Published online Jan 7, 2011. doi: 10.3748/wjg.v17.i1.69
Revised: August 10, 2010
Accepted: August 17, 2010
Published online: January 7, 2011
AIM: To develop lymph node metastasis (LNM)-associated biomarkers for colorectal cancer (CRC) using quantitative proteome analysis.
METHODS: Differences in protein expression between primary CRC with LNM (LNM CRC) and without LNM (non-LNM CRC) were assessed using methyl esterification stable isotope labeling coupled with 2D liquid chromatography followed by tandem mass spectrometry (2D-LC-MS/MS). The relationship to clinicopathological parameters and prognosis of candidate biomarkers was examined using an independent sample set.
RESULTS: Forty-three proteins were found to be differentially expressed by at least 2.5-fold in two types of CRC. S100A4 was significantly upregulated in LNM CRC compared with non-LNM CRC, which was confirmed by Western blotting, immunohistochemistry and real-time quantitative polymerase chain reaction. Further immunohistochemistry on another 112 CRC cases showed that overexpression of S100A4 frequently existed in LNM CRC compared with non-LNM CRC (P < 0.001). Overexpression of S100A4 was significantly associated with LNM (P < 0.001), advanced TNM stage (P < 0.001), increased 5-year recurrence rate (P < 0.001) and decreased 5-year overall survival rate (P < 0.001). Univariate and multivariate analyses indicated that S100A4 expression was an independent prognostic factor for recurrence and survival of CRC patients (P < 0.05).
CONCLUSION: S100A4 might serve as a powerful biomarker for LNM and a prognostic factor in CRC.
- Citation: Huang LY, Xu Y, Cai GX, Guan ZQ, Sheng WQ, Lu HF, Xie LQ, Lu HJ, Cai SJ. S100A4 over-expression underlies lymph node metastasis and poor prognosis in colorectal cancer. World J Gastroenterol 2011; 17(1): 69-78
- URL: https://www.wjgnet.com/1007-9327/full/v17/i1/69.htm
- DOI: https://dx.doi.org/10.3748/wjg.v17.i1.69
Colorectal cancer (CRC) is the third most prevalent human cancer worldwide, with 1 million estimated new cases annually, of which, about 50% die[1]. CRC frequently migrates through the lymphatic route, depositing tumor cells into local lymph nodes, namely lymph node metastasis (LNM). The status of the local lymph nodes delivers crucial information concerning cancer staging, prognosis, and clinical decision making, on the understanding that the existence of LNM notably reduces the chance of CRC survival[2]. Unfortunately, the mechanisms related to LNM remain poorly understood at present because LNM is a complicated process that involves cancer cell detachment from the primary tumor, migration, invasion, adhesion and implantation in the new environment. A variety of dysregulated molecules play a significant role in this highly sophisticated process[3,4]. Therefore, LNM-related investigations have attracted much attention.
Clinicopathological features such as poorly differentiated cancer, depth of wall penetration, lymphovascular invasion, and tumor size are considered to be associated with CRC with LNM (LNM CRC)[5,6]; however, these characteristics are still insufficient to predict the existence of LNM. In order to improve the diagnosis and prognosis of CRC, there is an urgent need to identify specific tumor molecular markers to recognize patients with LNM, which can define a subset of CRC patients who could benefit from rational management.
It is presently in progress to develop new strategies for the identification of cancer-related molecular markers. Proteomics, the emerging technology that examines the overall characteristics of the expressed proteins, has identified many differential proteins associated with tumor development and progression in various diseases[7-10]. The recent development of proteomic technology, which presents better sensitivity than conventional gel-based strategies - coupling stable isotope labeling with liquid chromatography followed by tandem mass spectrometry (LC-MS/MS) - introduces a powerful approach to accurate qualitative and quantitative proteomic analysis of clinical samples, as successfully applied to research on difference cancers[11-14]. This procedure can provide new opportunities to develop biomarkers associated with LNM for CRC.
In the present study, we employed the combination of methyl esterification stable isotope labeling and 2D-LC-MS/MS to perform an accurate quantitative analysis. A total of 43 proteins were identified that were differently expressed by at least 2.5-fold, including S100A4, which was significantly upregulated in LNM CRC compared with non-LNM CRC. S100A4 was recently reported in association with LNM in several studies that attracted our interest[15-17]. Meanwhile, there have been a limited number of similar studies on the association of S100A4 with CRC. As a result, we focused our attention on S100A4. After confirmation by Western blotting, immunohistochemistry and real-time quantitative polymerase chain reaction (PCR), we further investigated the relationship between S100A4 expression and the lymph node metastatic phenotype of CRC, and determined its prognostic value on another independent set of 112 CRC cases.
A total of 144 colorectal carcinoma samples were collected after obtaining informed consent in our hospital (Fudan University Shanghai Cancer Center, Shanghai, China). None of the patients received chemotherapy or radiotherapy before surgery. Resected specimens were reviewed by two senior pathologists according to the criteria described in the American Joint Committee on Cancer Cancer Staging Manual (6th edition, 2002)[18]. For the screening and confirmation study, 32 primary CRC tissue samples that were obtained from patients who underwent curative resection in 2009 were collected and divided into two groups of LNM CRC and non-LNM CRC, with 16 cases in each group. The number of lymph nodes retrieved was not less than 12 in the non-LNM CRC. None of them had distant metastasis. The fresh colorectal tumor tissues were obtained immediately after surgery, washed twice with chilled phosphate buffered saline (PBS), immediately stored in liquid nitrogen and at -80°C in our tissue bank until further use. The detailed clinical data of these patients is provided in Table 1. For the S100A4 expression study, paraffin-embedded tissues in another independent set of 112 primary CRC samples between January and August 2004 were used for immunohistochemistry assessment. Ethical approval was obtained from the Cancer Center Research Ethics Committee.
| Patient No. | TNM | Sex | Age (yr) | Location |
| Non-LNM CRC | ||||
| 1 | T2N0M0 | Male | 73 | Colon |
| 2 | T4N0M0 | Male | 44 | Rectum |
| 3 | T2N0M0 | Male | 49 | Colon |
| 4 | T3N0M0 | Female | 53 | Colon |
| 5 | T3N0M0 | Male | 54 | Colon |
| 6 | T4N0M0 | Female | 52 | Colon |
| 7 | T3N0M0 | Male | 69 | Rectum |
| 8 | T3N0M0 | Female | 59 | Colon |
| 9 | T4N0M0 | Male | 66 | Colon |
| 10 | T2N0M0 | Male | 73 | Colon |
| 11 | T2N0M0 | Female | 72 | Rectum |
| 12 | T3N0M0 | Male | 68 | Rectum |
| 13 | T2N0M0 | Male | 71 | Colon |
| 14 | T4N0M0 | Male | 68 | Colon |
| 15 | T3N0M0 | Male | 59 | Rectum |
| 16 | T2N0M0 | Female | 43 | Rectum |
| LNM CRC | ||||
| 1 | T4N2M0 | Male | 41 | Colon |
| 2 | T3N1M0 | Male | 60 | Rectum |
| 3 | T2N1M0 | Male | 63 | Rectum |
| 4 | T2N2M0 | Female | 37 | Rectum |
| 5 | T4N1M0 | Female | 40 | Colon |
| 6 | T4N2M0 | Male | 80 | Colon |
| 7 | T4N1M0 | Male | 32 | Colon |
| 8 | T4N2M0 | Female | 65 | Colon |
| 9 | T3N1M0 | Female | 49 | Colon |
| 10 | T2N1M0 | Female | 47 | Rectum |
| 11 | T2N2M0 | Female | 55 | Rectum |
| 12 | T4N1M0 | Male | 86 | Colon |
| 13 | T3N1M0 | Female | 71 | Colon |
| 14 | T3N2M0 | Male | 79 | Colon |
| 15 | T3N1M0 | Male | 70 | Colon |
| 16 | T4N1M0 | Male | 56 | Colon |
Frozen samples were crushed to powder in liquid nitrogen, and dissolved in lysis buffer [7 mol/L urea, 2 mol/L thiourea, 100 mmol/L DTT and 1 × protease inhibitor cocktail (Roche, Penzberg, Germany)] by continuous vortex at 4°C for 1 h. After centrifugation at 15 000 r/min for 45 min at 4°C, equal amounts of each sample from LNM CRC and non-LNM CRC groups were pooled together. One hundred micrograms of proteins from each sample pool were reduced with 10 mmol/L dithiothreitol (60 min, 56°C) and alkylated with 12 mmol/L iodoacetamide in darkness (45 min, 37°C), followed by digestion with 1:20 (w/w) ratio of trypsin (Promega, Madison, WI, USA) overnight at 37°C. The lyophilized peptides from LNM CRC sample were tagged with d0-methanolic HCl, whereas those from non-LNM CRC samples were labeled with d3-methanolic HCl (Sigma-Aldrich, St. Louis, MO, USA) as previously described[13,19]. Briefly, 100 μL of d0-methanolic HCl or d3-methanolic HCl (created by dropwise addition of 160 μL acetyl chloride with stirring to 1 mL d0-methanol or d3-methanol) was added to the corresponding sample. The reaction was allowed to proceed for 2 h at room temperature. After lyophilization and re-dissolving in 100 μL 5% acetonitrile in 0.1% formic acid, the two peptides were mixed, followed by desalination using Sep-Pak Vac C18 (Waters, Milford, MA, USA). They were lyophilized again and re-dissolved in 40 μL of the above-mentioned solution. Subsequently, the resultant sample was separated by a 2D microcapillary HPLC system, followed by MS/MS analysis using an LTQ Orbitrap (Thermo Fisher, San Jose, CA, USA). LC solvent gradients were controlled by the chameleon 6.5 (Dionex, Amsterdam, The Netherlands). The MS scan was operated in the data-dependent mode to switch automatically between MS and MS/MS acquisition. Fragment ion selection was based on ion intensity (above 10 counts) and charge state (+2, +3).
Bioworks 3.3.1 was used to generate the peaklists of all acquired MS/MS spectra, which were then automatically searched against the International Protein Index human protein sequence database, version 3.43 using SEQUEST (University of Washington, licensed to Thermo Fisher), with a 95% confidence level. Except for 57 Da reductive alkylation modification on Cys, static modifications were set on Asp, Glu, C-terminal (+14 Da and +17 Da for light and heavy isotope labeling, respectively). The mass tolerance of the peptides and fragment ions was 10 ppm and 1.0 Da, respectively. The identified peptides were further analyzed with two computer software programs, PeptideProphet and ProteinProphet. PeptideProphet with a probability score of 0.9 and ProteinProphet with a probability score of 0.95 were used to ensure an overall false-positive rate below 0.005. Quantification of the ratio of each protein was achieved using the Xpress program. Proteins with expression fold change > 2.5, P < 0.05 were defined as differentially expressed proteins.
The same protein samples for screening were used for Western blotting. Briefly, 30-μg protein samples from each case were separated by 10% SDS-PAGE and subsequently transferred to PVDF membranes. The membranes were incubated with rabbit polyclonal antibody against S100A4 (1:1000 dilution; Abcam, Cambridge, UK) and then incubated with a horseradish-peroxidase-conjugated secondary antibody (1:100 dilution; Proteintech, Chicago, IL, USA). β-actin was detected simultaneously as a loading control (anti-β-actin, 1:1000 dilution; Kangchen, Beijing, China). All blots were visualized using an ECL detection system (Amersham, Arlington Heights, IL, USA )and quantitated by densitometry using an LAS-3000 imager.
S100A4 expression was examined immunohistochemically using paraffin-embedded tissues. In brief, 4-μm-thick tissue sections were heated in 6.5 mmol/L citrate buffer (pH 6.0) at 100°C for 28 min, and incubated with antibody against S100A4 (1:200 dilution). Immunostaining was performed employing the DAKO En-Vision System (Dako Diagnostics, Zug, Switzerland). In the negative control group, PBS was used instead of primary antibody. S100A4 expression was scored by two independent experienced pathologists. Each sample was graded according to the intensity and extent of staining as described previously[11]. The intensity of staining was scored as 0 (no staining), 1 (weak staining), and 2 (strong staining). The extent of staining was based on the percentage of positive tumor cells: 0 (no staining), 1 (1%-25%), 2 (26%-50%), 3 (51%-75%), and 4 (76%-100%). The final score was assessed by summarizing the results of intensity and extent of staining. The case was considered negative if the final score was 0 or 1 (-) or 2 or 3 (±), and positive if the score was 4 or 5 (+) or 6 or 7 (++). In most cases, the two reviewers provided consistent results. Any inconsistencies were resolved by discussion to achieve a consensus score.
Total tissue RNA was extracted using the Rneasy Mini Kit (Qiagen, Valencia, CA, USA). Real-time quantitative PCR analysis was performed according to the manufacturer’s instructions (the Quant SYBR Green PCR Kit, TIANGEN BIOTECH, Beijing, China). β-actin was applied as an internal control. The primers for β-actin (205 bp) were 5'-TGACGTGGACATCCGCAAAG-3' (sense) and 5'-CTGGAAGGTGGACAGCGAGG-3' (antisense). The primers for S100A4 (185 bp) were 5'-GCCCTGGATGTGATGGTGT-3' (sense) and 5'-TCGTTGTCCCTGTTGCTGTC-3' (antisense). Each assay was done in triplicate, and the average was calculated. For relative quantification, 2-ΔΔCt was calculated and used as an indication of the relative expression levels.
The Student t test was used to evaluate the differences in S100A4 expression between LNM CRC and non-LNM CRC. The χ2 test was used to assess the relationships between S100A4 expression and clinicopathological factors. The cumulative recurrence and survival probability were estimated using the Kaplan-Meier method, and differences were calculated by log-rank test. Prognostic factors were determined using Cox regression analysis. The recurrence-free and overall survival times were calculated from the first resection of the primary tumor to first evidence of recurrence or to death from any cause, respectively. The diagnosis of recurrence was based on the typical features presented on computed tomography/magnetic resonance imaging and elevated serum carcinoembryonic antigen. All P values were two-sided, and P < 0.05 was considered to be significant. Statistical analyses were performed using SPSS 13.0 software.
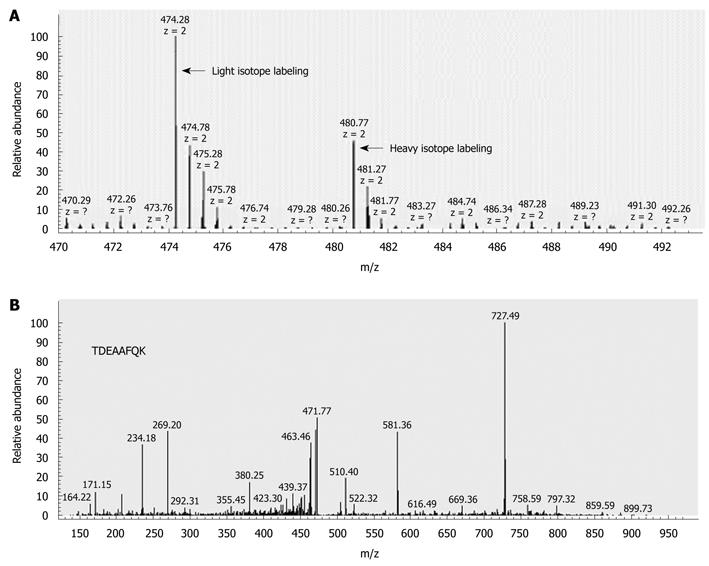
To perform accurate quantitative analysis, we compared the protein expression profiles between LNM CRC and non-LNM CRC using methyl esterification stable isotope labeling combined with 2D-LC-MS/MS. The quantitative differential expression of 644 proteins was identified, after calibration with β-casein. Significantly, 43 of these (6.7%) proteins were displayed differentially (at least 2.5-fold) in LNM CRC compared with non-LNM CRC. Among these 43 proteins, 28 were found to be upregulated in LNM CRC (Table 2), and 15 were downregulated (Table 2). The identification of S100A4 significantly upregulated in LNM CRC is shown in Figure 1 as an example. These differentially expressed proteins formed the possible protein profiles associated with LNM in CRC.
| Accession No. | Protein name | Protein ratio1 | SD | Peptide | Coverage rate (%) |
| Upregulated | |||||
| IPI00171494 | Isoform 2 of cytoplasmic dynein 2 heavy chain 1 | 21.8 | 0 | 1 | 2.4 |
| IPI00103253 | Isoform 5 of pyrin and hin domain-containing protein 1 | 8.68 | 2.85 | 19 | 4.9 |
| IPI00298520 | Putative uncharacterized protein dkfzp686m09245 | 7.39 | 0 | 1 | 2.5 |
| IPI00027194 | Syntaxin-18 | 6.30 | 0 | 1 | 3.9 |
| IPI00410639 | Isoform 2 of fch and double sh3 domains protein 2 | 5.25 | 1.03 | 5 | 2.1 |
| IPI00300631 | Scaffold attachment factor B1 | 4.69 | 0 | 1 | 1.51 |
| IPI00009236 | Isoform α of caveolin-1 | 4.42 | 3.75 | 2 | 3.0 |
| IPI00216654 | Isoform β of nucleolar phosphoprotein P130 | 4.35 | 0 | 1 | 1.6 |
| IPI00004273 | RNA binding motif protein 25 | 3.87 | 0 | 1 | 1.5 |
| IPI00239077 | Histidine triad nucleotide-binding protein 1 | 3.73 | 0.38 | 5 | 7.91 |
| IPI00010320 | Chromobox protein homolog 1 | 3.56 | 1.90 | 4 | 1.8 |
| IPI00010274 | Isoform 1 of tryptase α-1 precursor | 3.52 | 0.09 | 2 | 9.0 |
| IPI00010414 | Pdz and lim domain protein 1 | 3.41 | 0 | 1 | 4.0 |
| IPI00014852 | Isoform 1 of phosphoglucomutase-like protein 5 | 3.24 | 1.07 | 4 | 11.1 |
| IPI00000156 | Ligase III, DNA, ATP-dependent isoform β precursor | 3.10 | 0.60 | 2 | 2.1 |
| IPI00102821 | Isoform 1 of proapoptotic caspase adapter protein precursor | 3.09 | 0.40 | 2 | 36.1 |
| IPI00032313 | Protein S100-A4 | 3.04 | 0.69 | 2 | 7.9 |
| IPI00008750 | Metallothionein-1H | 2.95 | 0.11 | 2 | 27.9 |
| IPI00216153 | 40S Ribosomal protein S15 | 2.94 | 0.57 | 4 | 9.0 |
| IPI00654777 | Eukaryotic translation initiation factor 3 subunit 5 | 2.90 | 0.24 | 2 | 5.1 |
| IPI00024933 | 60S Ribosomal protein L12 | 2.86 | 1.02 | 11 | 14.4 |
| IPI00025366 | Citrate synthase, mitochondrial precursor | 2.85 | 0.47 | 4 | 5.9 |
| IPI00007928 | Pre-mRNA-processing-splicing factor 8 | 2.81 | 0 | 1 | 1.3 |
| IPI00024976 | Mitochondrial import receptor subunit tom22 homolog | 2.78 | 0 | 1 | 8.5 |
| IPI00577039 | Annexin A2 | 2.77 | 0.32 | 2 | 3.6 |
| IPI00062151 | Similar to 60s ribosomal protein L15 | 2.70 | 0.37 | 6 | 6.5 |
| IPI00219757 | Glutathione S-transferase P1 | 2.55 | 0.37 | 2 | 9.1 |
| IPI00396437 | Isoform 2 of drebrin-like protein | 2.51 | 0.45 | 5 | 2.8 |
| Downregulated | |||||
| IPI00023673 | Galectin-3-binding protein precursor | 0.09 | 0 | 1 | 2.2 |
| IPI00011062 | Isoform 1 of carbamoyl-phosphatesynthase , mitochondrial precursor | 0.10 | 0 | 1 | 1.0 |
| IPI00030279 | Isoform 1 of zinc finger ran-binding domain-containing protein 3 | 0.11 | 0 | 1 | 7.6 |
| IPI00219682 | Erythrocyte band 7 integral membrane protein | 0.20 | 0 | 1 | 9.8 |
| IPI00433499 | Rhomboid, veinlet-like 6 isoform 1 | 0.20 | 0 | 1 | 2.1 |
| IPI00000230 | Tropomyosin 1 α chain isoform 2 | 0.26 | 0.06 | 32 | 51.8 |
| IPI00029631 | Enhancer of rudimentary homolog | 0.27 | 0 | 1 | 7.7 |
| IPI00009950 | Vesicular integral-membrane protein vip36 precursor | 0.30 | 0 | 1 | 3.4 |
| IPI00384603 | Isoform 2 of ribonuclease p protein subunit P21 | 0.30 | 0.11 | 13 | 10.7 |
| IPI00177543 | Peptidylglycine α-amidating monooxygenase isoform A | 0.31 | 0.08 | 2 | 8.1 |
| IPI00000330 | Isoform 1 of uncharacterized protein c9orf80 | 0.35 | 0 | 1 | 17.6 |
| IPI00298949 | Cyclin G-associated kinase | 0.35 | 0 | 1 | 1.3 |
| IPI00008964 | RAS-related protein rab-1B | 0.37 | 0 | 1 | 7.5 |
| IPI00013860 | 3-Hydroxyisobutyrate dehydrogenase, mitochondrial precursor | 0.37 | 0 | 1 | 5.4 |
| IPI00296053 | Isoform mitochondrial of fumarate hydratase, mitochondrial precursor | 0.39 | 0.18 | 1 | 2.4 |
We extended the experiments to confirm the differential expression of S100A4 in the same samples described above.
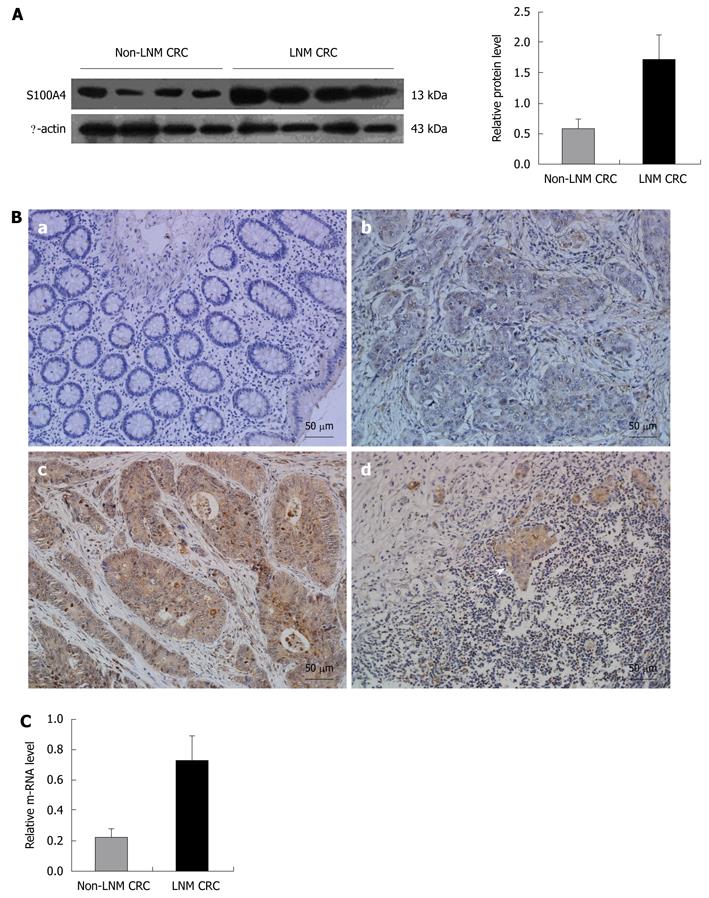
Thirty micrograms of total proteins from LNM CRC and non-LNM CRC were analyzed via Western blotting. The expression of S100A4 was dramatically higher in LNM CRC compared with non-LNM CRC (P < 0.001). A representative Western blotting result is presented in Figure 2A.
To confirm upregulation of S100A4 at the protein level, we used immunohistochemistry to evaluate S100A4 expression in situ. In normal mucosa, there was no immunoreactivity in cells. In non-LNM CRC, there was weak staining in cancer cells. In LNM CRC, there was notable brown staining in both primary and matched metastatic lymph node cancer cells. Positive staining was present mainly in the cytoplasm and/or nucleus of cancer cells (Figure 2B).
Real-time quantitative PCR revealed that S100A4 mRNA level was higher in LNM CRC than in non-LNM CRC (P < 0.001, Figure 2C), which is consistent with the trend at the protein level.
To detect the relationship between S100A4 expression and clinicopathological features and whether S100A4 could be a prognostic factor in predicting clinical outcomes of CRC patients, we evaluated S100A4 expression in an additional archived 112 CRC samples. In the group of 53 LNM CRC samples, 83% were positive for S100A4 expression, whereas 16.9% of the 59 non-LNM CRC samples had positive expression.
After division of these patients into S100A4-positive and S100A4-negative groups, statistical analysis revealed that positive expression of S100A4 was significantly associated with LNM, and advanced TNM stage (P < 0.001). However, no significant correlations were observed between S100A4 expression and other clinicopathological parameters of sex, age, tumor size, tumor differentiation and tumor location (Table 3).
| Clinicopathological factors | n | S100A4 expression | P value1 | |
| Negative | Positive | |||
| Sex | ||||
| Male | 59 | 32 | 27 | 0.252 |
| Female | 53 | 23 | 30 | |
| Age (yr) | ||||
| ≤ 60 | 76 | 39 | 37 | 0.497 |
| > 60 | 36 | 16 | 20 | |
| Tumor size (cm) | ||||
| ≤ 5 | 74 | 32 | 42 | 0.083 |
| > 5 | 38 | 23 | 15 | |
| Tumor location | ||||
| Colon | 47 | 23 | 24 | 0.975 |
| Rectum | 65 | 32 | 33 | |
| Tumor differentiation2 | ||||
| I-II | 82 | 43 | 39 | 0.244 |
| III-IV | 30 | 12 | 18 | |
| Tumor status2 | ||||
| T1-2 | 34 | 18 | 16 | 0.592 |
| T3-4 | 78 | 37 | 41 | |
| Lymph node metastasis2 | ||||
| N0 | 59 | 49 | 10 | < 0.001 |
| N1-2 | 53 | 9 | 44 | |
| TNM stage2 | ||||
| I-II | 57 | 46 | 11 | < 0.001 |
| III-IV | 55 | 9 | 46 | |
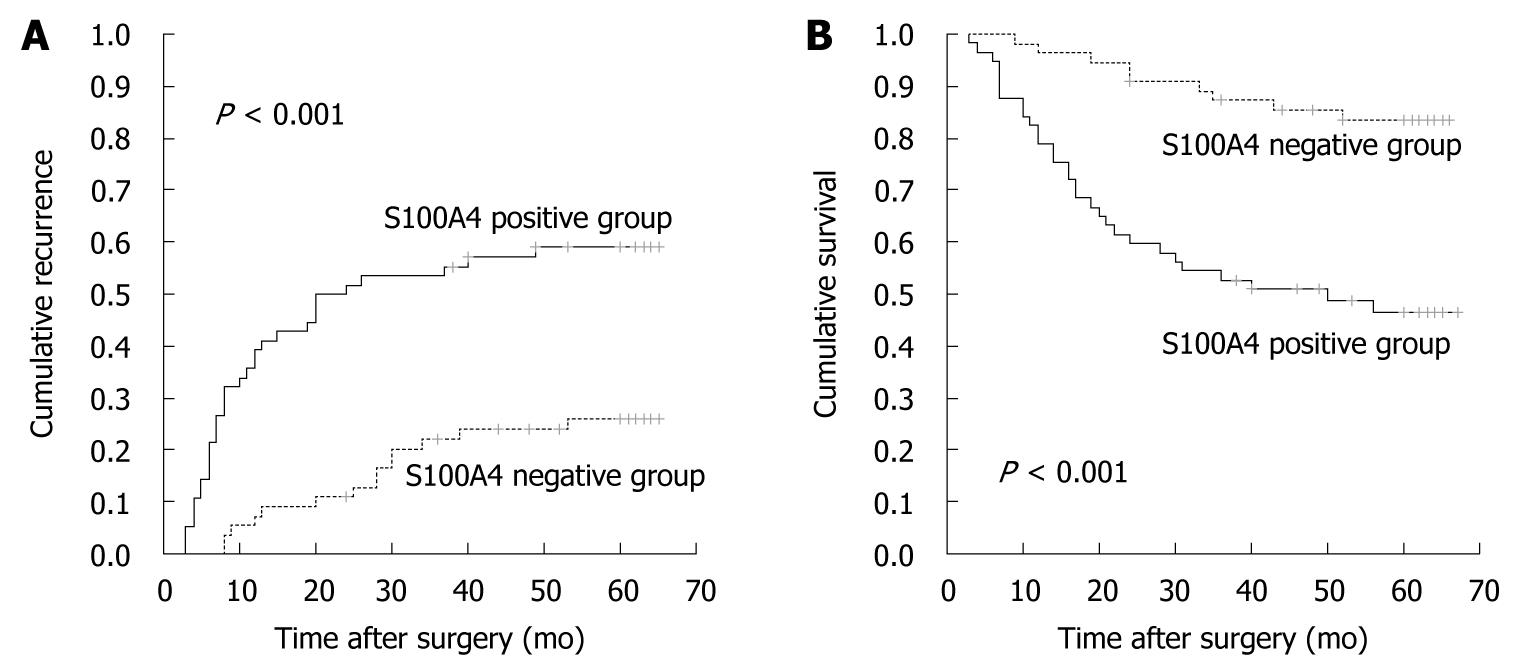
Furthermore, we found that patients with S100A4-positive CRC had significantly poorer prognosis than those with S100A4-negative CRC. The 5-year cumulative recurrence rate was significantly higher in patients with S100A4-positive CRC than in the S100A4-negative group (P < 0.001, Figure 3A). The 5-year cumulative survival rate in patients with S100A4-positive CRC was much lower than in those with S100A4-negative CRC (P < 0.001, Figure 3B). Univariate analyses revealed that LNM, TNM stage and S100A4 expression were associated with recurrence and overall survival. In multivariate analysis, LNM, TNM stage and S100A4 expression were also independent prognostic factors of recurrence and overall survival (P < 0.05, Table 4).
| Variables | Recurrence | Survival | ||
| HR (95% CI) | P value | HR (95% CI) | P value | |
| Univariate analysis | ||||
| Sex | ||||
| Male/female | 0.872 (0.490-1.549) | 0.640 | 0.857 (0.452-1.624) | 0.636 |
| Age (yr) | ||||
| ≤ 60/> 60 | 1.517 (0.842-2.732) | 0.165 | 1.842 (0.967-3.508) | 0.063 |
| Tumor size (cm) | ||||
| ≤ 5/> 5 | 0.881 (0.165-0.685) | 0.687 | 0.880 (0.444-1.745) | 0.715 |
| Tumor location | ||||
| Colon/rectum | 0.823 (0.463-1.464) | 0.508 | 0.920 (0.483-1.752) | 0.799 |
| Tumor differentiation | ||||
| I-II/III-IV | 1.234 (0.660-2.305) | 0.511 | 1.171 (0.581-2.361) | 0.659 |
| Tumor status | ||||
| T1-2/T3-4 | 0.886 (0.484-1.620) | 0.693 | 1.026 (0.517-2.033) | 0.942 |
| Lymph node metastasis | ||||
| N0/N1-2 | 2.727 (1.510-4.923) | 0.001 | 2.852 (1.463-5.559) | 0.002 |
| TNM stage | ||||
| I-II/III-IV | 3.560 (1.940-6.534) | < 0.001 | 3.393 (1.680-6.850) | < 0.001 |
| S100A4 expression | ||||
| Negative/positive | 3.666 (1.929-6.964) | < 0.001 | 4.154 (1.963-8.792) | < 0.001 |
| Multivariate analysis | ||||
| LNM | ||||
| N0/N1-2 | 0.205 (0.055-0.769) | 0.019 | 0.193 (0.044-0.857) | 0.031 |
| TNM stage | ||||
| I-II/III-IV | 8.915 (2.081-38.189) | 0.003 | 9.057 (1.705-48.108) | 0.010 |
| S100A4 expression | ||||
| Negative/positive | 2.454 (1.056-5.705) | 0.037 | 2.888 (1.131-7.379) | 0.027 |
Metastasis remains one of the major challenges in management of CRC patients. LNM is the most common form of metastasis in CRC. To develop LNM-associated biomarkers for CRC, we employed the quantitative proteomic strategy of methyl esterification stable isotope labeling coupled with 2D-LC-MS/MS. A total of 644 proteins were identified, including 43 that were differentially expressed by at least 2.5-fold between LNM CRC and non-LNM CRC. We found many of the 43 proteins that possibly participate in the biological processes associated with tumor metastasis, such as cell motility and adhesion, migration, and signal transduction.
Among the upregulated proteins, annexin A2, one of the calcium- and phospholipid-binding proteins, has been widely reported in various cancers with the regulation of cell growth, motility, invasion and signaling pathways[20]. The increase in caveolin-1 performs the functions of signal transduction, cell transformation and anti-apoptotic activity[21]. Moreover, caveolin-1 has been found to be overexpressed in several multidrug-resistant cancer cell lines[22,23]. In addition, some downregulated proteins identified in our study have also been observed to possess similar biological effects, including galectin-3-binding protein[24] and cyclin-G-associated kinase[25].
Previously, Pei et al[26] have carried out a proteomic study on 10 CRC samples using conventional 2D electrophoresis coupled with MALDI-TOF-MS, and have reported a pattern of four differentially expressed proteins potentially associated with LNM. In contrast, our results revealed up to 43 proteins that were differentially expressed by at least 2.5-fold. Annexin A2 and glutathione S-transferase P1 both correlated with LNM in our study and that of Pei et al Heat shock protein-27 and liver fatty acid binding protein (L-FABP) were found to correlate with LNM by Pei et al, but not in our study, whereas 41 proteins identified in our study were not listed by Pei et al This discordance is probably due to the different clinical background of the samples included in the studies, and the different proteomic strategies used. It is required to analyze systemically and integrate all the complementary data from various institutions into a common databank to elucidate exactly the molecular background of CRC. In addition, Pang et al[27] have identified and confirmed six differentially expressed proteins (e- fatty acid binding protein 5, methylcrotonoyl Coenzyme A carboxylase 2, pyrophosphatase 2, synaptotagmin-like protein 2, Ezrin, and smooth muscle protein) that are associated with LNM in prostate cancer by DIGE-based proteome analysis. However, there was no concordance between the results in that study and our study, which is probably mainly due to the different cancers and proteomic approaches included.
Recently, several studies have shown that S100A4 is an important factor relevant to progression and prognosis in various human cancers, such as thyroid[28], breast[29], pancreatic[30], bladder[31], gastric[17] and colorectal[32] cancer. In particular, several studies have revealed that overexpression of S100A4 strongly indicates the presence of LNM[15-17], which agrees with our original study aim. However, similar investigations have been limited between S100A4 expression and LNM in CRC. In view of the above reasons, S100A4, one of the significantly upregulated proteins identified in LNM CRC compared with non-LNM CRC, which has been confirmed at the protein and mRNA levels, attracted our attention and interest.
S100A4, also known as 18A2/mts1, CAPL, PEL-98, 42A, p9Ka, and metastasin, belongs to the S100 super-family of calcium-binding proteins[33]. S100A4-mediated calcium signaling plays a major role in crucial biological functions that influence various aspects of cell physiology, including proliferation and apoptosis, and differentiation and morphogenesis. It is also significantly involved in cell adhesion and motility, and cancer invasion and metastasis[34,35]. A large body of evidence suggests that S100A4 is involved in cell metastatic phenotype by modulating the cytoskeletal dynamics, cadherin/catenin complex cytoskeletal linkage, CD44/cytoskeletal linkage, and extracellular-matrix-associated proteolytic enzyme. Furthermore, S100A4 can participate in the activation of the matrix metalloproteinase/tissue inhibitor of metalloproteinase system and angiogenic factor vascular endothelial growth factor, which in turn can lead to tumor neovascularization[36,37].
To study further the relationship between S100A4 expression and the LNM phenotype of CRC, and determine whether S100A4 could be a prognostic factor in predicting clinical outcomes of CRC patients, we examined an additional 112 archived CRC samples for S100A4 expression. We found that the elevation in S100A4 expression level was significantly correlated with LNM and advanced TNM stage, which suggests that S100A4 plays an important part in the progression of CRC from a localized to lymph node metastatic disease. In addition, patients with S100A4-positive CRC had an increasing risk of recurrence and significantly reduced overall survival. Univariate and multivariate analyses indicated that S100A4 expression is a powerful independent prognostic factor for recurrence and overall survival in CRC, which indicates the considerable prognostic value of S100A4 expression.
In conclusion, our current study employed a quantitative proteome analysis to profile the differently expressed proteins associated with LNM in CRC. S100A4 was identified and confirmed to be significantly overexpressed in LNM CRC. Further evaluation in an independent sample set has suggested that S100A4 acts as a powerful biomarker for LNM and prognosis in CRC. However, many questions remain to be answered with respect to the cellular function of S100A4 and how it exerts its influence on metastatic progression, with further investigations on our part in progress. We also identified a number of proteins besides S100A4 that might provide a more profound insight into the mechanism of LNM in CRC and merit further research.
Colorectal cancer (CRC) is one of the most prevalent cancers worldwide, and it is estimated that half of the patients die from cancer annually. Lymph node metastasis (LNM) is the most common form of metastasis although the mechanism is largely unknown. Studies on metastasis of CRC were performed in order to improve the diagnosis and prognosis.
LNM is a complicated process that involves a variety of dysregulated molecules playing a significant role. Increasingly, it has become a hot research topic to employ proteome analysis to identify proteins associated with tumor development and progression in various diseases.
To date, there has been a limited number of studies regarding specific tumor molecular markers associated with LNM in CRC. In this study, the authors employed more sensitive proteome analysis than conventional strategies to identify a set of differently expressed proteins associated with LNM. Furthermore, the authors confirmed the significant correlation between overexpression of S100A4 and LNM, advanced TNM stage, increased recurrence rate and decreased overall survival rate.
By identifying the protein S100A4 as being associated with LNM, the authors evaluated the biological features and prognosis in CRC, which could improve our understanding of CRC, and provide a scientific basis for the application of S100A4 inhibitors in the treatment of CRC.
Proteome: all proteins that derive from the genome of cells, a tissue or an organism. It is a dynamic collection that reflects both the intrinsic genetic information of the cell and the impact of its immediate environment. Compared with gene analysis, proteome analysis can provide a more accurate view of the biological status and be expected to be more useful for evaluating, for example, disease development, progression and response to treatment.
The authors performed also an extensive proteomics analysis, and identified 43 proteins that were differentially regulated in metastatic cancer, including 16 proteins that were upregulated even more than S100A4. The results indicate that overexpression of S100A4 could be used as biomarker for LNM in CRC. However, none of the latter candidate biomarkers were further examined in terms of prognostic value.
| 1. | Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74-108. |
| 2. | O'Connell JB, Maggard MA, Ko CY. Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging. J Natl Cancer Inst. 2004;96:1420-1425. |
| 3. | Royston D, Jackson DG. Mechanisms of lymphatic metastasis in human colorectal adenocarcinoma. J Pathol. 2009;217:608-619. |
| 4. | Sundar SS, Ganesan TS. Role of lymphangiogenesis in cancer. J Clin Oncol. 2007;25:4298-4307. |
| 5. | Ricciardi R, Madoff RD, Rothenberger DA, Baxter NN. Population-based analyses of lymph node metastases in colorectal cancer. Clin Gastroenterol Hepatol. 2006;4:1522-1527. |
| 6. | Fang W, Fan B, Xiong B. Analysis of pathological risk factors for lymph node metastasis in colorectal cancer. Hepatogastroenterology. 2009;56:663-666. |
| 7. | Alfonso P, Núñez A, Madoz-Gurpide J, Lombardia L, Sánchez L, Casal JI. Proteomic expression analysis of colorectal cancer by two-dimensional differential gel electrophoresis. Proteomics. 2005;5:2602-2611. |
| 8. | Suehara Y, Kondo T, Seki K, Shibata T, Fujii K, Gotoh M, Hasegawa T, Shimada Y, Sasako M, Shimoda T. Pfetin as a prognostic biomarker of gastrointestinal stromal tumors revealed by proteomics. Clin Cancer Res. 2008;14:1707-1717. |
| 9. | Farina A, Dumonceau JM, Frossard JL, Hadengue A, Hochstrasser DF, Lescuyer P. Proteomic analysis of human bile from malignant biliary stenosis induced by pancreatic cancer. J Proteome Res. 2009;8:159-169. |
| 10. | Liu YF, Xiao ZQ, Li MX, Li MY, Zhang PF, Li C, Li F, Chen YH, Yi H, Yao HX. Quantitative proteome analysis reveals annexin A3 as a novel biomarker in lung adenocarcinoma. J Pathol. 2009;217:54-64. |
| 11. | Bai DS, Dai Z, Zhou J, Liu YK, Qiu SJ, Tan CJ, Shi YH, Huang C, Wang Z, He YF. Capn4 overexpression underlies tumor invasion and metastasis after liver transplantation for hepatocellular carcinoma. Hepatology. 2009;49:460-470. |
| 12. | Niu D, Sui J, Zhang J, Feng H, Chen WN. iTRAQ-coupled 2-D LC-MS/MS analysis of protein profile associated with HBV-modulated DNA methylation. Proteomics. 2009;9:3856-3868. |
| 13. | Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol. 2002;20:301-305. |
| 14. | Ibarrola N, Kalume DE, Gronborg M, Iwahori A, Pandey A. A proteomic approach for quantitation of phosphorylation using stable isotope labeling in cell culture. Anal Chem. 2003;75:6043-6049. |
| 15. | Min HS, Choe G, Kim SW, Park YJ, Park do J, Youn YK, Park SH, Cho BY, Park SY. S100A4 expression is associated with lymph node metastasis in papillary microcarcinoma of the thyroid. Mod Pathol. 2008;21:748-755. |
| 16. | Lee OJ, Hong SM, Razvi MH, Peng D, Powell SM, Smoklin M, Moskaluk CA, El-Rifai W. Expression of calcium-binding proteins S100A2 and S100A4 in Barrett's adenocarcinomas. Neoplasia. 2006;8:843-850. |
| 17. | Wang YY, Ye ZY, Zhao ZS, Tao HQ, Chu YQ. High-level expression of S100A4 correlates with lymph node metastasis and poor prognosis in patients with gastric cancer. Ann Surg Oncol. 2010;17:89-97. |
| 18. | Greene FL, Page DL, Fleming ID, Fritz AG, Balch CM, Haller DG, Morrow M. AJCC Cancer Staging Manual. 6th ed. New York: Springer-Verlag 2002; 113-124. |
| 19. | Hunt DF, Yates JR 3rd, Shabanowitz J, Winston S, Hauer CR. Protein sequencing by tandem mass spectrometry. Proc Natl Acad Sci USA. 1986;83:6233-6237. |
| 20. | Rescher U, Gerke V. Annexins--unique membrane binding proteins with diverse functions. J Cell Sci. 2004;117:2631-2639. |
| 21. | Williams TM, Lisanti MP. The Caveolin genes: from cell biology to medicine. Ann Med. 2004;36:584-595. |
| 22. | Lavie Y, Fiucci G, Liscovitch M. Up-regulation of caveolae and caveolar constituents in multidrug-resistant cancer cells. J Biol Chem. 1998;273:32380-32383. |
| 23. | Yang CP, Galbiati F, Volonte D, Horwitz SB, Lisanti MP. Upregulation of caveolin-1 and caveolae organelles in Taxol-resistant A549 cells. FEBS Lett. 1998;439:368-372. |
| 24. | Hancq S, Salmon I, Brotchi J, Gabius HJ, Heizmann CW, Kiss R, Decaestecker C. Detection of S100B, S100A6 and galectin-3 ligands in meningiomas as markers of aggressiveness. Int J Oncol. 2004;25:1233-1240. |
| 25. | Zhang L, Gjoerup O, Roberts TM. The serine/threonine kinase cyclin G-associated kinase regulates epidermal growth factor receptor signaling. Proc Natl Acad Sci USA. 2004;101:10296-10301. |
| 26. | Pei H, Zhu H, Zeng S, Li Y, Yang H, Shen L, Chen J, Zeng L, Fan J, Li X. Proteome analysis and tissue microarray for profiling protein markers associated with lymph node metastasis in colorectal cancer. J Proteome Res. 2007;6:2495-2501. |
| 27. | Pang J, Liu WP, Liu XP, Li LY, Fang YQ, Sun QP, Liu SJ, Li MT, Su ZL, Gao X. Profiling protein markers associated with lymph node metastasis in prostate cancer by DIGE-based proteomics analysis. J Proteome Res. 2010;9:216-226. |
| 28. | Zou M, Al-Baradie RS, Al-Hindi H, Farid NR, Shi Y. S100A4 (Mts1) gene overexpression is associated with invasion and metastasis of papillary thyroid carcinoma. Br J Cancer. 2005;93:1277-1284. |
| 29. | Rudland PS, Platt-Higgins A, Renshaw C, West CR, Winstanley JH, Robertson L, Barraclough R. Prognostic significance of the metastasis-inducing protein S100A4 (p9Ka) in human breast cancer. Cancer Res. 2000;60:1595-1603. |
| 30. | Ai KX, Lu LY, Huang XY, Chen W, Zhang HZ. Prognostic significance of S100A4 and vascular endothelial growth factor expression in pancreatic cancer. World J Gastroenterol. 2008;14:1931-1935. |
| 31. | Matsumoto K, Irie A, Satoh T, Ishii J, Iwabuchi K, Iwamura M, Egawa S, Baba S. Expression of S100A2 and S100A4 predicts for disease progression and patient survival in bladder cancer. Urology. 2007;70:602-607. |
| 32. | Gongoll S, Peters G, Mengel M, Piso P, Klempnauer J, Kreipe H, von Wasielewski R. Prognostic significance of calcium-binding protein S100A4 in colorectal cancer. Gastroenterology. 2002;123:1478-1484. |
| 33. | Marenholz I, Volz A, Ziegler A, Davies A, Ragoussis I, Korge BP, Mischke D. Genetic analysis of the epidermal differentiation complex (EDC) on human chromosome 1q21: chromosomal orientation, new markers, and a 6-Mb YAC contig. Genomics. 1996;37:295-302. |
| 34. | Parker C, Whittaker PA, Usmani BA, Lakshmi MS, Sherbet GV. Induction of 18A2/mts1 gene expression and its effects on metastasis and cell cycle control. DNA Cell Biol. 1994;13:1021-1028. |
| 35. | Garrett SC, Varney KM, Weber DJ, Bresnick AR. S100A4, a mediator of metastasis. J Biol Chem. 2006;281:677-680. |
| 36. | Sherbet GV. Metastasis promoter S100A4 is a potentially valuable molecular target for cancer therapy. Cancer Lett. 2009;280:15-30. |
| 37. | Helfman DM, Kim EJ, Lukanidin E, Grigorian M. The metastasis associated protein S100A4: role in tumour progression and metastasis. Br J Cancer. 2005;92:1955-1958. |
Peer reviewers: Mark De Ridder, MD, PhD, Professor, Dienst Radiotherapie, UZ Brussel, Vrije Universiteit Brussel, Laarbeeklaan 101, B-1090 Brussel, Belgium; Shashi Bala, PhD, Post Doctoral Associate, Department of Medicine, LRB 270L, 364 Plantation street, UMass Medical School, Worcester, MA 01605, United States
S- Editor Shi ZF L- Editor Kerr C E- Editor Lin YP