Published online Mar 14, 2011. doi: 10.3748/wjg.v17.i10.1276
Revised: December 10, 2010
Accepted: December 17, 2010
Published online: March 14, 2011
AIM: To isolate and identify the biological characteristics of human colon cancer stem cells (SW1116 cells) and further study their proteome.
METHODS: SW1116 cells were isolated and cultured with a serum-free medium (SFM). Sphere formation was assayed to observe the formation of colon cancer stem cell spheres. SW1116 cells were inoculated into a serum-containing medium for observing their differentiation characteristics. Proliferation curve and cross-resistance of SW1116 cells to different drugs were detected by MTT. Percentage of SP cells in SW1116 cells was detected with Hoechst33342 staining. Telomerase activity in SW1116cells was checked by polymerase chain reaction (PCR)-enzyme linked immunosorbent assay. Expressions of stem cell relevant genes and proteins were detected by reverse transcription-PCR and Western blot, respectively. Total protein was isolated from SW1116 cells by two-dimensional gel electrophoresis (2-DE) and differentially expressed proteins were identified by tandem mass spectrometry (MALDI-TOF/TOF).
RESULTS: The isolated SW1116 cells presented as spheroid and suspension growths in SFM with a strong self-renewal, proliferation, differentiation and drug-resistance ability. The percentage of SP cells in SW1116 cells was 38.9%. The SW1116 cells co-expressed the CD133 and CD29 proteins. The telomerase activity in SW1116 cells was increased. The expressions of different stem cell relevant genes and proteins were detected. The proteomic analysis showed that the 26 protein spots were differently expressed in SW1116 cells and 10 protein spots were identified as ubiquitin fusion-degradation 1-like protein, nuclear chloride channel protein, tubulin β, Raichu404X, stratifin, F-actin capping protein α-1 subunit, eukaryotic translation elongation factor 1 delta isoform 2, hypothetical protein, glyceraldehyde-3-phosphate dehydrogenase and guanine nucleotide binding protein β polypeptide 2-like 1, respectively.
CONCLUSION: SW1116 cells are biologically characterized by self-renewal, proliferation and differentiation, and the differently expressed proteins in SW1116 cells may be essential for isolating cancer stem cells.
- Citation: Zou J, Yu XF, Bao ZJ, Dong J. Proteome of human colon cancer stem cells: A comparative analysis. World J Gastroenterol 2011; 17(10): 1276-1285
- URL: https://www.wjgnet.com/1007-9327/full/v17/i10/1276.htm
- DOI: https://dx.doi.org/10.3748/wjg.v17.i10.1276
Colon cancer is the leading cause of cancer-related death in developed countries[1]. Although its incidence has significantly reduced in most Asian countries, the incidence of colorectal cancer in Japan is increasing due to westernized diets. More information on factors influencing the increase, progression, and metastasis of colon cancer will lead to the development of its novel diagnostic and treatment methods. One of these factors is stem cells, but their function in the early stage of cancer is not completely understood.
Stem cells are defined as cells able to perpetuate themselves through self-renewal and to generate mature cells of a particular tissue through differentiation. They play an important role in the maintenance of organ homeostasis and may be responsible for tumorigenesis and contribute to resistance to cancer therapy[2]. Since the identification and characterization of cancer stem cells (CSC) in hematological malignancies, an increasing number of studies have described CSC in solid tumors such as ovarian tumor[3], colon tumor[4], lung tumor[5], breast tumor[6], liver tumor[7], melanoma[8] and pancreatic tumor[9], raising that the cancer stem cell hypothesis can be applied to all neoplastic systems. CSC are more important than other tumor cells because they are capable of self-renewing, differentiating, and maintaining tumor growth and heterogeneity, thus playing an important role in both tumorigenesis and therapeutics. However, research has been hampered by the lack of distinct molecular markers for CSC.
The most recent research findings have extended the criteria for CSC to human colon cancers, suggesting that colon carcinoma is organized in a hierarchical fashion, where only a CD133+ small subset of cells with self-renewal properties are able to recapitulate the bulk tumor population[4,10]. CD133+ cells within colon carcinoma can be propagated in vitro as spheroid culture retains the tumorigenic capacity under these conditions. The high resistance of CD133+ cells to apoptosis induced by chemotherapeutic drugs is additionally consistent with the cancer stem cell hypothesis, whereas the number of CSC is particularly resistant to death-induced signals. In addition to CD133- based identification of colon cancer stem cells, Dalerba and co-workers[11] have recently reported an alternative protocol for the isolation of human colon CSC by exploiting the surface phenotype EpCAMhigh/CD44+/CD166+. In their study, EpCAM and CD44 antigens were selected on the basis of their previously described expression in human breast cancer stem cells. CD166 is known as a mesenchymal stem cell marker and its increased expression in colon cancer is associated with a poor clinical outcome[12]. In this study, a comparative proteomic analysis of a human colon CSC line was performed to find more specific phenotypic markers for colon cancer.
Human colon cancer cells (SW1116 cells) were purchased from Shanghai Institute of Life Science, Chinese Academy of Sciences. TeloTAGGG Telomerase polymerase chain reaction (PCR) ELISAPLUS kit was purchased from Roche Molecular Biochemicals (Basel, Switzerland). Chemiluminescent detection kit was from SuperArray Bioscience (Frederick, MD, USA). Bio-Rad protein assay kit and silver stain plus™ kit were from Bio-Rad (Hercules, CA, USA).
Human SW1116 cells were maintained in RPMI-1640 medium (Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (Gibco BRL, USA), 1 × 105 U/L penicillin G and 100 mg/L streptomycin in an atmosphere containing 5% CO2 at 37°C. Adherent SW1116 cells were dissociated to single cell suspensions and seeded in serum-free medium (SFM). After spheres of SW1116 cells were formed, proliferation and differentiation potentials of SW1116 cells were observed. SW1116 cells were isolated and maintained in a SFM (DMEM/F12 medium) containing 20 μg/L human recombinant epidermal growth factor (EGF; Invitrogen, Carlsbad, CA, USA), 20 μg/L human recombinant basic fibroblast growth factor (bFGF; Invitrogen, Carlsbad, CA, USA), 2 mmol/L L-glutamine, 4 U/L insulin, 1 × 105 U/L penicillin G, and 100 mg/L streptomycin.
Primary spheres of SW1116 cells were dissociated to single cell suspensions and inoculated in ultra-low attachment 96-well plates (Corning Life Sciences, Acton, MA) (100 cells per well) supplemented with 200 μL SFM. Then, 25 μL SFM per well was added every 2 d. The number of spheres of SW1116 cells in each well was evaluated 14 d after culture.
Two days after primary culture, SW1116 cells were plated in 24-well culture plates with 10% FBS and cultured with FBS-supplemented medium every two days. Differentiation potentials of SW1116 cells were observed under a microscope.
SW1116 cells were seeded onto 35 mm Petri dishes at a density of 1 × 104. Cultured SW1116 cells were stained with trypan blue and counted in triplicate under a microscope for 6 wk.
SW1116 cells were seeded onto 96-well plates at a density of 5 × 103 per well. Twenty four hours after drugs were added at different concentrations, SW1116 cells were exposed to drugs for 2 d. Then MTT (5 mg/mL) was added and the plates were incubated at 37°C for 4 h before 100 μL 100% dimethyl sulfoxide was added to each well. The optical density of SW1116 cells in each well was detected with a microplate reader (Bio-Rad, Model 550) at a wavelength of 570 nm. The survival time of SW1116 cells was calculated as OD value of OM- exerted cells/OD value of mocked-cells × 100%.
Stained SW1116 cells at a concentration of 1 × 106 cells per 100 μL buffer contained PBS at pH 7.2, 0.5% BSA, and 2 mmol/L EDTA. Antibodies against CD133 (anti-CD133-PE, BD Pharmingen, Franklin Lakes, USA) and CD29 (anti-CD29-FITC, Chemicon, Billerica, MA, USA) were used. Antibody was incubated at 4°C for 30 min. FACS analysis was done using a FACS calibur flow cytometer (Becton Dickinson).
SW1116 cells were maintained in a SFM containing 5 mg/L Hoechst 33342, cultured in an atmosphere containing 5% CO2 at 37°C for 90 min, harvested, trypsinized and fixed with 4% methanol for 20 min. SP cells were counted under a fluorescence microscope.
To quantitatively detect changes in telomerase activity, SW1116 cells were assayed with a telomerase PCR-enzyme linked immunosorbent assay (ELISA) kit according to its manufacturer’s instructions. After PCR-ELISA, telomerase activity was detected using a microplate reader (Bio-Rad, Model 550) and recorded as the absorbance units. Relative telomerase activity (RTA) within different samples was obtained according to the following equation. RTA = [(AS - AS0)/AS,IS]/[(ATS8 - ATS8.0)/ATS8,IS] × 100 Where AS is the absorbance of sample, AS0 is the absorbance of heat- or RNase-treated sample, AS,IS is the absorbance of internal standard (IS) of the sample, ATS8 is the absorbance of control template (TS8), ATS8.0 is the absorbance of lysis buffer, and ATS8.0 is the absorbance of IS of the TS8.
Total RNA was isolated from SW1116 cells using Trizol reagent (Invitrogen, Carlsbad, CA, USA) following its manufacturer’s instructions. First strand cDNA was synthesized using M-MLV for reverse transcription (RT-PCR) in a 24 μL solution containing 2 μL Oligo(dT)18 (500 μg/mL) (Sangon Co., Shanghai, China), 1 μg total RNA, 2 μL dNTP (10 mmol/L) (Sangon Co., Shanghai, China) and 7 μL DEPC water. Then, 5 μL 5 × first-strand buffer, 6 μL DEPC water, 1 μL RNasin ribonuclease inhibitor (40 U/μL) (Hua Mei Co., Guangzhou, China) were added and incubated at 42°C for 2 min. One μL M-MLV reverse transcriptase (Hua Mei Co., Guangzhou, China, 200 U) was added, the solution was placed into water bath at 42°C for 50 min and then at 70°C for 15 min. A 50 μL solution containing 1 μL Taq DNA polymerase (Hua Mei Co., Guangzhou, China), 5 μL 10 × buffer, 1 μL dNTP (10 mmol/L), 2 μL primer (10 μmmol/L) and 1 μL cDNA (0.1 μg/μL) was used for PCR. PCR amplification was performed in a thermal cycler (Perkin-Elmer Co., USA). The PCR products were analyzed and photographed with a gel documentation system (FR-200, Shanghai Fu Ri Bio Co., China). RT-PCR primers were designed and synthesized by Sangon Co, Shanghai, China (Table 1).
| Genes | Primer sequences | Products |
| GAPDH | 5’-TTGGTATCGTGGAAGGACTCA-3’ | 270 bp |
| 5’-TGTCATCATATTTGGCAGGTT-3’ | ||
| CD133 | 5’-TGGGGCTGCTGTTTATTATTCT-3’ | 194 bp |
| 5’-TGCCACAAAACCATAGAAGATG-3’ | ||
| CD29 | 5’-GGAAAACGGCAAATTGTCAG-3’ | 600 bp |
| 5’-TTGGGGTTGCACTCACACAC-3’ | ||
| Mus-1 | 5’-GGCTTCGTCACTTACATGGACCAGGCG-3’ | 542 bp |
| 5’-GGAAACTGGTAGGTGTAG-3’ | ||
| ABCG2 | 5’-GGGTTCTCTTCTTCCTGACGACC-3’ | 398 bp |
| 5’-TGGTTGTGAGATTGACCAACAGACC-3’ | ||
| TERT | 5’-CGGAAGAGTGTCTGGAGCAA-3’ | 145 bp |
| 5-GGATGAAGCGGAGTCTGGA-3 | ||
| Oct-4 | 5’-GACAACAATGAGAACCTTCAGA-3’ | 218 bp |
| 5’-CTGGCGCCGGTTACAGAACCA-3’ | ||
| Sca-1 | 5’-AACCATATTTGCCTTCCCGTCT-3’ | 135 bp |
| 5’-CCAGGTGCTGCCTCCAGTG-3’ |
SW1116 cell extract was prepared using a lysing buffer containing 20 mmol/L Tris-HCl at pH 7.5, 0.1% Triton X, 0.5% sodium deoxycholate, 1 mmol/L phenylmethylsulfonyl fluoride, 10 μg/mL aprotinin, and 10 μg/mL leupeptin and centrifuged (12 000 ×g) at 4°C. Total protein concentration was measured by BCA assay. Cellular extracts containing 50 μg total protein were subjected to 10% SDS-PAGE, and then transferred electrophoretically to polyvinylidene difluoride membranes (Invitrogen, Carlsbad, CA, USA). Blots were probed at 4°C overnight with primary antibodies in 5% milk/TBST. The antibodies used for Western blotting were CD133, CD29, Musashi-1, ABCG2, TERT and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Santa Cruz Biotechnology, Santa Cruz, CA).
Cultured SW1116 cells were harvested, washed with PBS, and lysed in a lysis buffer containing 8 mol/L urea, 4% CHAPS, 40 mmol/L Tris, 65 mmol/L DTT, 2% Bio-Lytes+ and centrifuged at 25 000 ×g for 1 h at 4°C. Protein concentrations were measured by a modified Bradford assay. All samples were stored at -80°C for electrophoresis. Two-DE was performed using the PROTEAN IEF and PROTEAN xi II systems (Bio-Rad, Hercules, CA, USA). Total protein (80 mg) was run in an IEF system using a 17 cm pH 3 - 10 ReadyStrip (Bio-Rad, Hercules, CA, USA). The total Vh was 47 000-52 000. Following IEF separation, gel strips were equilibrated with buffer I containing 6 mol/L urea, 30% glycerol, 2% SDS, 1% DTT, followed by buffer II (DTT was replaced with 2.5% IAA), each for 15 min. The equilibrated gel strips were placed on top of a 12% T slab gel and sealed with 0.5% agarose. SDS-PAGE was performed for 30 min at a constant current of 10 mA and then at 25 mA until bromophenol blue reached the bottom of gels. The separated proteins were visualized with silver diamine-staining. For preparative 2-DE, 400 μg of total proteins was separated as described above. The total Vh of IEF was 90 000-120 000. Proteins were detected with modified silver-staining compatible with MS analysis. Experiments (from cell culture to 2-DE) were performed in triplicate.
Silver-stained 2-DE gels were scanned at an optical resolution of 84.7 μm perpixel using a GS-710 imaging densitometer (Bio-Rad, Hercules, CA, USA). Digitized images were analyzed with the PD Quest 7.1 software package (Bio-Rad, Hercules, CA, USA). Following spot detection, a matchset including the three batches of SW1116 cells was built. A reference gel was selected from one of the SW1116 gels, and unmatched protein spots of the member gels were automatically added to the reference gel. The raw quantity of each spot in a member gel was divided by the total quantity of valid spots in the gel. Quantitative analysis of SW1116 gels was performed by Student’s t-test.
Protein spots were excised from gels, destained and washed until the gels became clear. The spots were kept in 0.2 mol/L NH4HCO3 for 20 min, dried by lyophilization, and digested overnight in 12.5 ng/mL trypsin in 0.1 mol/L NH4HCO3. Peptides were extracted three times with 50% ACN (Fisher, Fair Lawn, New Jersey, USA), 0.1% TFA (Merck, Schuchardt, Hohenbrunn, Germany) and dried in vacuum. The peptide mixture was dissolved in 0.1% TFA and desalted using a C18 ZipTip (Millipore, Bedford, MA). The eluted peptides in 0.1% TFA/50% ACN mixed with an equal volume of 0.1% TFA/30% ACN saturated with CHCA solution were applied onto the target, air-dried and analyzed by Ultraflex III matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF)/TOF mass spectrometry (MS; Bruker, Bremen, Germany). The extraction voltage was set at 20 kV and the cut-off mass value was set at 500. Tandem mass spectrometry (MS/MS) spectra were used to search protein identity from NCBI human database using Mascot search engine (http://www.matrixscience.com).
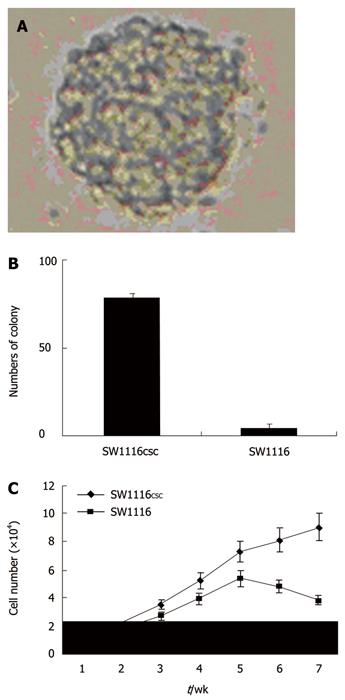
Serum-free culture condition was used to isolate and maintain SW1116 cells. SFM could maintain an undifferentiated stem cell state, and addition of bFGF and EGF induced the proliferation of multipotent, self-renewing, and expandable colon stem cells. Within 24-48 h of primary culture, a minority fraction of SW1116 cells that demonstrated growth into clonally derived spheres was yielded (Figure 1A). Primary sphere formation assay showed that the frequency of SW1116 cells was 1.9% ± 0.3%. The majority of the remaining cancer SW1116 cells exhibited adherence, loss of proliferation, and subsequent differentiation, whereas tumor spheres remained non-adherent, continuous proliferation and expansion in tumor cell culture over time. When plated in a medium with 10% FBS, SW1116 cells exhibited adherent growth phenomena. After 72 h, no difference was observed in cellular volume or shape of SW1116 cells.
The self-renewing capacity of SW1116 cell spheres was assayed by dissociating primary tumor spheres, and plating SW1116 cells at serial dilutions down to 1 cell/well. Nearly all the dissociated primary tumor spheres were able to form secondary tumor spheres, exhibiting a self-renewing ability. SW1116 cells at a density of 100 cells/well generated a greater mean number of secondary tumor spheres (87.4 ± 5.1) than that (1.9 ± 0.3) of SW1116 cells (Figure 1B).
Difference was also detected in proliferation rate of SW1116 cells. The number of SW1116 cells was calculated during the first 7 wk after seeding. A difference was observed in growth rate of SW1116 cells (Figure 1C). SW1116 cells grew slowly and showed a growth inhibition after 5 wk.
Chemosensitivity of SW1116csc and SW1116 cells to chemotherapeutic drugs was detected by MTT, which showed that the resistance of SW1116 cells to paclitaxel, adriamycin, etoposide, cytarabine, fluorouracil, cisplatin and mitomycin was 15.6-fold, 6.8-fold, 4.7-fold, 3.6-fold, 2.2-fold, 1.6-fold and 1.5-fold higher than that of SW1116csc (Table 2), indicating that SW1116 cells are more sensitive than SW1116csc to therapeutic drugs.
| IC50 (mg/L) | |||
| Drugs | SW1116csc | SW1116 cells | Times (fold) |
| Paclitaxel | 23.4 ± 1.8 | 1.5 ± 0.2 | 15.6 |
| Adriamycin | 29.7 ± 2.1 | 4.4 ± 0.4 | 6.8 |
| Etoposide | 9.8 ± 0.3 | 2.1 ± 0.2 | 4.7 |
| Cytarabine | 34.2 ± 2.5 | 9.6 ± 0.7 | 3.6 |
| Fluorouracil | 57.7 ± 3.8 | 26.5 ±1.5 | 2.2 |
| Cisplatin | 11.6 ± 0.9 | 7.3 ± 0.3 | 1.6 |
| Mitomycin | 0.67 ± 0.04 | 0.46 ± 0.03 | 1.5 |
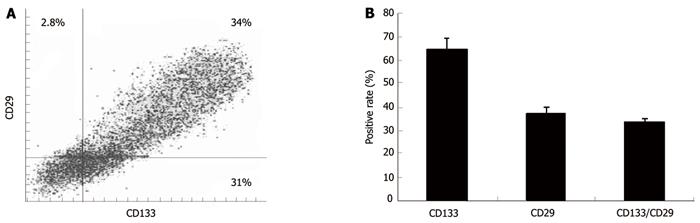
FACS analysis showed that 65% and 36.8% of SW1116 cells were positive for CD133 and for CD29 protein, respectively. SW1116 cells (34%) co-expressed CD133 and CD29, suggesting that the in vitro propagated spheroid cells can express both markers (Figure 2A and B).
After cultured for 30 d, SW1116csc and SW1116 cells were stained with Hoechst 33 342 to analyze differences in the SP proportion. The data showed that SW1116csc contained 38.9% ± 7.5% of Hoechst 33342-stained dull cells, while SW1116 cells contained only 1.2% ± 0.3% of Hoechst 33 342-stained dull cells.
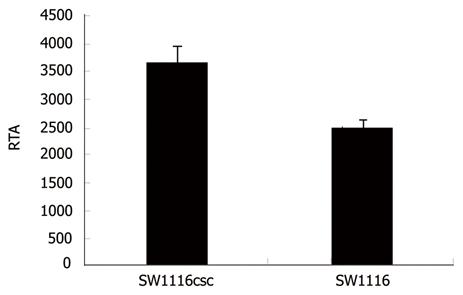
Telomerase reactivation is essential for stabilization of telomere length in attaining cellular immortality and telomerase is activated in human CSC. In this study, the RTA of SW1116csc and SW1116 cells was 3 674 ± 287 and 2 518 ± 140, respectively, indicating that the telomerase activity of SW1116csc is higher than that of SW1116 cells (Figure 3, P < 0.01).
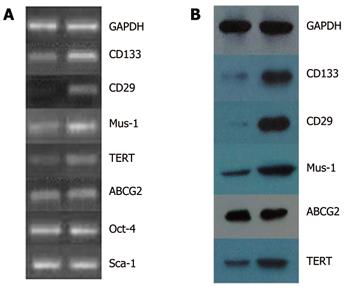
RT-PCR showed that the expressions of CD133, CD29, Musashi-1, ABCG2, TERT genes increased significantly in SW1116 cells, while no change occurred in expressions of Oct-4 and Sca-1 gene in SW1116csc and SW1116 cells (Figure 4A). Western blot showed that the expressions of CD133, CD29, Musashi-1, ABCG2, and TERT proteins in SW1116csc and SW1116 cells were increased at transcriptional level (Figure 4B).
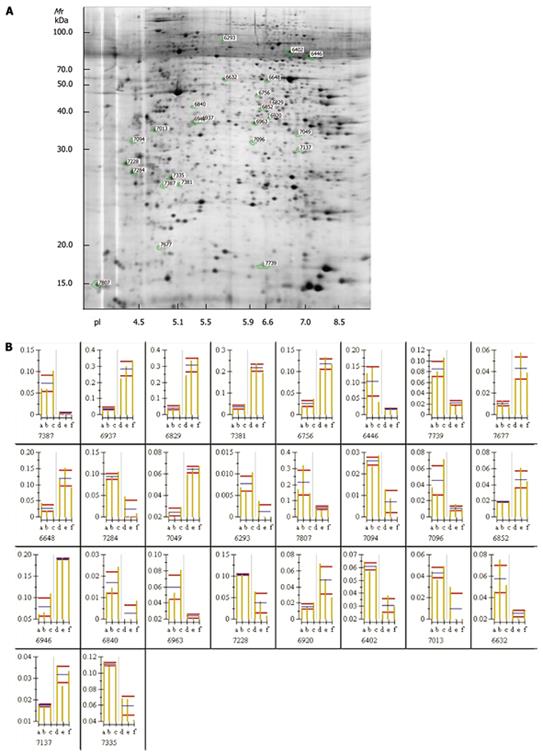
The silver-stained 2-DE gels of proteomes expressed in SW1116csc and SW1116cells were presented. The protein spots detected in gels of total protein were 2 115 ± 137 and 2 133 ± 153, respectively. One of the gels was selected as a reference gel. Student’s t-test showed that the volume of 26 protein spots was significantly changed in gels (Figure 5A, P < 0.05). The expression levels were increased and decreased, respectively, in 15 and 11 out of the 26 protein spots of SW1116 cells (Figure 5B).
Ten out of the differently expressed protein spots were chosen and identified as ubiquitin fusion-degradation 1 like protein, nuclear chloride channel protein, tubulin β, Raichu404X, stratifin, F-actin capping protein α-1 subunit, eukaryotic translation elongation factor 1 delta isoform 2, hypothetical protein, glyceraldehyde-3-phosphate dehydrogenase and guanine nucleotide binding protein β polypeptide 2-like 1, respectively, as detected by Western blotting (Table 3).
| Protein | Protein description index | NCBI ID | Theoretical Mr | Theoretical pI | Protein coverage(%) | Summary score |
| 7387 | stratifin | gi | 5454052 | 27774 | 4.68 | 3 | 41 |
| 6402 | Raichu404X | gi | 14595132 | 85016 | 6.45 | 10 | 102 |
| 6963 | Ubiquitin fusion-degradation 1 like protein | gi | 1654346 | 39170 | 6.04 | 7 | 58 |
| 7013 | eukaryotic translation elongation factor 1 delta isoform 2 | gi | 25453472 | 31121 | 4.9 | 8 | 119 |
| 6840 | tubulin beta | gi | 223429 | 50223 | 4.67 | 6 | 79 |
| 7335 | nuclear chloride channel protein | gi | 4588526 | 27249 | 5.02 | 10 | 54 |
| 7049 | glyceraldehyde-3-phosphate dehydrogenase | gi | 31645 | 36202 | 8.26 | 8 | 143 |
| 6946 | F-actin capping protein alpha-1 subunit | gi | 5453597 | 32923 | 5.45 | 9 | 84 |
| 6852 | hypothetical protein | gi | 51476996 | 40696 | 6.43 | 7 | 68 |
| 7137 | guanine nucleotide binding protein beta polypeptide 2-like 1 | gi | 21619296 | 35077 | 7.6 | 6 | 125 |
Cancer is composed of heterogeneous cells. A cancer stem cell concept means that cancer cells exhibit a hierarchy and a small number of cancer cells are maintained as cancer stem cells able to renew and differentiate. Therefore, presumably all cancers come from stem cells and these cancer stem cells may be associated with metastasis, treatment resistance, and recurrence. Biologically distinct and relatively rare populations of tumor-initiating cells have been detected by several methods and markers in a variety of cancers. Putative breast cancer tumorigenic cells and CD44+ CD24-/ low ESA+ cells are able to drive tumor formation when a few hundred cells are injected into the mammary fat pad of NOD/SCID mice[13]. In addition, different populations of cancer cells derived from primary head and neck squamous cell carcinomas display a tumorigenic potential. A minority number of CD44+ cells give rise to new tumors in vivo[14]. Pancreatic cancer cells with the CD44+ CD24+ ESA+ phenotype have stem cell properties and exhibit a 100-fold higher tumorigenic potential than nontumorigenic cancer cells[15]. In this study, colon CSC were found in the CD133+ CD29+ fraction and colon cancer stem cells with this phenotype were biologically characterized by self-renewal, proliferation and differentiation.
CD133 (prominin-1, PROM1) is a 5-transmembrane glycoprotein of 865 amino acids with a total molecular weight of 120 kDa. It has been shown that CD133 antigen is expressed in undifferentiated endothelial progenitor cells[16], hematopoietic stem cells[17], fetal brain stem cells[18], embryonic epithelium[19], prostatic epithelial stem cells[20], and myogenic cells[21]. CD133 has also been found on cancer stem or tumor-initiating cells in cancers such as retinoblastoma[22], teratocarcinoma[23], leukemia[24], brain tumor[25], hepatocellular carcinoma[26], and colon cancer[27]. It was reported that a small number of CD133+ cells from primary colon cancer tissue have a tumorigenic potential in immunodeficient mice[4], indicating that CD133+ cells in colon cancer tissue have a high tumorigenic ability.
Integrin is a non-covalently-bound heterodimeric cell adhesion molecule that links ECM to cytoskeleton. β1 integrin family (β1 integrins, CD29), the largest group of integrins, is composed of a β1 and one of the 12 α subunits and functions predominantly as cell-ECM adhesion. Both subunits have single hydrophobic transmembrane domains and transmit information from ECM context surrounding cells into outside-in signaling cells, while the extracellular binding activity of integrin is regulated from the inside of cells (inside-out signaling). It was reported that integrins are involved in many biological processes such as cell growth, differentiation, migration, and death[28], suggesting that CD29 (β1 integrin) is a new stem cell marker for colon cancer.
In mouse small intestine and human colon, Musashi-1 (Mus-1), a mammalian RNA-binding protein associated with the maintenance of neural stem cell state and its differentiation, has been found only in the lower third crypt, with a distribution that is compared in terms of cell position with the theoretical distribution of potential stem cells in the intestinal epithelium[29]. It has been shown that Mus-1 can activate the Notch signaling pathway by suppressing the translation of the Notch inhibitor m-Numb[30]. The Numb protein is asymmetrically distributed in neural progenitor cells in Drosophila and a similar asymmetrical distribution can maintain the intestinal epithelium stem cell compartment. However, interaction of Mus-1 with Notch, Delta and Tcf-4 (which appears to be intimately involved in intestinal stem cell maintenance) together with the wide variety of stem cells that express Mus-1[31,32], suggest that this protein plays a general role in regulation of stem cell maintenance and differentiation, thus representing distinct progenitor cells.
In this study, the expression of stem cell genes (Musashi-1, TERT, ABCG2, Oct-4, Sca-1) was detected by RT-PCR, which showed that the expression levels of Musashi-1, TERT, ABCG2 genes were significantly increased in SW1116 cells, indicating that the expression of these genes in SW1116 cells is up-regulated at transcriptional level and that SW1116 cells can express stem cell genes and proteins biologically characterized by self-renewal, proliferation and differentiation.
Telomerase is a ribonucleoprotein that extends the telomeric ends of chromosomes to counterbalance their natural shortening due to incomplete DNA replication in eukaryotic cells. It has been demonstrated that telomerase is activated in 90% of malignant tumors, but is stringently repressed in normal somatic cells[33,34], displaying that telomerase reactivation is a critical step in carcinogenesis. In this study, the activity of telomerase was increased in SW1116 cells, which is essential for the stabilization of telomere length in attaining cellular immortality.
In this study, proteomics of SW1116 cells was used to identify more specific phenotypic markers of colon CSC and elucidate the mechanism underlying their self-renewal and differentiation[26]. Differential protein spots were found and 10 proteins were identified. Among the differentially expressed proteins, some may be essential for isolation and identification of colon CSC.
Ubiquitin fusion degradation 1 L (UFD1L) is a human homologue of the yeast ubiquitin fusion degradation 1 (Ufd1) gene. In yeast, Ufd1 protein is involved in a degradation pathway for ubiquitin fused products (UFD pathway). The biochemical role of UFD1L protein in human cells is unknown. Velazquez-Fernandez D[35] used microarray to study the expression profiling of adrenocortical neoplasms, and found that UFD1L is a molecular signature of malignancy. In this study, the UFD1L protein was intensively expressed in SW1116csc and hardly expressed in SW1116 cells, indicating that the degradation pathway for ubiquitin fused products is active in colon CSC.
Stratifin is a member of 14-3-3 protein family, a highly conserved group of proteins consisting 7 isoforms involved in numerous crucial intracellular functions such as cell cycle and apoptosis, regulation of signal transduction pathways, cellular trafficking, cell proliferation and differentiation, cell survival, protein folding and processing. In eukaryotes, peptide chain elongation is mediated by elongation factors, EF-1 and EF-2. EF-1 is composed of a nucleotide-binding protein EF-1α, and a nucleotide exchange protein complex EF-1β gamma. Elongation factors are highly conserved among different species and may be involved in functions other than protein synthesis, such as organization of the mitotic apparatus, signal transduction, developmental regulation, ageing and transformation. Increased expression levels of stratifin and EF-1 delta in SW1116 cells may be related to cell proliferation and differentiation.
In this study, the expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was down-regulated in SW1116 cells. GAPDH, a multifunctional protein with defined functions in numerous subcellular processes, plays an integral role in glycolysis. New investigations are needed to establish the primary role of GAPDH in a variety of critical nuclear pathways apart from its already recognized role in apoptosis. The new roles of GAPDH include its requirement for transcriptional control of histone gene expression[36], its essential function in nuclear membrane fusion, its necessity for recognition of fraudulently incorporated nucleotides in DNA, and its mandatory participation in maintenance of telomere structure. To undertake these new functions, GAPDH is recruited to the nuclei in S phase or its intracellular distribution is regulated as a function of drug exposure. Further study is needed to explore the functions of GAPDH in SW1116 cells.
In conclusion, SW1116 cells and CD133+/CD29+ fraction in human colon cancer cells are biologically characterized by self-renewal, proliferation and differentiation. SW1116csc is also more chemoresistant than SW1116 cells. CD133, CD29 and Mus-1 may be used in isolation and identification of colon cancer stem cells.
Cancer stem cell hypothesis is currently at the centre of a rapidly evolving field, involving a change of perspective in development and treatment of cancers. However, research has been hampered by the lack of distinct molecular markers of cancer stem cells.
Since the identification and characterization of cancer stem cells (CSC) in hematological malignancies, an increasing number of studies have described CSC in solid tumors such as ovarian tumor, colon tumor, lung tumor, breast tumor, liver tumor, melanoma and pancreatic tumor, raising that the cancer stem cell hypothesis can be applied to all neoplastic systems.
In our study, human CSC were isolated, which presented as spheroid and suspension growths in serum-free medium, with a strong ability of self-renewal, proliferation, differentiation and drug-resistance. Proteomic analysis showed that 26 differentially expressed protein spots were detected and 10 protein spots were chosen and identified.
The results of this study have important implications for future cancer treatment. The CSC hypothesis infers that if the CSC were eliminated, the tumor would simply regress due to cell differentiation and death. It is possible to treat patients with aggressive, non-resectable tumors and prevent their metastasis by selectively targeting CSC.
Cancer stem cells are a sub-population of cancer cells that possess characteristics associated with normal stem cells, such as self renewal and differentiation into multiple cell types. CSC are tumorigenic while the bulk of cancer cells are non-tumorigenic. CSC persist in tumors as a distinct population and cause relapse and metastasis by giving rise to new tumors.
The authors identified the biological characteristics and proteome of human colon cancer stem cells. The results are interesting and may be essential for CSC isolation and characterization.
| 1. | Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ. Cancer statistics, 2006. CA Cancer J Clin. 2006;56:106-130. |
| 2. | Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105-111. |
| 3. | Peng S, Maihle NJ, Huang Y. Pluripotency factors Lin28 and Oct4 identify a sub-population of stem cell-like cells in ovarian cancer. Oncogene. 2010;29:2153-2159. |
| 4. | Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111-115. |
| 5. | Eramo A, Lotti F, Sette G, Pilozzi E, Biffoni M, Di Virgilio A, Conticello C, Ruco L, Peschle C, De Maria R. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008;15:504-514. |
| 6. | Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592-603. |
| 7. | Ji J, Yamashita T, Budhu A, Forgues M, Jia HL, Li C, Deng C, Wauthier E, Reid LM, Ye QH. Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells. Hepatology. 2009;50:472-480. |
| 8. | Schatton T, Murphy GF, Frank NY, Yamaura K, Waaga-Gasser AM, Gasser M, Zhan Q, Jordan S, Duncan LM, Weishaupt C. Identification of cells initiating human melanomas. Nature. 2008;451:345-349. |
| 9. | Ji Q, Hao X, Zhang M, Tang W, Yang M, Li L, Xiang D, Desano JT, Bommer GT, Fan D. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS One. 2009;4:e6816. |
| 10. | Todaro M, Alea MP, Di Stefano AB, Cammareri P, Vermeulen L, Iovino F, Tripodo C, Russo A, Gulotta G, Medema JP. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell. 2007;1:389-402. |
| 11. | Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci USA. 2007;104:10158-10163. |
| 12. | Weichert W, Knösel T, Bellach J, Dietel M, Kristiansen G. ALCAM/CD166 is overexpressed in colorectal carcinoma and correlates with shortened patient survival. J Clin Pathol. 2004;57:1160-1164. |
| 13. | Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983-3988. |
| 14. | Prince ME, Sivanandan R, Kaczorowski A, Wolf GT, Kaplan MJ, Dalerba P, Weissman IL, Clarke MF, Ailles LE. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci USA. 2007;104:973-978. |
| 15. | Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030-1037. |
| 16. | Peichev M, Naiyer AJ, Pereira D, Zhu Z, Lane WJ, Williams M, Oz MC, Hicklin DJ, Witte L, Moore MA. Expression of VEGFR-2 and AC133 by circulating human CD34(+) cells identifies a population of functional endothelial precursors. Blood. 2000;95:952-958. |
| 17. | Yin AH, Miraglia S, Zanjani ED, Almeida-Porada G, Ogawa M, Leary AG, Olweus J, Kearney J, Buck DW. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood. 1997;90:5002-5012. |
| 18. | Uchida N, Buck DW, He D, Reitsma MJ, Masek M, Phan TV, Tsukamoto AS, Gage FH, Weissman IL. Direct isolation of human central nervous system stem cells. Proc Natl Acad Sci USA. 2000;97:14720-14725. |
| 19. | Corbeil D, Röper K, Hellwig A, Tavian M, Miraglia S, Watt SM, Simmons PJ, Peault B, Buck DW, Huttner WB. The human AC133 hematopoietic stem cell antigen is also expressed in epithelial cells and targeted to plasma membrane protrusions. J Biol Chem. 2000;275:5512-5520. |
| 20. | Richardson GD, Robson CN, Lang SH, Neal DE, Maitland NJ, Collins AT. CD133, a novel marker for human prostatic epithelial stem cells. J Cell Sci. 2004;117:3539-3545. |
| 21. | Torrente Y, Belicchi M, Sampaolesi M, Pisati F, Meregalli M, D'Antona G, Tonlorenzi R, Porretti L, Gavina M, Mamchaoui K. Human circulating AC133(+) stem cells restore dystrophin expression and ameliorate function in dystrophic skeletal muscle. J Clin Invest. 2004;114:182-195. |
| 22. | Maw MA, Corbeil D, Koch J, Hellwig A, Wilson-Wheeler JC, Bridges RJ, Kumaramanickavel G, John S, Nancarrow D, Röper K. A frameshift mutation in prominin (mouse)-like 1 causes human retinal degeneration. Hum Mol Genet. 2000;9:27-34. |
| 23. | Miraglia S, Godfrey W, Yin AH, Atkins K, Warnke R, Holden JT, Bray RA, Waller EK, Buck DW. A novel five-transmembrane hematopoietic stem cell antigen: isolation, characterization, and molecular cloning. Blood. 1997;90:5013-5021. |
| 25. | Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396-401. |
| 26. | Yin S, Li J, Hu C, Chen X, Yao M, Yan M, Jiang G, Ge C, Xie H, Wan D. CD133 positive hepatocellular carcinoma cells possess high capacity for tumorigenicity. Int J Cancer. 2007;120:1444-1450. |
| 27. | O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106-110. |
| 29. | Potten CS, Booth C, Tudor GL, Booth D, Brady G, Hurley P, Ashton G, Clarke R, Sakakibara S, Okano H. Identification of a putative intestinal stem cell and early lineage marker; musashi-1. Differentiation. 2003;71:28-41. |
| 30. | Okano H, Imai T, Okabe M. Musashi: a translational regulator of cell fate. J Cell Sci. 2002;115:1355-1359. |
| 31. | Siddall NA, McLaughlin EA, Marriner NL, Hime GR. The RNA-binding protein Musashi is required intrinsically to maintain stem cell identity. Proc Natl Acad Sci USA. 2006;103:8402-8407. |
| 32. | Nagata H, Akiba Y, Suzuki H, Okano H, Hibi T. Expression of Musashi-1 in the rat stomach and changes during mucosal injury and restitution. FEBS Lett. 2006;580:27-33. |
| 33. | Hiyama E, Gollahon L, Kataoka T, Kuroi K, Yokoyama T, Gazdar AF, Hiyama K, Piatyszek MA, Shay JW. Telomerase activity in human breast tumors. J Natl Cancer Inst. 1996;88:116-122. |
| 34. | Kyo S, Kanaya T, Ishikawa H, Ueno H, Inoue M. Telomerase activity in gynecological tumors. Clin Cancer Res. 1996;2:2023-2028. |
Peer reviewer: Ned Abraham, Senior Lecturer, Coffs Harbour Health Campus, Faculty of Medicine University of New South Wales, Coffs Harbour, POB 2244, NSW 2450, Australia.
S- Editor Tian L L- Editor Kerr C E- Editor Ma WH