small-cell lung cancer and is correlated with pN and the stage of the disease[84]. In a breast-cancer metastatic model using iTRAQ technology, UBE2N was downregulated when comparing cells with the most metastatic potential and non-metastatic cells[85]. On the other hand, it was observed in a 2-DE experiment to be overexpressed in HBV-associated liver cancer[46], which is consistent with our results. It has already been shown to be differentially expressed in GC[86]; however, it has not been validated whether it is up- or downregulated. UBE2N-dependent Lys63-mediated polyubiquitination regulates processes that often enhance cell survival in response to certain forms of stress[87], therefore, our result supports its implication in the regulation of similar processes in gastric cancerogenesis.
TALDO1 is an almost ubiquitous cofactor-less enzyme of the pentose phosphate pathway[88]. Its activity is tissue specific and, in the brain, it is selectively expressed in oligodendrocytes, thus connecting it to different autoimmune diseases, such as multiple sclerosis[89]. Its expression is developmentally controlled[88].
TALDO1 is the rate-limiting enzyme of the non-oxidative part of the pentose phosphate pathway[88] that catalyzes the reversible transfer of a three-carbon unit between various sugar phosphates (from ketose to aldose sugar phosphates). It has a role in regulating the balance between the two branches of the pentose phosphate pathway and its overall output, as measured by NADPH and glutathione production, and thus influences the sensitivity to cell-death signals[90].
When comparing tumor and normal TALDO1, its activity is increased in neoplastic liver[91]. Its gene is highly expressed in a subgroup of patients with squamous cell carcinoma of the head and neck[60]. Furthermore, it is upregulated in late-stage human colon-cancer tissue[76] and in the sera of colorectal cancer patients[92]. In metastatic, compared to non-metastatic GC cell lines[93] and in GC tissue[86], TALDO1 was overexpressed, as shown by 2-DE. However, in both studies, again, no validation was performed. All these studies are consistent with our results, which were also validated by immunoblotting. We also found that TALDO1 correlated with pN status at stages pN0 and pN3. A higher TALDO1 expression in the tumor tissue could reflect an increased metabolism of glucose for the synthesis of nucleic acids in malignant cells[92].
TPT1 is a ubiquitously expressed and highly conserved protein. It is associated with various cellular processes, such as cell-cycle progression, release of histamine and various interleukins, apoptosis, malignant transformation, and tumor reversion[94,95]. Very recently, it was also discovered as a glucose-regulated protein, important for the survival of pancreatic beta cells[96].
It has been implicated in cancer, although it is not tumor-specific. It is upregulated in various tumor tissue cell lines when compared to normal tissue cell lines[97], in breast[98] and colon cancer[99]. As for the gastric tissue, TPT1 has been reported as cDNA present in libraries only from normal gastric tissues[100]. In our study, TPT1 was not validated as generally differentially expressed in the whole group of samples. Instead, its expression was location-correlated; TPT1 was upregulated in gastric adenocarcinoma from the cardia/gastroesophageal border. In contrast to the general worldwide decline of GC rates, an increasing incidence of gastric cardia cancer has been observed in several countries[1]. This suggests that it is a distinct clinical entity[26]. Therefore, it is possible that TPT1 is implemented only in gastric cardia/gastroesophageal border carcinogenesis.
Comparison of tumor and adjacent, non-tumor gastric tissues by means of proteome analysis, including differential 2-DE coupled to MS analysis, revealed 30 protein alterations. Some of the differentially expressed proteins had already been observed in GC in previous studies, which supports the reliability of our analysis. Several other proteins were found with the same trend of differential expression in other types of cancer, which could suggest that they are commonly involved in carcinogenesis. The high mortality rate from GC is due to delayed detection and surgical resection at advanced stages of the disease. A breakthrough in the early diagnosis of GC has not occurred yet and there are currently very few markers that are clinically in use; however, advances in proteomic research are facilitating the identification of novel diagnostic, prognostic, or therapeutic biomarkers. It is apparent that a collection of protein biomarkers will be necessary for reliable cancer detection and monitoring, as single biomarkers often have an inadequate predictive value[101]. There is, therefore, a need for the expedited development of new, validated biomarkers to be added to the list of clinically relevant tumor-associated proteins in the proteome databases of gastric tissue and cell lines. To the best of our knowledge, we are the first to observe aberrant expression of MRPL12 in gastric adenocarcinoma, and, in addition, aberrant expression of PACAP, GSTM3, SEPT2, UBE2N, TALDO1 and TPT1 for the gastric cardia/esophageal border were validated in gastric adenocarcinoma, also for the first time. Future experiments are planned to use these biomarkers in the design of a combinatory microarray and to translate the obtained results to blood samples, so the proteins would ultimately be useful as biomarkers for early detection.
We would like to thank Imagif, La Plateforme du Vivant, France, for their major contribution to the MS analysis.
Gastric cancer (GC) is the fourth most common cancer worldwide. Despite declining incidence rates, it is still the second leading cause of cancer death and thus remains a major health problem. The majority of patients are diagnosed at an advanced stage when the 5-year survival rate is very low.
Current diagnosis is invasive, whereas blood biomarkers lack sensitivity and specificity. This study investigated proteome changes in gastric cancerous vs non-cancerous tissue in the hope of discovering new biomarker candidates. It reports the validation of seven aberrantly expressed proteins, one of them being reported as a novel candidate biomarker.
This is believed to be the first study that reports aberrant expression of 39S ribosomal protein L12 in gastric adenocarcinoma, and for the first time, validates aberrant expression of plasma cell-induced resident endoplasmic reticulum protein, glutathione S-transferase mu 3, septin-2, ubiquitin-conjugating enzyme E2 N, transaldolase, and translationally controlled tumor protein for the cardia/gastroesophageal border at the protein level.
Future work will be focused on using the validated biomarkers in the design of a diagnostic protein microarray and on translating the research to blood samples, so that the proteins would ultimately be useful as biomarkers for early detection. By showing their differential expression, this work might contribute to the efforts to understand GC carcinogenesis, as well as present a set of possible diagnostic biomarkers for gastric adenocarcinoma.
Proteomic analysis was done in the present study. And several potential biomarkers were selected as an important protein for gastric cancer carcinogenesis. These fields are very important for further developments of the clinical treatment in patients with various malignancies including gastric cancer.
| 1. | Jemal A, Center MM, DeSantis C, Ward EM. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol Biomarkers Prev. 2010;19:1893-1907. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1754] [Cited by in RCA: 1914] [Article Influence: 119.6] [Reference Citation Analysis (1)] |
| 2. | Menges M. Gastric cancer: Where is the place for the surgeon, the oncologist and the endoscopist today? World J Gastrointest Oncol. 2011;3:10-13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 9] [Cited by in RCA: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 3. | Cappetta A, Lonardi S, Pastorelli D, Bergamo F, Lombardi G, Zagonel V. Advanced gastric cancer (GC) and cancer of the gastro-oesophageal junction (GEJ): focus on targeted therapies. Crit Rev Oncol Hematol. 2012;81:38-48. [PubMed] |
| 4. | Takayama S, Wakasugi T, Funahashi H, Takeyama H. Strategies for gastric cancer in the modern era. World J Gastrointest Oncol. 2010;2:335-341. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 7] [Cited by in RCA: 12] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 5. | Sturgeon CM, Duffy MJ, Hofmann BR, Lamerz R, Fritsche HA, Gaarenstroom K, Bonfrer J, Ecke TH, Grossman HB, Hayes P. National Academy of Clinical Biochemistry Laboratory Medicine Practice Guidelines for use of tumor markers in liver, bladder, cervical, and gastric cancers. Clin Chem. 2010;56:e1-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 141] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 6. | Lottspeich F. Introduction to proteomics. Methods in Molecular Biology. New York: Humana Press 2009; 3-10. |
| 7. | Chen G, Gharib TG, Huang CC, Taylor JM, Misek DE, Kardia SL, Giordano TJ, Iannettoni MD, Orringer MB, Hanash SM. Discordant protein and mRNA expression in lung adenocarcinomas. Mol Cell Proteomics. 2002;1:304-313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 710] [Cited by in RCA: 753] [Article Influence: 31.4] [Reference Citation Analysis (0)] |
| 8. | Plymoth A, Hainaut P. Proteomics beyond proteomics: toward clinical applications. Curr Opin Oncol. 2011;23:77-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 9. | May C, Brosseron F, Chartowski P, Schumbrutzki C, Schoenebeck B, Marcus K. Instruments and methods in proteomics. Methods in Molecular Biology. New York: Humana Press 2011; 3-26. |
| 10. | O'Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007-4021. [PubMed] |
| 11. | Rabilloud T, Chevallet M, Luche S, Lelong C. Two-dimensional gel electrophoresis in proteomics: Past, present and future. J Proteomics. 2010;73:2064-2077. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 322] [Cited by in RCA: 295] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 12. | Ryu JW, Kim HJ, Lee YS, Myong NH, Hwang CH, Lee GS, Yom HC. The proteomics approach to find biomarkers in gastric cancer. J Korean Med Sci. 2003;18:505-509. [PubMed] |
| 13. | Kim YS, Hwang SY, Oh S, Sohn H, Kang HY, Lee JH, Cho EW, Kim JY, Yoo JS, Kim NS. Identification of target proteins of N-acetylglucosaminyl-transferase V and fucosyltransferase 8 in human gastric tissues by glycomic approach. Proteomics. 2004;4:3353-3358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 14. | Melle C, Ernst G, Schimmel B, Bleul A, Kaufmann R, Hommann M, Richter KK, Daffner W, Settmacher U, Claussen U. Characterization of pepsinogen C as a potential biomarker for gastric cancer using a histo-proteomic approach. J Proteome Res. 2005;4:1799-1804. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 47] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 15. | Stroumbouli PK, Lazaris AC, Patsouris ES, Kalofoutis A, Nonni A, Tseleni-Balafouta S. Analysis of cytoplasmic polypeptides expression in gastric cancer and correlation with pathologic parameters. Cancer therapy. 2006;4:223-230. |
| 16. | Kang HJ, Koh KH, Yang E, You KT, Kim HJ, Paik YK, Kim H. Differentially expressed proteins in gastrointestinal stromal tumors with KIT and PDGFRA mutations. Proteomics. 2006;6:1151-1157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 33] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 17. | Li W, Li JF, Qu Y, Chen XH, Qin JM, Gu QL, Yan M, Zhu ZG, Liu BY. Comparative proteomics analysis of human gastric cancer. World J Gastroenterol. 2008;14:5657-5664. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 20] [Cited by in RCA: 18] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 18. | Mortz E, Krogh TN, Vorum H, Görg A. Improved silver staining protocols for high sensitivity protein identification using matrix-assisted laser desorption/ionization-time of flight analysis. Proteomics. 2001;1:1359-1363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 19. | Yoshihara T, Kadota Y, Yoshimura Y, Tatano Y, Takeuchi N, Okitsu H, Umemoto A, Yamauchi T, Itoh K. Proteomic alteration in gastic adenocarcinomas from Japanese patients. Mol Cancer. 2006;5:75. [PubMed] |
| 20. | Nishigaki R, Osaki M, Hiratsuka M, Toda T, Murakami K, Jeang KT, Ito H, Inoue T, Oshimura M. Proteomic identification of differentially-expressed genes in human gastric carcinomas. Proteomics. 2005;5:3205-3213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 92] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 21. | Wu C, Luo Z, Chen X, Wu C, Yao D, Zhao P, Liu L, Shi B, Zhu L. Two-dimensional differential in-gel electrophoresis for identification of gastric cancer-specific protein markers. Oncol Rep. 2009;21:1429-1437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 12] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 22. | Zhang J, Kang B, Tan X, Bai Z, Liang Y, Xing R, Shao J, Xu N, Wang R, Liu S. Comparative analysis of the protein profiles from primary gastric tumors and their adjacent regions: MAWBP could be a new protein candidate involved in gastric cancer. J Proteome Res. 2007;6:4423-4432. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 23. | Ebert MP, Krüger S, Fogeron ML, Lamer S, Chen J, Pross M, Schulz HU, Lage H, Heim S, Roessner A. Overexpression of cathepsin B in gastric cancer identified by proteome analysis. Proteomics. 2005;5:1693-1704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 83] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 24. | Chen CD, Wang CS, Huang YH, Chien KY, Liang Y, Chen WJ, Lin KH. Overexpression of CLIC1 in human gastric carcinoma and its clinicopathological significance. Proteomics. 2007;7:155-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 90] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 25. | He QY, Cheung YH, Leung SY, Yuen ST, Chu KM, Chiu JF. Diverse proteomic alterations in gastric adenocarcinoma. Proteomics. 2004;4:3276-3287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 119] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 26. | Cheng Y, Zhang J, Li Y, Wang Y, Gong J. Proteome analysis of human gastric cardia adenocarcinoma by laser capture microdissection. BMC Cancer. 2007;7:191. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 27. | Jang JS, Cho HY, Lee YJ, Ha WS, Kim HW. The differential proteome profile of stomach cancer: identification of the biomarker candidates. Oncol Res. 2004;14:491-499. [PubMed] |
| 28. | Lim BH, Cho BI, Kim YN, Kim JW, Park ST, Lee CW. Overexpression of nicotinamide N-methyltransferase in gastric cancer tissues and its potential post-translational modification. Exp Mol Med. 2006;38:455-465. [PubMed] |
| 29. | Wang KJ, Wang RT, Zhang JZ. Identification of tumor markers using two-dimensional electrophoresis in gastric carcinoma. World J Gastroenterol. 2004;10:2179-2183. [PubMed] |
| 30. | Chen J, Kähne T, Röcken C, Götze T, Yu J, Sung JJ, Chen M, Hu P, Malfertheiner P, Ebert MP. Proteome analysis of gastric cancer metastasis by two-dimensional gel electrophoresis and matrix assisted laser desorption/ionization-mass spectrometry for identification of metastasis-related proteins. J Proteome Res. 2004;3:1009-1016. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 59] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 31. | Huang Q, Huang Q, Chen W, Wang L, Lin W, Lin J, Lin X. Identification of transgelin as a potential novel biomarker for gastric adenocarcinoma based on proteomics technology. J Cancer Res Clin Oncol. 2008;134:1219-1227. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 43] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 32. | Yoshikawa Y, Mukai H, Hino F, Asada K, Kato I. Isolation of two novel genes, down-regulated in gastric cancer. Jpn J Cancer Res. 2000;91:459-463. [PubMed] |
| 33. | Toback FG, Walsh-Reitz MM, Musch MW, Chang EB, Del Valle J, Ren H, Huang E, Martin TE. Peptide fragments of AMP-18, a novel secreted gastric antrum mucosal protein, are mitogenic and motogenic. Am J Physiol Gastrointest Liver Physiol. 2003;285:G344-G353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 59] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 34. | Walsh-Reitz MM, Huang EF, Musch MW, Chang EB, Martin TE, Kartha S, Toback FG. AMP-18 protects barrier function of colonic epithelial cells: role of tight junction proteins. Am J Physiol Gastrointest Liver Physiol. 2005;289:G163-G171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 57] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 35. | Hnia K, Notarnicola C, de Santa Barbara P, Hugon G, Rivier F, Laoudj-Chenivesse D, Mornet D. Biochemical properties of gastrokine-1 purified from chicken gizzard smooth muscle. PLoS One. 2008;3:e3854. |
| 36. | Rippa E, La Monica G, Allocca R, Romano MF, De Palma M, Arcari P. Overexpression of gastrokine 1 in gastric cancer cells induces fas-mediated apoptosis. J Cell Physiol. 2010;Dec 28; Epub ahead of print. [PubMed] |
| 37. | Oien KA, McGregor F, Butler S, Ferrier RK, Downie I, Bryce S, Burns S, Keith WN. Gastrokine 1 is abundantly and specifically expressed in superficial gastric epithelium, down-regulated in gastric carcinoma, and shows high evolutionary conservation. J Pathol. 2004;203:789-797. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 82] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 38. | Nardone G, Martin G, Rocco A, Rippa E, La Monica G, Caruso F, Arcari P. Molecular expression of Gastrokine 1 in normal mucosa and in Helicobacter pylori-related preneoplastic and neoplastic gastric lesions. Cancer Biol Ther. 2008;7:1890-1895. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 39. | Moss SF, Lee JW, Sabo E, Rubin AK, Rommel J, Westley BR, May FE, Gao J, Meitner PA, Tavares R. Decreased expression of gastrokine 1 and the trefoil factor interacting protein TFIZ1/GKN2 in gastric cancer: influence of tumor histology and relationship to prognosis. Clin Cancer Res. 2008;14:4161-4167. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 78] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 40. | Marty L, Fort P. A delayed-early response nuclear gene encoding MRPL12, the mitochondrial homologue to the bacterial translational regulator L7/L12 protein. J Biol Chem. 1996;271:11468-11476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 41. | Marty L, Taviaux S, Fort P. Expression and human chromosomal localization to 17q25 of the growth-regulated gene encoding the mitochondrial ribosomal protein MRPL12. Genomics. 1997;41:453-457. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 42. | Wang Z, Cotney J, Shadel GS. Human mitochondrial ribosomal protein MRPL12 interacts directly with mitochondrial RNA polymerase to modulate mitochondrial gene expression. J Biol Chem. 2007;282:12610-12618. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 80] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 43. | Shutt TE, Shadel GS. A compendium of human mitochondrial gene expression machinery with links to disease. Environ Mol Mutagen. 2010;51:360-379. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 49] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 44. | Litonin D, Sologub M, Shi Y, Savkina M, Anikin M, Falkenberg M, Gustafsson CM, Temiakov D. Human mitochondrial transcription revisited: only TFAM and TFB2M are required for transcription of the mitochondrial genes in vitro. J Biol Chem. 2010;285:18129-18133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 162] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 45. | Lexander H, Palmberg C, Auer G, Hellström M, Franzén B, Jörnvall H, Egevad L. Proteomic analysis of protein expression in prostate cancer. Anal Quant Cytol Histol. 2005;27:263-272. [PubMed] |
| 46. | Li C, Tan YX, Zhou H, Ding SJ, Li SJ, Ma DJ, Man XB, Hong Y, Zhang L, Li L. Proteomic analysis of hepatitis B virus-associated hepatocellular carcinoma: Identification of potential tumor markers. Proteomics. 2005;5:1125-1139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 93] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 47. | Chen Y, Cairns R, Papandreou I, Koong A, Denko NC. Oxygen consumption can regulate the growth of tumors, a new perspective on the Warburg effect. PLoS One. 2009;4:e7033. |
| 48. | Hasegawa S, Furukawa Y, Li M, Satoh S, Kato T, Watanabe T, Katagiri T, Tsunoda T, Yamaoka Y, Nakamura Y. Genome-wide analysis of gene expression in intestinal-type gastric cancers using a complementary DNA microarray representing 23,040 genes. Cancer Res. 2002;62:7012-7017. [PubMed] |
| 49. | Shimizu Y, Meunier L, Hendershot LM. pERp1 is significantly up-regulated during plasma cell differentiation and contributes to the oxidative folding of immunoglobulin. Proc Natl Acad Sci USA. 2009;106:17013-17018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 56] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 50. | van Anken E, Pena F, Hafkemeijer N, Christis C, Romijn EP, Grauschopf U, Oorschot VM, Pertel T, Engels S, Ora A. Efficient IgM assembly and secretion require the plasma cell induced endoplasmic reticulum protein pERp1. Proc Natl Acad Sci USA. 2009;106:17019-17024. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 72] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 51. | Flach H, Rosenbaum M, Duchniewicz M, Kim S, Zhang SL, Cahalan MD, Mittler G, Grosschedl R. Mzb1 protein regulates calcium homeostasis, antibody secretion, and integrin activation in innate-like B cells. Immunity. 2010;33:723-735. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 99] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 52. | Hagmann ML, Karl J, Kloeckner J, Roessler M, Tacke M; Roche Diagnostich GmbH, F. Hoffmann-La Roche AG, Hagmann ML, Karl J, Kloeckner J, Roessler M, Tacke M. PACAP as a marker for cancer. International patent. 2010;. |
| 53. | Katoh M, Katoh M. MGC29506 gene, frequently down-regulated in intestinal-type gastric cancer, encodes secreted-type protein with conserved cysteine residues. Int J Oncol. 2003;23:235-241. [PubMed] |
| 54. | Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2571] [Cited by in RCA: 2745] [Article Influence: 130.7] [Reference Citation Analysis (3)] |
| 55. | Patskovsky YV, Huang MQ, Takayama T, Listowsky I, Pearson WR. Distinctive structure of the human GSTM3 gene-inverted orientation relative to the mu class glutathione transferase gene cluster. Arch Biochem Biophys. 1999;361:85-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 24] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 56. | Martínez C, Martín F, Fernández JM, García-Martín E, Sastre J, Díaz-Rubio M, Agúndez JA, Ladero JM. Glutathione S-transferases mu 1, theta 1, pi 1, alpha 1 and mu 3 genetic polymorphisms and the risk of colorectal and gastric cancers in humans. Pharmacogenomics. 2006;7:711-718. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 65] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 57. | Tatemichi M, Iwasaki M, Sasazuki S, Tsugane S. Association between polymorphisms in glutathione S-transferase Mu3 and IgG titer levels in serum against Helicobacter pylori. J Hum Genet. 2009;54:557-563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 58. | Coles BF, Chen G, Kadlubar FF, Radominska-Pandya A. Interindividual variation and organ-specific patterns of glutathione S-transferase alpha, mu, and pi expression in gastrointestinal tract mucosa of normal individuals. Arch Biochem Biophys. 2002;403:270-276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 55] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 59. | Verhulst ML, van Oijen AH, Roelofs HM, Peters WH, Jansen JB. Antral glutathione concentration and glutathione S-transferase activity in patients with and without Helicobacter pylori. Dig Dis Sci. 2000;45:629-632. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 63] [Reference Citation Analysis (1)] |
| 60. | Chung CH, Parker JS, Karaca G, Wu J, Funkhouser WK, Moore D, Butterfoss D, Xiang D, Zanation A, Yin X. Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell. 2004;5:489-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 508] [Cited by in RCA: 498] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 61. | He QY, Zhu R, Ren Y, Tam PK, Chiu JF. Serological protein profiling of neuroblastoma by ProteinChip SELDI-TOF technology. J Cell Biochem. 2005;95:165-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 62. | Cortón M, Botella-Carretero JI, López JA, Camafeita E, San Millán JL, Escobar-Morreale HF, Peral B. Proteomic analysis of human omental adipose tissue in the polycystic ovary syndrome using two-dimensional difference gel electrophoresis and mass spectrometry. Hum Reprod. 2008;23:651-661. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 87] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 63. | Zimmermann U, Junker H, Krämer F, Balabanov S, Kleist B, Kammer W, Nordheim A, Walther R. Comparative proteomic analysis of neoplastic and non-neoplastic germ cell tissue. Biol Chem. 2006;387:437-440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 64. | Cao L, Yu W, Wu Y, Yu L. The evolution, complex structures and function of septin proteins. Cell Mol Life Sci. 2009;66:3309-3323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 58] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 65. | Kinoshita M, Kumar S, Mizoguchi A, Ide C, Kinoshita A, Haraguchi T, Hiraoka Y, Noda M. Nedd5, a mammalian septin, is a novel cytoskeletal component interacting with actin-based structures. Genes Dev. 1997;11:1535-1547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 282] [Cited by in RCA: 294] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 66. | Spiliotis ET, Kinoshita M, Nelson WJ. A mitotic septin scaffold required for Mammalian chromosome congression and segregation. Science. 2005;307:1781-1785. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 193] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 67. | Hu Q, Milenkovic L, Jin H, Scott MP, Nachury MV, Spiliotis ET, Nelson WJ. A septin diffusion barrier at the base of the primary cilium maintains ciliary membrane protein distribution. Science. 2010;329:436-439. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 434] [Cited by in RCA: 400] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 68. | Sidhaye VK, Chau E, Breysse PN, King LS. Septin-2 mediates airway epithelial barrier function in physiologic and pathologic conditions. Am J Respir Cell Mol Biol. 2011;45:120-126. [PubMed] |
| 69. | Nakahira M, Macedo JN, Seraphim TV, Cavalcante N, Souza TA, Damalio JC, Reyes LF, Assmann EM, Alborghetti MR, Garratt RC. A draft of the human septin interactome. PLoS One. 2010;5:e13799. |
| 70. | Russell SE, Hall PA. Do septins have a role in cancer? Br J Cancer. 2005;93:499-503. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 62] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 71. | Cerveira N, Correia C, Bizarro S, Pinto C, Lisboa S, Mariz JM, Marques M, Teixeira MR. SEPT2 is a new fusion partner of MLL in acute myeloid leukemia with t(2; 11)(q37; q23). Oncogene. 2006;25:6147-6152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 34] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 72. | Cerveira N, Santos J, Bizarro S, Costa V, Ribeiro FR, Lisboa S, Correia C, Torres L, Vieira J, Snijder S. Both SEPT2 and MLL are down-regulated in MLL-SEPT2 therapy-related myeloid neoplasia. BMC Cancer. 2009;9:147. |
| 73. | Khalil AA. Biomarker discovery: a proteomic approach for brain cancer profiling. Cancer Sci. 2007;98:201-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 105] [Article Influence: 5.5] [Reference Citation Analysis (1)] |
| 74. | Yu W, Ding X, Chen F, Liu M, Shen S, Gu X, Yu L. The phosphorylation of SEPT2 on Ser218 by casein kinase 2 is important to hepatoma carcinoma cell proliferation. Mol Cell Biochem. 2009;325:61-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 75. | Craven RA, Hanrahan S, Totty N, Harnden P, Stanley AJ, Maher ER, Harris AL, Trimble WS, Selby PJ, Banks RE. Proteomic identification of a role for the von Hippel Lindau tumour suppressor in changes in the expression of mitochondrial proteins and septin 2 in renal cell carcinoma. Proteomics. 2006;6:3880-3893. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 48] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 76. | Nibbe RK, Markowitz S, Myeroff L, Ewing R, Chance MR. Discovery and scoring of protein interaction subnetworks discriminative of late stage human colon cancer. Mol Cell Proteomics. 2009;8:827-845. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 87] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 77. | Kim DS, Hubbard SL, Peraud A, Salhia B, Sakai K, Rutka JT. Analysis of mammalian septin expression in human malignant brain tumors. Neoplasia. 2004;6:168-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 50] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 78. | van Wijk SJ, Timmers HT. The family of ubiquitin-conjugating enzymes (E2s): deciding between life and death of proteins. FASEB J. 2010;24:981-993. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 275] [Cited by in RCA: 320] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 79. | Hofmann RM, Pickart CM. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 1999;96:645-653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 628] [Cited by in RCA: 640] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 80. | Andersen PL, Zhou H, Pastushok L, Moraes T, McKenna S, Ziola B, Ellison MJ, Dixit VM, Xiao W. Distinct regulation of Ubc13 functions by the two ubiquitin-conjugating enzyme variants Mms2 and Uev1A. J Cell Biol. 2005;170:745-755. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 145] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 81. | Huen MS, Huang J, Yuan J, Yamamoto M, Akira S, Ashley C, Xiao W, Chen J. Noncanonical E2 variant-independent function of UBC13 in promoting checkpoint protein assembly. Mol Cell Biol. 2008;28:6104-6112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 38] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 82. | Nakada S, Tai I, Panier S, Al-Hakim A, Iemura S, Juang YC, O'Donnell L, Kumakubo A, Munro M, Sicheri F. Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature. 2010;466:941-946. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 259] [Cited by in RCA: 303] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 83. | Gez S, Crossett B, Christopherson RI. Differentially expressed cytosolic proteins in human leukemia and lymphoma cell lines correlate with lineages and functions. Biochim Biophys Acta. 2007;1774:1173-1183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 84. | Saviozzi S, Ceppi P, Novello S, Ghio P, Lo Iacono M, Borasio P, Cambieri A, Volante M, Papotti M, Calogero RA. Non-small cell lung cancer exhibits transcript overexpression of genes associated with homologous recombination and DNA replication pathways. Cancer Res. 2009;69:3390-3396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 57] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 85. | Ho J, Kong JW, Choong LY, Loh MC, Toy W, Chong PK, Wong CH, Wong CY, Shah N, Lim YP. Novel breast cancer metastasis-associated proteins. J Proteome Res. 2009;8:583-594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 103] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 86. | Liu R, Li Z, Bai S, Zhang H, Tang M, Lei Y, Chen L, Liang S, Zhao YL, Wei Y. Mechanism of cancer cell adaptation to metabolic stress: proteomics identification of a novel thyroid hormone-mediated gastric carcinogenic signaling pathway. Mol Cell Proteomics. 2009;8:70-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 57] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 87. | Scheper J, Guerra-Rebollo M, Sanclimens G, Moure A, Masip I, Gonzalez-Ruiz D, Rubio N, Crosas B, Meca-Cortes O, Loukili N. Protein-protein interaction antagonists as novel inhibitors of non-canonical polyubiquitylation. PLoS One. 2010;5:e11403. |
| 88. | Samland AK, Sprenger GA. Transaldolase: from biochemistry to human disease. Int J Biochem Cell Biol. 2009;41:1482-1494. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 84] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 89. | Banki K, Colombo E, Sia F, Halladay D, Mattson DH, Tatum AH, Massa PT, Phillips PE, Perl A. Oligodendrocyte-specific expression and autoantigenicity of transaldolase in multiple sclerosis. J Exp Med. 1994;180:1649-1663. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 105] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 90. | Banki K, Hutter E, Colombo E, Gonchoroff NJ, Perl A. Glutathione levels and sensitivity to apoptosis are regulated by changes in transaldolase expression. J Biol Chem. 1996;271:32994-33001. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 164] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 91. | Heinrich PC, Morris HP, Weber G. Behavior of transaldolase (EC 2.2.1.2) and transketolase (EC 2.2.1.1) Activities in normal, neoplastic, differentiating, and regenerating liver. Cancer Res. 1976;36:3189-3197. [PubMed] |
| 92. | Ma Y, Peng J, Huang L, Liu W, Zhang P, Qin H. Searching for serum tumor markers for colorectal cancer using a 2-D DIGE approach. Electrophoresis. 2009;30:2591-2599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 38] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 93. | Chen YR, Juan HF, Huang HC, Huang HH, Lee YJ, Liao MY, Tseng CW, Lin LL, Chen JY, Wang MJ. Quantitative proteomic and genomic profiling reveals metastasis-related protein expression patterns in gastric cancer cells. J Proteome Res. 2006;5:2727-2742. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 60] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 94. | Bommer UA, Thiele BJ. The translationally controlled tumour protein (TCTP). Int J Biochem Cell Biol. 2004;36:379-385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 326] [Cited by in RCA: 334] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 95. | Telerman A, Amson R. The molecular programme of tumour reversion: the steps beyond malignant transformation. Nat Rev Cancer. 2009;9:206-216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 149] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 96. | Diraison F, Hayward K, Sanders KL, Brozzi F, Lajus S, Hancock J, Francis JE, Ainscow E, Bommer UA, Molnar E. Translationally controlled tumour protein (TCTP) is a novel glucose-regulated protein that is important for survival of pancreatic beta cells. Diabetologia. 2011;54:368-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 38] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 97. | Li F, Zhang D, Fujise K. Characterization of fortilin, a novel antiapoptotic protein. J Biol Chem. 2001;276:47542-47549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 216] [Cited by in RCA: 227] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 98. | Deng SS, Xing TY, Zhou HY, Xiong RH, Lu YG, Wen B, Liu SQ, Yang HJ. Comparative proteome analysis of breast cancer and adjacent normal breast tissues in human. Genomics Proteomics Bioinformatics. 2006;4:165-172. [PubMed] |
| 99. | Friedman DB, Hill S, Keller JW, Merchant NB, Levy SE, Coffey RJ, Caprioli RM. Proteome analysis of human colon cancer by two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics. 2004;4:793-811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 275] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 100. | Kim NS, Hahn Y, Oh JH, Lee JY, Oh KJ, Kim JM, Park HS, Kim S, Song KS, Rho SM. Gene cataloging and expression profiling in human gastric cancer cells by expressed sequence tags. Genomics. 2004;83:1024-1045. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 101. | Rusling JF, Kumar CV, Gutkind JS, Patel V. Measurement of biomarker proteins for point-of-care early detection and monitoring of cancer. Analyst. 2010;135:2496-2511. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 476] [Cited by in RCA: 375] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
Peer reviewers: Joshua R Friedman, MD, PhD, Assistant Professor of Pediatrics, Division of Gastroenterology, Hepatology and Nutrition, The Children’s Hospital of Philadelphia, University of Pennsylvania School of Medicine, 3615 Civic Center Blvd., ARC 902G, Philadelphia, PA 19104, United States; Takaaki Arigami, MD, PhD, Department of Surgical Oncology and Digestive Surgery, Field of Oncology, Kagoshima University Graduate School of Medical and Dental Sciences, 8-35-1 Sakuragaoka, Kagoshima 891-0175, Japan; Michele Milella, MD, Division of Medical Oncology A, Regina Elena National Cancer Institute, Via Elio Chianesi, n. 53, 00144 Rome, Italy
S- Editor Tian L L- Editor Kerr C E- Editor Li JY