Published online Aug 7, 2011. doi: 10.3748/wjg.v17.i29.3420
Revised: February 26, 2011
Accepted: March 5, 2011
Published online: August 7, 2011
AIM: To investigate the relationship between late SV40 factor (LSF) and Notch signaling in the development and progress of hepatocellular carcinoma (HCC).
METHODS: Liver cancer tissue specimens from 25 patients were analyzed for Notch-1 and LSF expression by immunohistochemistry. The correlation between expression and the biological effects of Notch-1 and LSF were analyzed using genetic and pharmacological strategies in HCC cell lines and human normal cell lines, including hepatic stellate cells (HSC) and human embryonic kidney epithelial cells (HEK).
RESULTS: Immunohistochemistry showed that both Notch-1 and LSF were significantly upregulated in HCC samples (76%, 19/25, P < 0.0001 and 84%, 21/25, P < 0.0001, respectively) compared with non-cancer samples. Activation of Notch-1 by exogenous transfection of Notch1 intracellular domain increased LSF expression in HSC and HEK cells to levels similar to those seen in HepG2 cells. Furthermore, blocking Notch-1 activation with a γ-secretase inhibitor, DAPT, downregulated LSF expression in HepG2 cells. Additionally, a biological behavior assay showed that forced overexpression of LSF promoted HepG2 cell proliferation and invasion.
CONCLUSION: LSF is a key mediator of the Notch signaling pathway, suggesting that it might be a novel therapeutic target for the treatment of HCC.
- Citation: Fan RH, Li J, Wu N, Chen PS. Late SV40 factor: A key mediator of Notch signaling in human hepatocarcinogenesis. World J Gastroenterol 2011; 17(29): 3420-3430
- URL: https://www.wjgnet.com/1007-9327/full/v17/i29/3420.htm
- DOI: https://dx.doi.org/10.3748/wjg.v17.i29.3420
Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related death in the world[1]. In China, the incidence of HCC has nearly doubled over the past 2 decades, and it is now second only to lung cancer in terms of total deaths[2]. The high mortality rate of HCC is due to the fact that no systemic therapy is available for advanced cases of the disease[3]. A previous study suggested that deregulated Notch signaling plays a crucial role in the transformation and neoplastic proliferation of human malignancies[4]. We showed that the transcriptional factor, late SV40 factor (LSF), is overexpressed in the liver of > 80% of HCC patients compared with normal liver. However, the exact role played by aberrant Notch signaling in hepatocarcinogenesis has not been elucidated. Here, we identified LSF as a downstream mediator of Notch1 signaling and showed that LSF mediates, at least in part, Notch1-induced carcinogenesis.
Notch genes encode heterodimeric transmembrane receptors, which play a critical role in maintaining the balance between cell proliferation, differentiation, and apoptosis. Aberrant expression of wild-type Notch receptors, ligands and downstream targets has been reported in cervical carcinoma[5,6], lung[7], colon[8], head and neck[9] and renal carcinomas[10], acute myeloid leukemia[11] and Hodgkin’s and large-cell lymphomas[12]. Hence, aberrant Notch signaling may contribute to carcinogenesis.
LSF, also known as LBP-1c and TFCP2, is a ubiquitously expressed mammalian transcription factor that regulates diverse cellular and viral promoters[13,14]. A major cellular target of LSF is the thymidylate synthase gene, which encodes the rate limiting enzyme in the production of dTTP, required for DNA synthesis[15]. Deregulated LSF expression might facilitate entry into the G1/S phase of the cell cycle, promote DNA synthesis, stimulate transformation, and facilitate cancer cell survival. However, there is little evidence to suggest a potential role for LSF in HCC. Moreover, the relationship between Notch-1 and LSF in human hepatocarcinogenesis is unknown.
In this study, we identified LSF as a novel mediator of dysregulated Notch signaling, and showed that activation of Notch-1 in human Ras-transformed cells upregulated the expression of LSF. We also performed a detailed experimental analysis to elucidate the relationship between Notch signaling and LSF expression in hepatocarcinogenesis and to identify signaling pathways that may be targeted for the treatment of highly aggressive HCC.
Archived liver cancer specimens were obtained from the Department of Pathology, Nanjing First Hospital, from November 2008 to January 2010. Twenty-five patients (20 men and 5 women) undergoing surgery for HCC were enrolled in the study. All diagnoses were based on pathological and/or cytological evidence. Paraffin sections were tested by histopathological examination and immunohistochemistry. Histological classification was conducted according to the criteria set out by Edmonson and Steiner[16]. Ethical approval was obtained from the hospital and informed consent was given by all patients prior to sample collection.
The human HCC cell lines, HepG2, Bel-7402, and QGY-7703 were obtained from the Chinese Academy of Sciences (Institute of Shanghai Cell Biology and Chinese Type Culture Collection, China). Human embryonic kidney epithelial cells (HEK), hepatic stellate cells (HSC) and hepatocytes (L02 cells) were preserved in our laboratory. Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen, USA) containing 10% fetal bovine serum (FBS) (Wisent, Canada) and maintained in humidified air containing 5% CO2 at 37°C and 95% humidity. HepG2 cells and hepatocytes (negative controls) were treated with 50 μmol/L DAPT (Sigma, USA) to block Notch signaling.
Reverse transcription-polymerase chain reaction (RT-PCR) was carried out using human HCC cell RNA as the template (Table 1). After incubation with specific restriction endonucleases (BamHI and XhoI for hRAS, Takara, Japan; BglII and HindIII for Notch-ICD1, Takara; and BglII
| Primer | Sequence (5’-3’) | Product size (bp) | Temperature (°C) |
| Notch1-ICD | Forward: CTAAGATCTCCTGAGGGCTTCAAAGTGTC | 2375 | 63 |
| Reverse: GCGAATTCCTTGAAGGCCTCCGGAAT | |||
| H-Ras (ORF) | Forward: GAGGATCCATGACGGAATATAAGCTGG | 668 | 61 |
| Reverse: GTGAATTCTCAGGAGAGCACACACTTG | |||
| Hes-1 | Forward: ATGAACGAGGTGACCCGCTT | 443 | 62 |
| Reverse: CTGGAAGGTGACACTGCGTT | |||
| LSF (ORF) | Forward: GGAAAGATCTAGGATGGCCTGGGCTCTGAAG | 1547 | 59 |
| Reverse: CAAAGCTTGGGCACGAAACGCCGCACTCCT | |||
| LSF (promoter analysis) | Forward: CTAGGTACCCAACATGGTAAGATCCTGTCTCT | 2020 | 56 |
| Reverse: GGAGATCTTCTCATCCCTGCTTTCTGTTTCCT | |||
| GAPDH | Forward: GGTGGAGGTCGGAGTCAACGGA | 240 | 60 |
| Reverse: GAGGGATCTCGCTCCTGGAGGA |
and HindIII for LSF, Takara) the PCR products were inserted into the corresponding sites of the pEGFP-C1 vector. Notch-ICD1 (bp 5309-7655 bp of the Notch-1 intracellular domain), the open-reading frame of H-Ras, and LSF were cloned into pEGFP-C1 (Clontech, Palo Alto, California, USA) to generate the H-Ras-pEGFP-C1, NICD1-pEGFP-C1, LSF-pEGFP-C1 recombinant vectors (for expression of green fluorescent protein), respectively. Transfection efficiency was estimated from the percentage of GFP-positive cells.
Successful construction of the recombinant vectors was confirmed by enzyme digestion and gene sequencing. The recombinant vectors were then transiently transfected into HEK cells and stably transfected to HSC cells using Lipofectamine 2000 (Invitrogen). NICD1-pEGFP-C1 and LSF-pEGFP-C1 were stably transfected to HepG2 cells after screening with 450 mg/L G418 (Sigma).
Immunoblotting of cellular extracts was performed using antibodies to Notch1-ICD (Cell Signaling Technology, MA, USA), LSF (Abcam Incorporation, MA, USA), H-Ras (Oncogene Research Products, CA, USA) and β-actin (Santa Cruz Biotechnology, CA, USA). The secondary antibodies used were HRP-conjugated anti-rabbit and anti-mouse Ig (Santa Cruz Biotechnology). Cells were harvested using NE-PER™ Nuclear and Cytoplasmic Extraction Reagents (Pierce, Rockford, USA) and equal amounts of cellular proteins were separated on 8% SDS-PAGE gels. Proteins were transferred to PVDF membranes, and the blots were probed with primary antibodies to Notch1-ICD (intracellular domain) and LSF. β-actin was used as the internal control protein. Blots were washed 3 times for 5 min with phosphate buffered saline (PBS) containing 0.1% Tween-20, and incubated with the HRP-conjugated secondary antibody (1:5000) for 1 h. The membranes were washed as described above and the bands detected by enhanced chemiluminescence (Pierce, Rockford, USA).
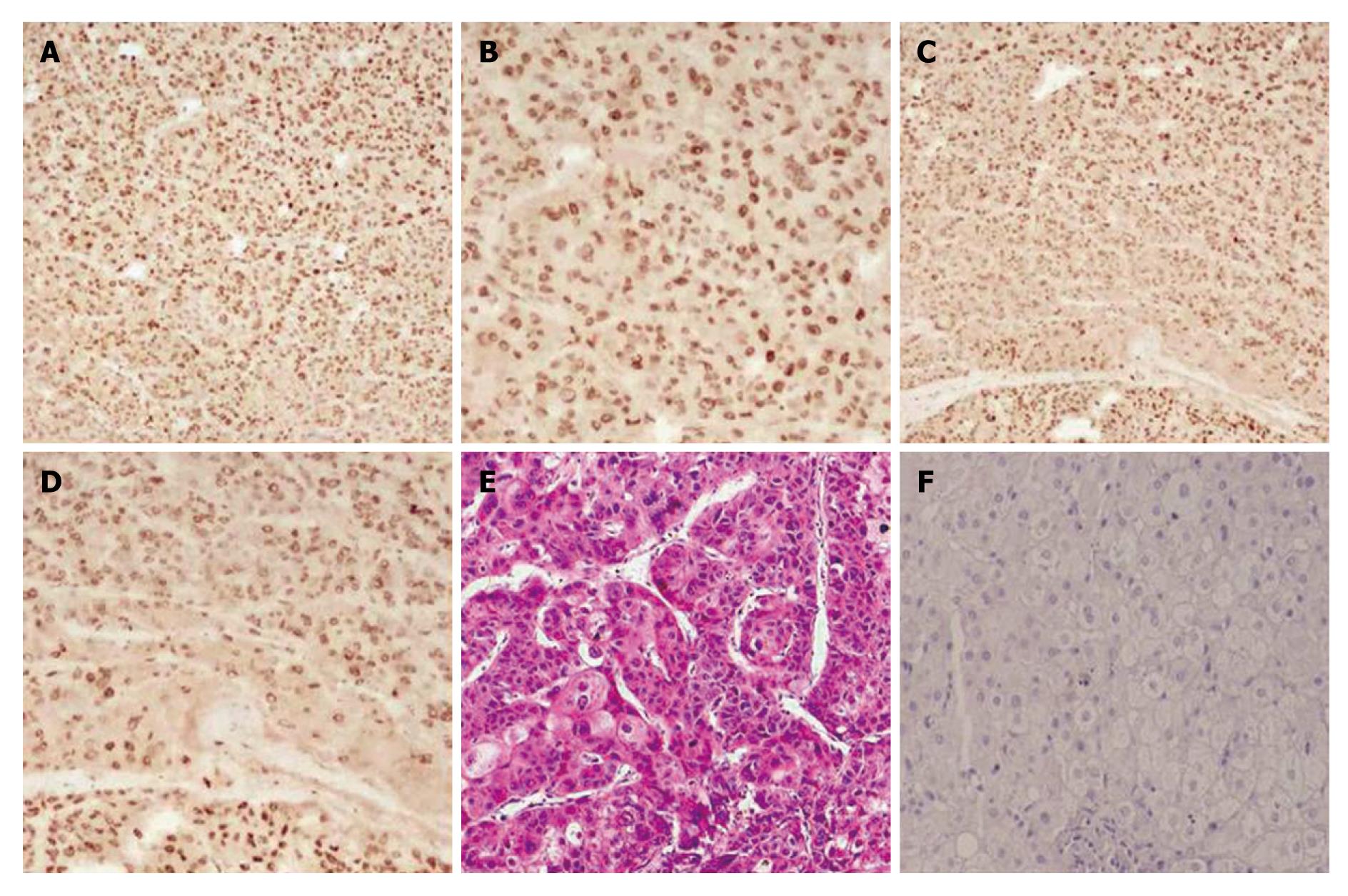
Immunohistochemistry was performed on formalin-fixed, paraffin-embedded surgical liver cancer specimens from 25 patients. Detection of the antigen-antibody complex was performed using the Mouse ABC staining kit (Santa Cruz Biotechnology, USA). Staining of sections was assessed in 10 consecutive fields (× 20 magnification) using a validated semi-quantitative scale where - denotes absence of staining; +/- denotes occasional, weak hepatocytic staining; + denotes staining of > 5% hepatocytes; ++ denotes staining of 6%-30% of hepatocytes, and +++ denotes staining of > 30% hepatocytes (high expression).
LSF DNA sequences between nucleotides (nt) 51 475 800
and 51 478 000 of GenBank accession GCF_000001305.13 were cloned into the pGL-3 vector (Premerger, USA) to generate LSF-pGL-3. Transient co-transfection into HSC was performed using a control plasmid (pSV-βgal) as an internal control in 96-well plates using the Lipofectamine 2000 transfection reagent. Dual luciferase activity was measured after 48 h using a kit from Promega (Madison, Wisconsin) and values were normalized for protein content and transfection efficiency (established from the percentage of GFP-positive cells).
The HepG2 cell cycle was assessed by flow cytometric analysis. Briefly, HepG2 cells were harvested and immediately fixed in 70% ethanol at 4°C overnight. Cells were then treated with 50 mg/L RNaseA (Sigma) for 30 min at 37°C and stained for 10 min with 50 mg/L PI (Sigma). Samples were then analyzed for their DNA content using a FACSAria Cell Cytometer (BD Biosciences, San Jose, CA, USA). Data were analyzed using Cell Quest software (BD Biosciences).
To assess cell growth, HepG2 cells infected with LSF-pEGFP-C (1 × 106 cells/mL) were seeded into 25 cm2 plates. The cells were cultured for 24 h in the serum-free DMEM supplemented with insulin (10 mg/mL) for synchronization. The cells were then washed three times in PBS (pH 7.4), digested with 0.25% trypsin, and prepared as a single-cell suspension. The cells were then plated into 96-well plates at a concentration of 2 × 103 cells/mL. The cells were cultured at 37°C in 5% CO2 and 95% humidity for 24, 48, 72, and 96 h. A thiazolyl blue test was performed to measure the absorbance at 492 nm. Four hours prior to each time point, MTT (20 μL) was added to each well at 37°C for 4 h. After removing the medium, 150 μL dimethylsulfoxide was added to each well and the A was measured using a microplate reader (Multiskan MK 3, Thermo, Germany).
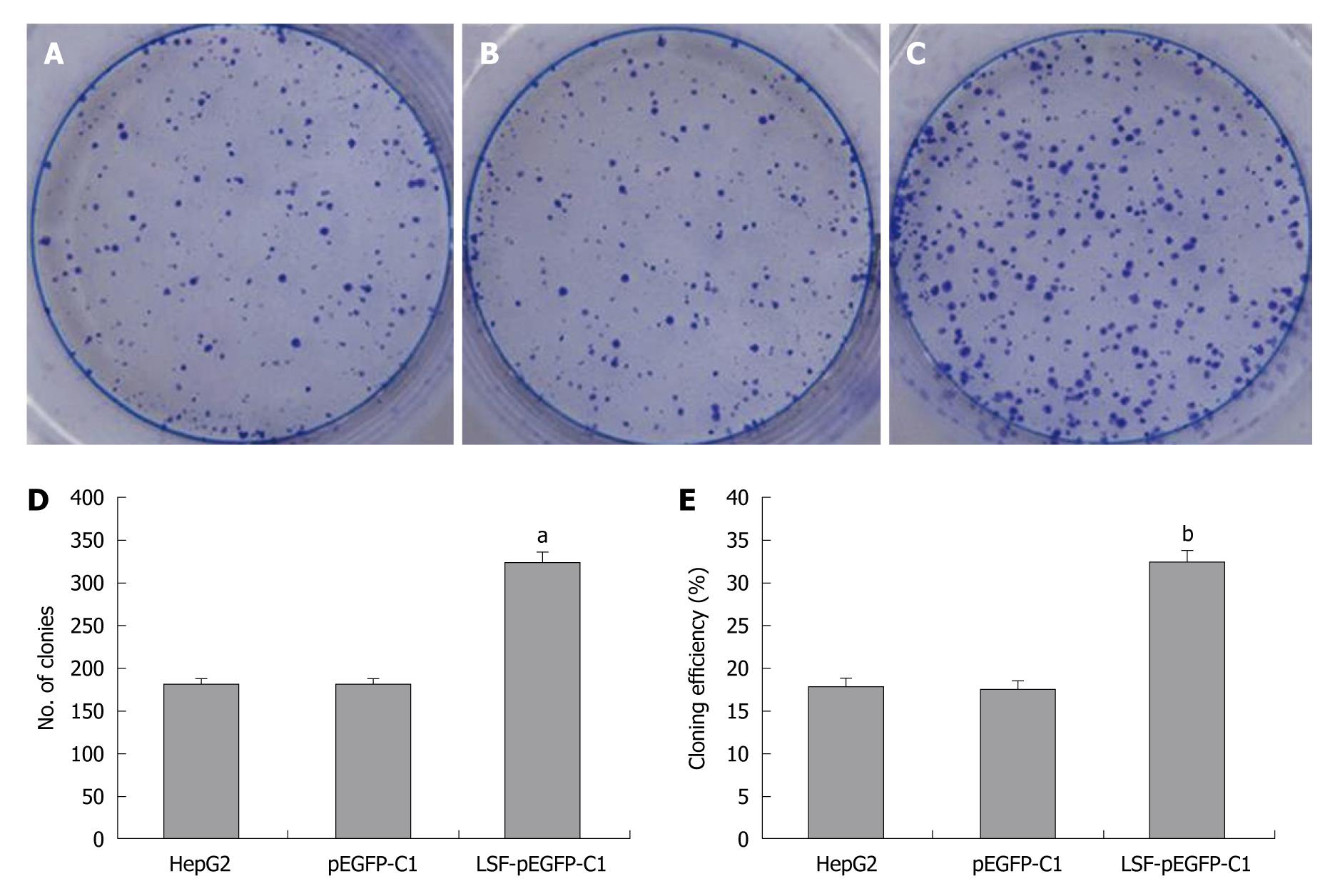
HepG2 cells were pre-treated by infection with LSF-pEGFP-C1. One thousand cells were seeded into six-well plates with 2 mL culture medium containing 10% FBS. After culture in DMEM containing 10% FBS at 37°C in a humidified, 5% CO2 atmosphere the colonies were counted. Cells were washed twice with PBS, stained with Giemsa, and colonies containing > 50 cells were counted. The cloning efficiency (%) = (the number of clones ⁄ the number of seed cells) × 100%.
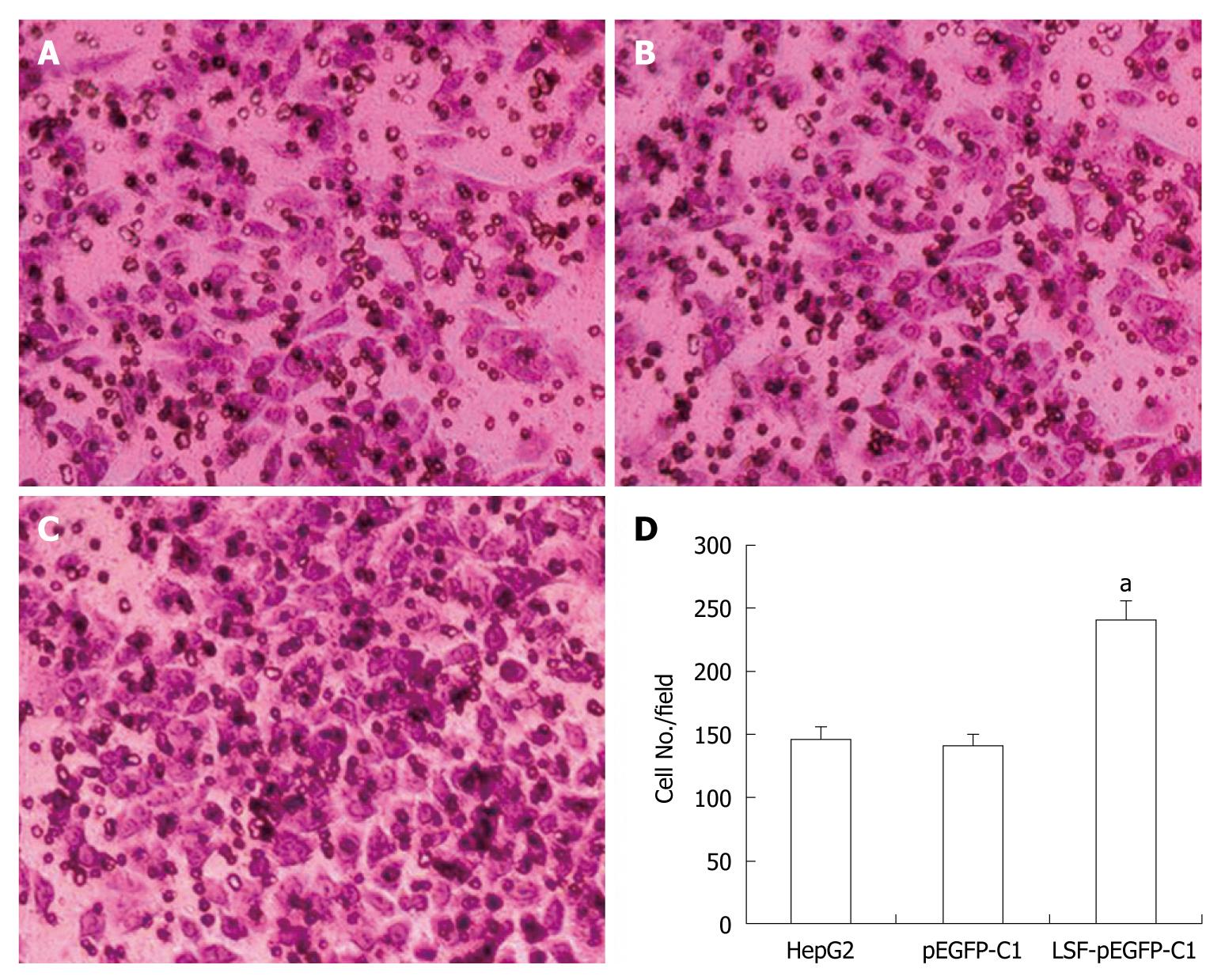
Cell invasion assays were performed in 6.5 mm Transwells (8.0 μm pore size) (Coring Incorporation, USA). Prior to each experiment, the polycarbonate filters were coated with diluted Matrigel (BD Bioscience). Untreated and treated HepG2 cells were added to the coated filters (5 × 104 cells/filter) in 200 μL of serum-free DMEM in triplicate wells. DMEM medium containing 10% FBS was added to the lower chambers. After 24 h at 37°C in a 5% CO2 and 95% humidity incubator, the non-invading cells on the upper surface of the filter were wiped off using a cotton swab. Invading cells were fixed in 95% alcohol for 10 min and stained with Giemsa for 12 min. The number of cells in five randomly selected fields was counted under a microscope (× 200 magnification).
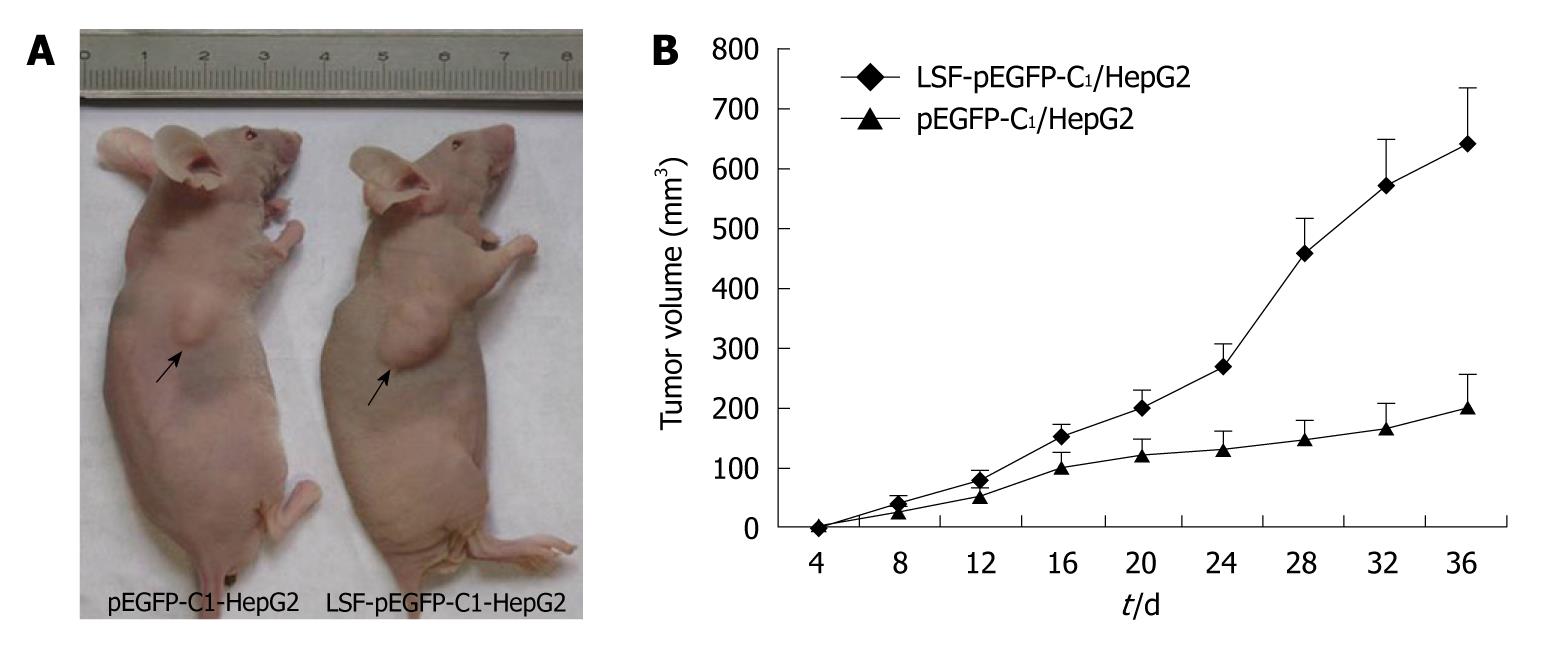
For the in vivo tumorigenicity assays, SCID/NOD mice (Shanghai, China) were housed under specific pathogen-free conditions. Mice (n = 8) were injected subcutaneously in the left flank with 3 × 106 HepG2 cells transfected with LSF-pEGFP-C1 or with an empty vector. Tumor growth was measured every 3-4 d in a 3-dimensional fashion using a caliper. All animal studies were conducted under IACUC-approved protocols at Southeast University, Nanjing, China.
All results were expressed as the mean ± SD or as percentages where appropriate. Significant differences were tested using SPSS 12.0 (SPSS Inc., Chicago, IL, USA) and a 2-tailed t-test or Fisher’s exact test. P < 0.05 was determined to be statistically significant.
Notch-1 interacts with many downstream effectors that regulate complex cytoplasmic signaling networks. We studied the expression of Notch1-ICD and LSF in 25 cases of human primary HCC using immunochemistry (Figure 1). Notch1-ICD was detectable in 19/25 cases (Figure 1A and B). Twenty-one out of 25 cancers were positive for LSF (Figure 1C and D). Liver cancer specimens stained with hematoxylin and eosin are shown in Figure 1E. Normal human liver tissue specimens were also labeled with Notch1 and LSF antibodies. The results showed negative expression in normal tissue (Figure 1F). Tumors positive for Notch1-ICD showed strong, condensed nuclear staining for LSF.
Next, we investigated the correlation between Notch1-ICD and LSF expression and HCC stage. As shown in Table 2, high expression of Notch1-ICD and LSF was observed during the advanced pathological stages of HCC [Notch1-ICD was positive in 3/8 (37.5%) of stage I/II, and in 16/17 (94.1%) of stage III/IV tumors, P = 0.006. LSF was positive in 4/8 (50%) of stage I/II and in 17/17 (100%) of stage III/IV tumors, P = 0.006. Notch1-ICD and LSF were both positive in 2/8 (25%) of stage I/II stage and 14/17 (82.5%) of stage III/IV tumors, P = 0.01]. However, there was no correlation with patient gender, age, histological type and cellular differentiation. Although the expression of Notch1-ICD and LSF was more frequent in HCC samples [Notch1-ICD was positive in 15/25 (60%); LSF was positive in 16/25 (64%); and Notch1-ICD and LSF were both positive in 13/17 (76.5%)] than in the other histological types, no statistically significant difference was found (P = 0.059, P = 0.081 and P = 0.359, respectively).
| Patients | n | Notch1 | LSF2 | Notch1 and LSF3 | P value |
| Gender | 0.5621,1.0002, 0.3123 | ||||
| Male | 20 | 16 | 17 | 14 | |
| Female | 5 | 3 | 4 | 2 | |
| Age (yr) | 0.1941, 0.5482, 0.0753 | ||||
| < 50 | 7 | 4 | 5 | 2 | |
| ≥ 50 | 18 | 15 | 16 | 13 | |
| Histological type | 0.0591, 0.0812, 0.3593 | ||||
| HCC | 17 | 15 | 16 | 13 | |
| Bile duct cell carcinoma | 1 | 0 | 0 | ||
| Nodular hepatocellular carcinoma | 6 | 4 | 5 | 4 | |
| Other | 1 | 0 | 0 | ||
| Cellular differentiation | 0.1371, 0.2942, 1.0003 | ||||
| Well/moderately | 18 | 12 | 14 | 11 | |
| Poor/undifferentiated | 7 | 7 | 7 | 5 | |
| Stage | 0.0061, 0.0062, 0.013 | ||||
| I/II | 8 | 3 | 4 | 2 | |
| III/IV | 17 | 16 | 17 | 14 |
Taken together, these results suggest that activation of Notch-1 signaling and elevated LSF expression play a key role in the pathogenesis of HCC.
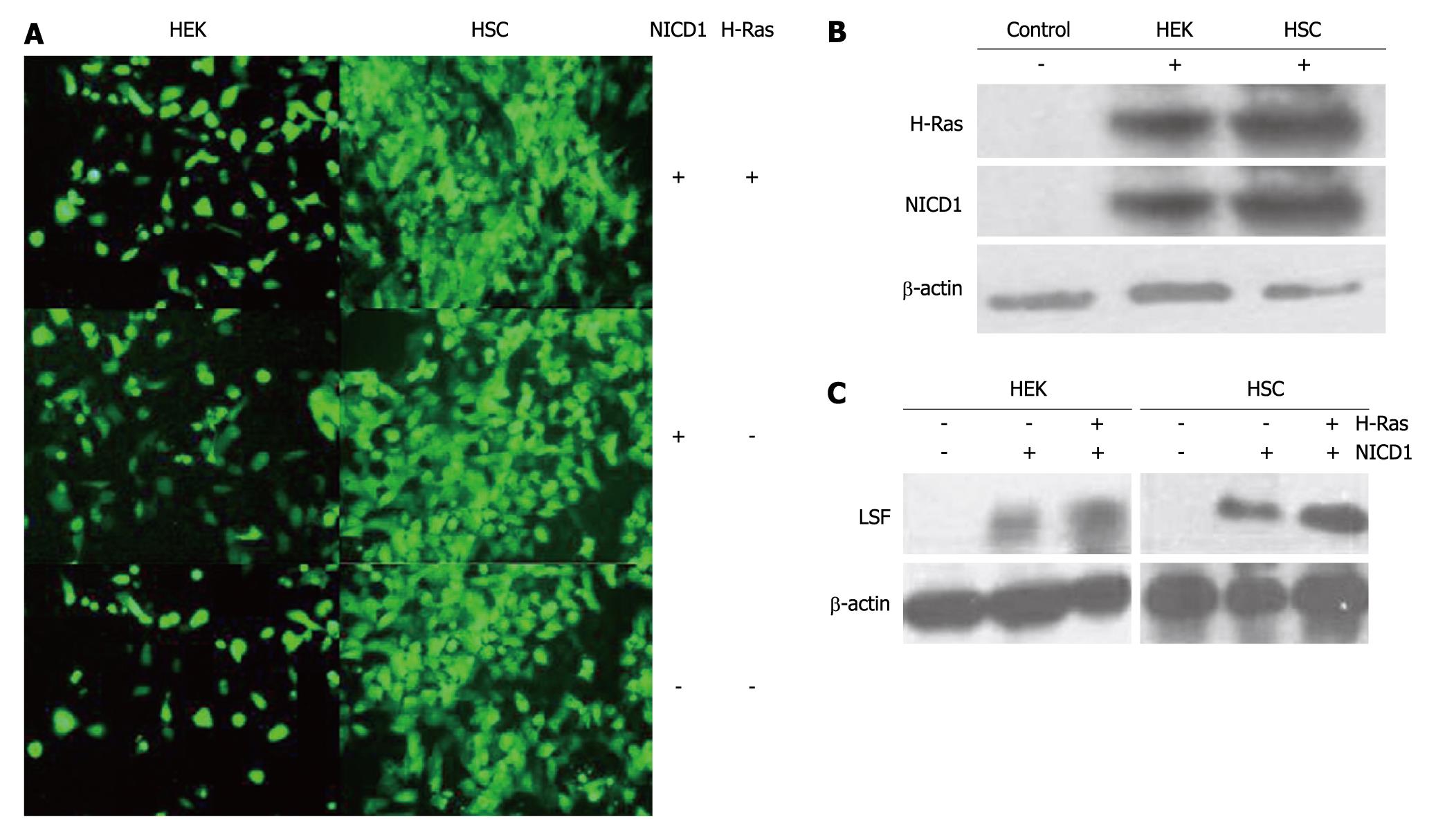
LSF is an important mammalian transcription factor that binds cellular promoters modulated by cell growth signals[13,14]. In this study, we examined the role of LSF and Notch-1 in 2 human H-Ras-transformed cell lines, HSC and HEK (Figure 2A). Western blotting with antibodies against intracellular Notch-1 revealed one major band with an apparent molecular mass of 110kD, corresponding to the intracellular cleavage product, Notch1-ICD. Western blotting also detected H-Ras (21kD, Figure 2B). Western blotting with antibodies to LSF revealed one major band with an apparent molecular mass of 63kD (Figure 2C). These results show that cells expressing exogenous Notch1-ICD can augment the expression of LSF protein. Furthermore, we observed that LSF was robustly up-regulated in both HSC and HEK cells after co-transfection of H-Ras-pEGFP-C1 and NICD1-pEGFP-C1. This suggests that LSF is involved in Notch signaling and establishes a regulatory role for LSF in the pathogenesis of HCC.
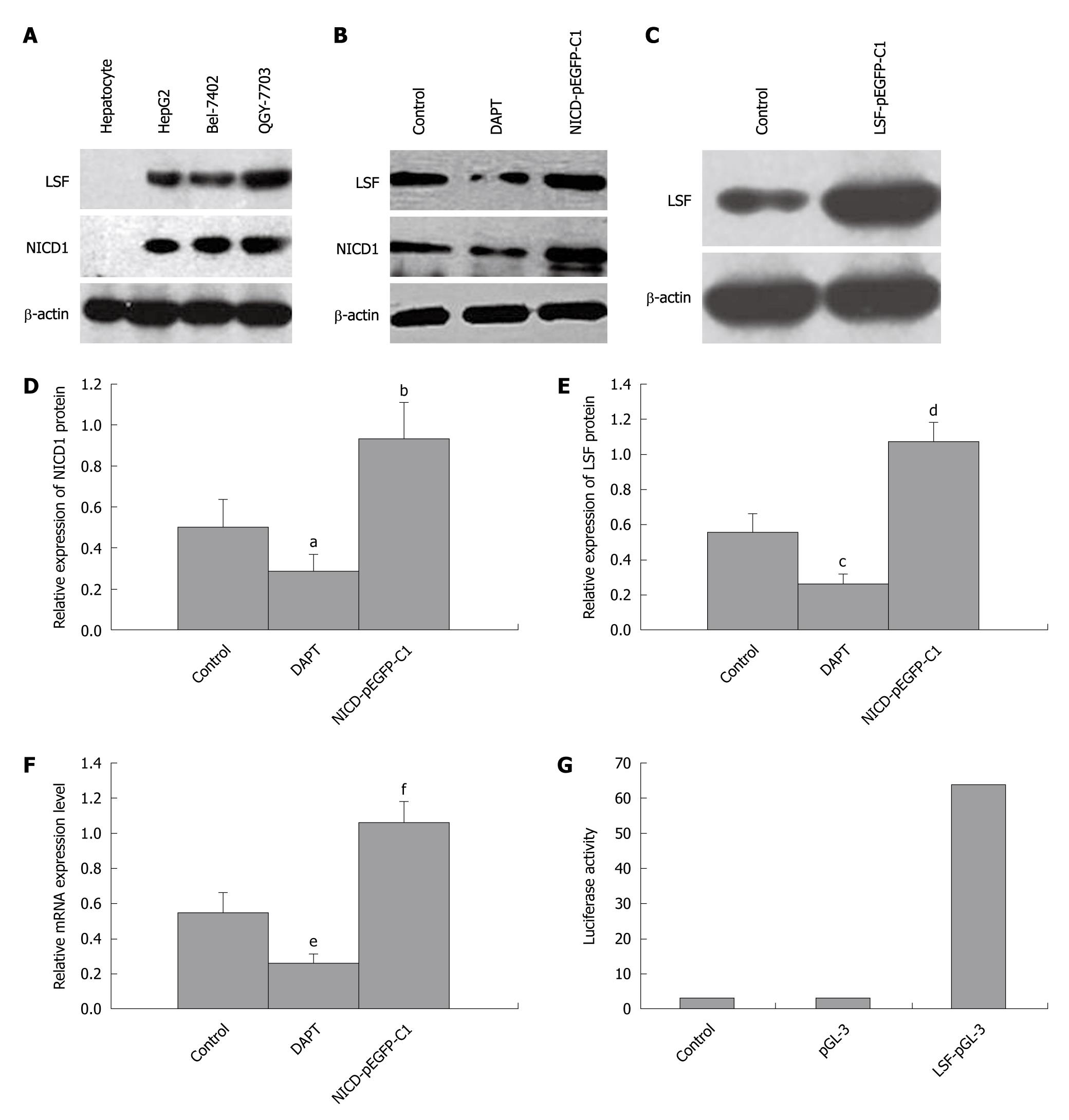
We next explored the relationship between Notch-1 and LSF in HCC cell lines from human primary tumors using pharmacological and genetic approaches. Western blotting analysis of Notch1-ICD and LSF expression in 3 HCC cell lines showed that Notch1-ICD and LSF levels were upregulated (hepatocytes were used as a negative control, Figure 3A). HepG2 cells were then treated with DAPT, which blocks endogenous Notch-1 activation. This resulted in decreased expression of Notch1-ICD and downregulation of LSF (Figure 3B). Next, NICD1-pEGFP-C1 was transfected into HepG2 cells expressing constitutively activated Notch-1 (Figure 3B). LSF-pEGFP-C1 was also introduced into HepG2 cells (Figure 3C). The expression levels of Notch-ICD and LSF protein were both elevated (Figure 3B, 3C, 3D, 3E). Hes-1 mRNA levels were also increased in NICD1-pEGFP-C1/HepG2 cells compared with that in the controls (untreated HepG2 cells) (Figure 3F and Table 1). We determined that perturbed Notch signaling significantly upregulated LSF expression in the HCC cell lines. We confirmed these results by stably introducing Notch1-ICD into HSC cells and by transient transfection in HEK cells as described above. Activation of LSF-dependent transcription is an indicator of LSF activity. The promoter assay showed that LSF reporter activity in HSC transfected with NICD1-pEGFP-C1 was directly correlated with LSF protein levels (Figure 3G), showing an approximately 60-fold increase above that in the controls. Taken together, these data show that LSF is a novel mediator of dysregulated Notch signaling in HCC.
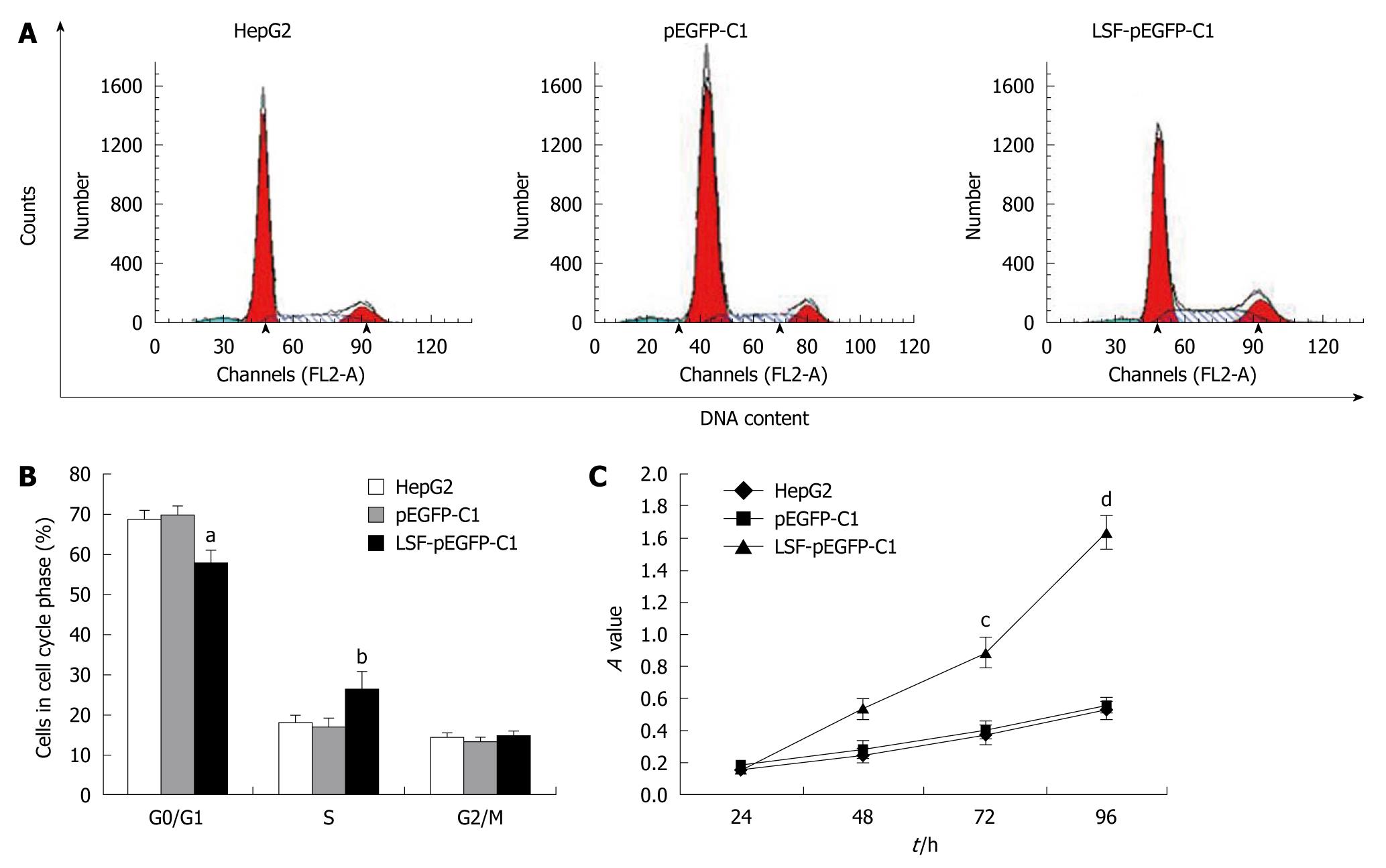
We next explored the biological role of LSF in the progression of HCC. As cell proliferation is closely linked to progression through the cell cycle, we analyzed the cell cycle of cultured cells by flow cytometry (Figure 4A). We first looked at the cell cycle in HepG2 cells transfected with LSF-pEGFP-C1. The data showed a decreasing proportion of cells in G0/G1 and an increasing proportion of cells in S phase compared with the controls (Figure 4B, P < 0.01). No obvious differences were observed at G2/M in any of the groups. The MTT assay showed that forced overexpression of LSF promoted proliferation and growth above that in untreated HepG2 cells (Figure 4C, P < 0.01), while there was no statistical significance between the growth of HepG2 cells and pEGFP-C1/HepG2 cells. However, the colony formation assay showed that the number of HepG2 cell clones overexpressing LSF was significantly increased compared with that in the controls (Figure 5A, 5B, 5C, 5D, P < 0.05); a similar result was shown when calculating the clone efficiency (Figure 5E, P < 0.05). However, in this study, the invasion assay showed that the invasive ability of LSF-transfected HepG2 cells was increased compared with that of the controls (Figure 6, P < 0.05). Taken together these observations suggest that LSF accelerates the development and progression of HCC.
In light of the pro-proliferative effects of LSF in vivo, we tested whether LSF could promote the proliferation of HCC cells in vivo. The tumorigenicity of HepG2 cells was examined in eight mice inoculated with either the empty vector or with the exogenous LSF expression vector. Representative tumor growth curves are shown in Figure 7A. The mean tumor volume was significantly larger in LSF-transfected nude mice than in those transfected with the empty vector (Figure 7B, P < 0.01).
HCC is a highly aggressive cancer for which there is no currently available effective treatment and it is the third most frequent cause of cancer deaths[17,18]. Therefore, identifying signaling pathways that could be targeted to enhance the sensitivity of a therapy is of great importance. Mounting evidence shows that constitutively active Notch receptors induce proliferative activity[19-22] and that deregulated expression and/or activity of wild-type Notch receptors occurs frequently in human malignancies, including T-cell acute lymphoblastic leukemias[11,21-26], HCC[27,28], non-small cell lung cancer[29], breast cancers[30], colon cancer[8], and Hodgkin’s and large-cell lymphomas[12]. However, the possible molecular mechanisms underlying deregulated Notch signaling and its involvement in the development and progress of human HCC are unknown. Immunohistochemistry of liver cancer tissue specimens showed that Notch-ICD1 (76%, 19⁄25), LSF (84%, 21/25) and both LSF and Notch1-ICD (76.5%, 13/17) were abundantly expressed in HCC, but were either undetectable or only very occasionally expressed in non-tumor liver tissues. This suggests that their overexpression may be linked to cancer development and/or progression. Western blotting showed that cell lines derived from HCCs spontaneously overexpressed Notch-1 and LSF compared with normal hepatocytes. We also found increased LSF protein expression in HepG2 cells transfected with NICD1-pEGFP-C1. We used a γ-secretase inhibitor, DAPT, to block Notch signaling and found that, when Notch signaling was inhibited in HCC cell lines, LSF protein levels decreased in a time dependent manner. These observations suggest that overexpression of Notch1-ICD protein plays an important role in human hepatocarcinogenesis. However, we also observed a new phenomenon: that the expression of LSF increased along with upregulated Notch1-ICD protein levels in HepG2 cells. So, we investigated the relationship between LSF and the Notch signaling pathway. We first performed a series of experiments involving 2 human tumor cell lines (HSC and HEK cells) expressing the human telomerase reverse transcriptase subunit and oncogenic H-Ras to explore the relationship between them. First, we determined that LSF expression was upregulated in NICD1-transformed human normal cell lines (such as HEK and HSC). Next, we showed that forced overexpression of exogenous Notch1-ICD in Ras-transformed cells robustly increased LSF expression. Furthermore, a promoter assay showed that LSF reporter activity in HSC transfected with NICD1-pEGFP-C1 was directly correlated with LSF protein levels. Thus, we speculate that LSF is a key mediator of the Notch signaling pathway.
LSF is directly targeted in diverse cell types by the MEK/ERK kinase pathway, which is central to growth factor signaling[31-33]. Recent data show that LSF acts as an oncogene in HCC[34]. However, any relationship between LSF signaling and its biological and tissue-specific functions is largely speculative at present. Our data suggest that LSF participates in dysregulated Notch signaling in human hepatocarcinogenesis. As described above, our observations also indicate that constitutively active Notch-1 increases LSF expression in liver cancer and in human HCC cell lines, such as HepG2. In fact, the exogenous Notch-1 activation we observed in normal human cells with upregulated LSF might underlie the molecular mechanisms involved in the pathogenesis of human HCC. In this study, the biological behavior assay suggested that LSF plays an important role in hepatocarcinogenesis. LSF is essential for cell cycle progression at the G1/S transition after reentry of quiescent cells into the cell cycle[13,14]. Our data suggest that LSF plays an important role in DNA synthesis and cell survival. Forced overexpression of LSF in HepG2 cells resulted in highly aggressive tumors in nude mice. Our results also showed that pathological activation of Notch signaling and elevation of LSF is necessary for neoplastic proliferation, which facilitates entry into G1/S phase of the cell cycle, promotes DNA synthesis, and functions as an anti-apoptotic factor. These observations indicate the need for detailed investigations into the role of Notch signaling and LSF and Ras mutations, increased expression of wild-type Ras isoforms, and other mechanisms involved in the formation of human HCC.
Plausible mechanisms that perturb Notch signaling mediated by LSF effects in human hepatocarcinogenesis involved in growth promotion and inhibition of apoptosis. Further study of how LSF affects Notch signaling pathways should be performed in deficient mice and/or in cell cultures in which signaling is knocked down. It would be highly informative to tease out the distinct contribution of Notch signaling to the overall function of LSF. In further studies, we will investigate the role of LSF in dysregulated Notch signaling in HCC in more detail.
The results of this study indicate that LSF is a novel mediator of Notch signaling and plays a crucial role in the neoplastic proliferation, invasion, development and progression of human HCC. Our observations place LSF among the key mediators of Notch-1 signaling, and suggest that it might be a novel therapeutic target for the treatment of highly aggressive HCC.
ACKNOWLEDGMENTS
We thank Dr. Xiao-Dong Bu and Xiao-Qiang Tian for HCC specimen collection and helpful suggestions and discussions and Dong-Feng Liu for producing the paraffin sections. We also thank Dr. Di Zhao and You-Wei Zhang for comments regarding the manuscript.
Although alterations in Notch1 and late SV40 factor (LSF) have been observed in hepatocellular carcinoma (HCC), little is known about their relationship during the development and progression of liver cancer. LSF has been identified as an important oncogene in human HCC. However, the mechanisms underlying tumorigenesis mediated by Notch1 and LSF are controversial and the relationship between them remains unclear.
The Notch family, including Notch1, is associated with the development and progression of HCC, and LSF acts as a key oncogene in this disease. However, the mechanism by which LSF is involved in Notch signaling in human hepatocarcinogenesis has not been addressed. In this study, the authors demonstrate the relationship between Notch signaling and LSF, and its biological role in human hepatocarcinogenesis.
Abnormal expression of Notch1 and LSF in human HCC suggests that LSF may interact with the Notch signaling pathway during human hepatocarcinogenesis. This is the first report showing that LSF is a downstream effector of dysregulated Notch signaling in human HCC. The in vitro and in vivo data suggest that LSF might be a potential therapeutic target.
This study identifies a signaling pathway involved in hepatocarcinogenesis that may be a novel therapeutic target for the treatment of HCC.
Notch was initially identified as being responsible for a specific phenotype that manifested as “notches” in the wing blades of Drosophila melanogaster. The Notch family comprises single-pass transmembrane proteins consisting of extracellular, transmembrane, and intracellular domains that correlate with different cellular functions, including cell-fate determination, tissue patterning and morphogenesis, cell differentiation, proliferation and death. LSF, also known as LBP-1c and TFCP2, regulates diverse cellular and viral promoters.
In this study, Fan et al show that LSF acts as a downstream mediator of the Notch signaling network, thus playing a crucial role in the neoplastic proliferation and invasion of human HCC.
| 1. | Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277-300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10002] [Cited by in RCA: 10466] [Article Influence: 654.1] [Reference Citation Analysis (2)] |
| 2. | Zhang Y, Song H, Miao Y, Wang R, Chen L. Frequent transcriptional inactivation of Kallikrein 10 gene by CpG island hypermethylation in non-small cell lung cancer. Cancer Sci. 2010;101:934-940. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 3. | Ferenci P, Fried M, Labrecque D, Bruix J, Sherman M, Omata M, Heathcote J, Piratsivuth T, Kew M, Otegbayo JA. World Gastroenterology Organisation Guideline. Hepatocellular carcinoma (HCC): a global perspective. J Gastrointestin Liver Dis. 2010;19:311-317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 212] [Cited by in RCA: 213] [Article Influence: 13.3] [Reference Citation Analysis (1)] |
| 4. | Weijzen S, Rizzo P, Braid M, Vaishnav R, Jonkheer SM, Zlobin A, Osborne BA, Gottipati S, Aster JC, Hahn WC. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nat Med. 2002;8:979-986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 408] [Cited by in RCA: 406] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 5. | Zagouras P, Stifani S, Blaumueller CM, Carcangiu ML, Artavanis-Tsakonas S. Alterations in Notch signaling in neoplastic lesions of the human cervix. Proc Natl Acad Sci U S A. 1995;92:6414-6418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 279] [Cited by in RCA: 301] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 6. | Daniel B, Rangarajan A, Mukherjee G, Vallikad E, Krishna S. The link between integration and expression of human papillomavirus type 16 genomes and cellular changes in the evolution of cervical intraepithelial neoplastic lesions. J Gen Virol. 1997;78:1095-1101. [PubMed] |
| 7. | Westhoff B, Colaluca IN, D’Ario G, Donzelli M, Tosoni D, Volorio S, Pelosi G, Spaggiari L, Mazzarol G, Viale G. Alterations of the Notch pathway in lung cancer. Proc Natl Acad Sci U S A. 2009;106:22293-22298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 297] [Cited by in RCA: 322] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 8. | Zhang Y, Li B, Ji ZZ, Zheng PS. Notch1 regulates the growth of human colon cancers. Cancer. 2010;116:5207-5218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 113] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 9. | Leethanakul C, Patel V, Gillespie J, Pallente M, Ensley JF, Koontongkaew S, Liotta LA, Emmert-Buck M, Gutkind JS. Distinct pattern of expression of differentiation and growth-related genes in squamous cell carcinomas of the head and neck revealed by the use of laser capture microdissection and cDNA arrays. Oncogene. 2000;19:3220-3224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 201] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 10. | Rae FK, Stephenson SA, Nicol DL, Clements JA. Novel association of a diverse range of genes with renal cell carcinoma as identified by differential display. Int J Cancer. 2000;88:726-732. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 11. | Okuhashi Y, Nara N, Tohda S. Effects of gamma-secretase inhibitors on the growth of leukemia cells. Anticancer Res. 2010;30:495-498. [PubMed] |
| 12. | Jundt F, Anagnostopoulos I, Förster R, Mathas S, Stein H, Dörken B. Activated Notch1 signaling promotes tumor cell proliferation and survival in Hodgkin and anaplastic large cell lymphoma. Blood. 2002;99:3398-3403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 297] [Cited by in RCA: 290] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 13. | Veljkovic J, Hansen U. Lineage-specific and ubiquitous biological roles of the mammalian transcription factor LSF. Gene. 2004;343:23-40. [PubMed] |
| 14. | Hansen U, Owens L, Saxena UH. Transcription factors LSF and E2Fs: tandem cyclists driving G0 to S? Cell Cycle. 2009;8:2146-2151. [PubMed] |
| 15. | Powell CM, Rudge TL, Zhu Q, Johnson LF, Hansen U. Inhibition of the mammalian transcription factor LSF induces S-phase-dependent apoptosis by downregulating thymidylate synthase expression. EMBO J. 2000;19:4665-4675. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 60] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 16. | EDMONDSON HA, STEINER PE. Primary carcinoma of the liver: a study of 100 cases among 48,900 necropsies. Cancer. 1954;7:462-503. [PubMed] |
| 17. | Thomas MB, Zhu AX. Hepatocellular carcinoma: the need for progress. J Clin Oncol. 2005;23:2892-2899. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 288] [Cited by in RCA: 305] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 18. | Grieco A, Pompili M, Caminiti G, Miele L, Covino M, Alfei B, Rapaccini GL, Gasbarrini G. Prognostic factors for survival in patients with early-intermediate hepatocellular carcinoma undergoing non-surgical therapy: comparison of Okuda, CLIP, and BCLC staging systems in a single Italian centre. Gut. 2005;54:411-418. [PubMed] |
| 19. | Giovannini C, Gramantieri L, Chieco P, Minguzzi M, Lago F, Pianetti S, Ramazzotti E, Marcu KB, Bolondi L. Selective ablation of Notch3 in HCC enhances doxorubicin’s death promoting effect by a p53 dependent mechanism. J Hepatol. 2009;50:969-979. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 75] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 20. | Moellering RE, Cornejo M, Davis TN, Del Bianco C, Aster JC, Blacklow SC, Kung AL, Gilliland DG, Verdine GL, Bradner JE. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462:182-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 591] [Cited by in RCA: 629] [Article Influence: 39.3] [Reference Citation Analysis (0)] |
| 21. | Weng AP, Nam Y, Wolfe MS, Pear WS, Griffin JD, Blacklow SC, Aster JC. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of notch signaling. Mol Cell Biol. 2003;23:655-664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 306] [Cited by in RCA: 304] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 22. | Roy M, Pear WS, Aster JC. The multifaceted role of Notch in cancer. Curr Opin Genet Dev. 2007;17:52-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 212] [Cited by in RCA: 225] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 23. | Palomero T, Sulis ML, Cortina M, Real PJ, Barnes K, Ciofani M, Caparros E, Buteau J, Brown K, Perkins SL. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007;13:1203-1210. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 731] [Cited by in RCA: 728] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 24. | Vilimas T, Mascarenhas J, Palomero T, Mandal M, Buonamici S, Meng F, Thompson B, Spaulding C, Macaroun S, Alegre ML. Targeting the NF-kappaB signaling pathway in Notch1-induced T-cell leukemia. Nat Med. 2007;13:70-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 266] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 25. | Masuda S, Kumano K, Suzuki T, Tomita T, Iwatsubo T, Natsugari H, Tojo A, Shibutani M, Mitsumori K, Hanazono Y. Dual antitumor mechanisms of Notch signaling inhibitor in a T-cell acute lymphoblastic leukemia xenograft model. Cancer Sci. 2009;100:2444-2450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 26. | Koch U, Radtke F. Notch and cancer: a double-edged sword. Cell Mol Life Sci. 2007;64:2746-2762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 259] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 27. | Ning L, Wentworth L, Chen H, Weber SM. Down-regulation of Notch1 signaling inhibits tumor growth in human hepatocellular carcinoma. Am J Transl Res. 2009;1:358-366. [PubMed] |
| 28. | Gao J, Chen Y, Wu KC, Liu J, Zhao YQ, Pan YL, Du R, Zheng GR, Xiong YM, Xu HL. RUNX3 directly interacts with intracellular domain of Notch1 and suppresses Notch signaling in hepatocellular carcinoma cells. Exp Cell Res. 2010;316:149-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 54] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 29. | Donnem T, Andersen S, Al-Shibli K, Al-Saad S, Busund LT, Bremnes RM. Prognostic impact of Notch ligands and receptors in nonsmall cell lung cancer: coexpression of Notch-1 and vascular endothelial growth factor-A predicts poor survival. Cancer. 2010;116:5676-5685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 30. | Rustighi A, Tiberi L, Soldano A, Napoli M, Nuciforo P, Rosato A, Kaplan F, Capobianco A, Pece S, Di Fiore PP. The prolyl-isomerase Pin1 is a Notch1 target that enhances Notch1 activation in cancer. Nat Cell Biol. 2009;11:133-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 143] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 31. | Saxena UH, Powell CM, Fecko JK, Cacioppo R, Chou HS, Cooper GM, Hansen U. Phosphorylation by cyclin C/cyclin-dependent kinase 2 following mitogenic stimulation of murine fibroblasts inhibits transcriptional activity of LSF during G1 progression. Mol Cell Biol. 2009;29:2335-2345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 17] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 32. | Pagon Z, Volker J, Cooper GM, Hansen U. Mammalian transcription factor LSF is a target of ERK signaling. J Cell Biochem. 2003;89:733-746. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 33. | Volker JL, Rameh LE, Zhu Q, DeCaprio J, Hansen U. Mitogenic stimulation of resting T cells causes rapid phosphorylation of the transcription factor LSF and increased DNA-binding activity. Genes Dev. 1997;11:1435-1446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 51] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 34. | Yoo BK, Emdad L, Gredler R, Fuller C, Dumur CI, Jones KH, Jackson-Cook C, Su ZZ, Chen D, Saxena UH. Transcription factor Late SV40 Factor (LSF) functions as an oncogene in hepatocellular carcinoma. Proc Natl Acad Sci U S A. 2010;107:8357-8362. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 69] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
Peer reviewer: Mara Massimi, PhD, Department of Basic and Applied Biology, University of L’Aquila, Via Vetoio-Coppito, L’Aquila, 67100, Italy
S- Editor Sun H L- Editor Cant MR E- Editor Zheng XM