Published online Sep 21, 2010. doi: 10.3748/wjg.v16.i35.4448
Revised: June 11, 2010
Accepted: June 18, 2010
Published online: September 21, 2010
AIM: To analyze the association between the p73 G4C14-to-A4T14 polymorphism (a.k.a., the GC/AT variation) and colorectal cancer risk and survival in the Korean population, and to evaluate the relationships between p73 polymorphism and the p73 protein expression or clinicopathological characteristics of colorectal cancer.
METHODS: Three hundred and eighty-three histologically confirmed cases and 469 healthy controls, recruited at one teaching hospital in Pusan, Korea from 2001 and 2007, were genotyped for p73 G4C14-to-A4T14 by PCR with confronting two-pair primers (PCR-CTPP) and the expression profile of p73 in cancer tissues (n = 383) was analyzed by immunohistochemistry.
RESULTS: Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated by unconditional logistic regression model adjusted for age and gender. Compared with the GC/GC genotypes, the GC/AT and AT/AT genotypes were significantly associated with colorectal cancer risk (GC/AT vs GC/GC: OR = 1.46, 95% CI: 1.10-1.94; AT/AT vs GC/GC: 1.72, 0.98-3.03; Ptrend = 0.01). When stratified by age and gender, the association was restricted to those less than 60 years of age (GC/AT or AT/AT vs GC/GC: 2.22, 1.39-3.55) and male (GC/AT or AT/AT vs GC/GC: 1.91, 1.31-2.77). The expression of p73 was associated with invasion depth (P = 0.003) and advanced Duke’s stage (P = 0.06) of colorectal cancer. The patients with the GC/GC genotype were associated with worse survival compared with those with the other genotypes (P = 0.02). However, no significant relationship was observed between the p73 G4C14-to-A4T14 polymorphism and p73 protein expression in cancer tissues.
CONCLUSION: Our results suggest that the p73 GC/AT polymorphism is associated with an increased colorectal cancer risk and survival in the Korean population.
-
Citation: Lee KE, Hong YS, Kim BG, Kim NY, Lee KM, Kwak JY, Roh MS.
p73 G4C14 to A4T14 polymorphism is associated with colorectal cancer risk and survival. World J Gastroenterol 2010; 16(35): 4448-4454 - URL: https://www.wjgnet.com/1007-9327/full/v16/i35/4448.htm
- DOI: https://dx.doi.org/10.3748/wjg.v16.i35.4448
Colorectal cancer is a multigenetic disease whereby biologically relevant single nucleotide polymorphisms might play roles as genetic susceptibility factors. One of the candidates is the p73 gene, which is structurally similar to p53 and localized to the 1p36 chromosomal region[1]. Functionally, p73 activates the promoters of several p53-responsive genes participating in cell-cycle control, DNA repair, and apoptosis and inhibits cell growth in a p53-like manner by inducing apoptosis or G1 cell-cycle arrest[2,3]. Increased p73 protein expression has been found in human malignancies associated with p53 mutations[4,5], suggesting a role of p73 in compensating for the loss of p53 function[4-6]. However, overexpression of wild-type p73 might have some p53-independent functions either as an oncogene or as a tumor suppressor gene in human tumorigenesis[7,8].
It is not known whether the alteration of p73 expression has any genetic basis such as sequence variations or polymorphisms. The two completely linked single nucleotide polymorphisms at positions 4 (G→A) and 14 (C→T) affect p73 function by altering gene expression[1]. This dinucleotide polymorphism lies just upstream of the initiating AUG in exon 2, a region that could form a stem-loop structure and modulate susceptibility to cancer[1,9,10]. There are several studies indicating that subjects with the p73 GC/AT polymorphism might have an increased risk for certain types of cancers, including colorectal cancer[11-13]. Other studies reported that overexpression of the p73 protein has been seen in benign and malignant tumors, including colorectal cancer, when compared with the matched normal tissues[14]. In addition, p73 overexpression was correlated with a poor prognosis in colorectal cancer[14], hepatocellular carcinoma[15], and breast cancer[16] in several studies. However, the genotype-phenotype relationship between the p73 polymorphism and p73 protein expression in colorectal cancer has not yet been studied.
In this context, we hypothesized that the p73 GC/AT polymorphism plays an important role in colorectal carcinogenesis and progression of colorectal cancer in the Korean population. We also evaluated the relationship between the p73 polymorphism and p73 protein expression.
Three hundred and eighty-three colorectal cancer cases and 469 cancer-free controls were recruited from the Dong-A University Medical Center between January 2001 and December 2007. All cancer cases were histopathologically confirmed and had no preoperative chemotherapy or radiotherapy. The clinical records and pathological reports were also obtained for cases. The HE-stained slides were reviewed in each case to confirm the original diagnosis, which was based on the WHO classification[17]. Postoperative pathological staging was determined according to the Dukes’ classification system for colorectal cancer[18]. The control subjects were randomly selected from a pool of non-cancer individuals of Busan city residents. These subjects were matched to cases by age (± 5 years) and area of residence.
This study was approved by the institutional review board and written informed consent was obtained from all subjects.
Immediately after surgical resection of colorectal cancer, tumor tissues and adjacent normal colon tissues were sampled by a pathologist and stored at -80°C. The genomic DNA was extracted from normal colorectal tissue (cancer cases) and blood samples (controls) using a Wizard genomic DNA purification kit (Promega, USA), according to the manufacturer’s instructions.
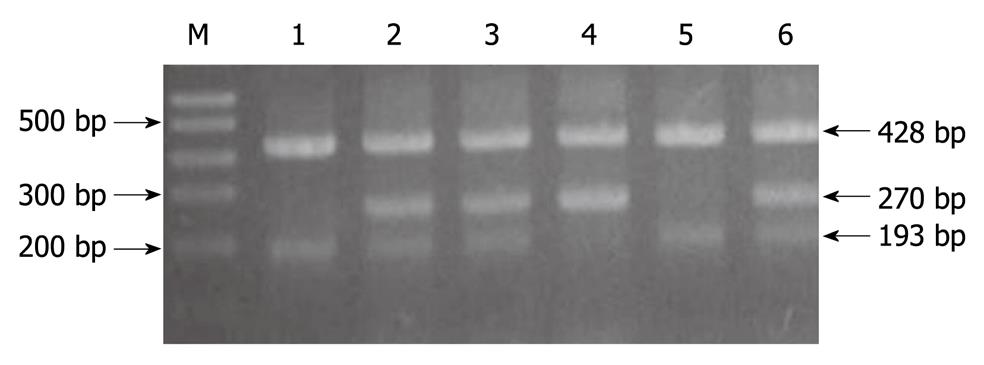
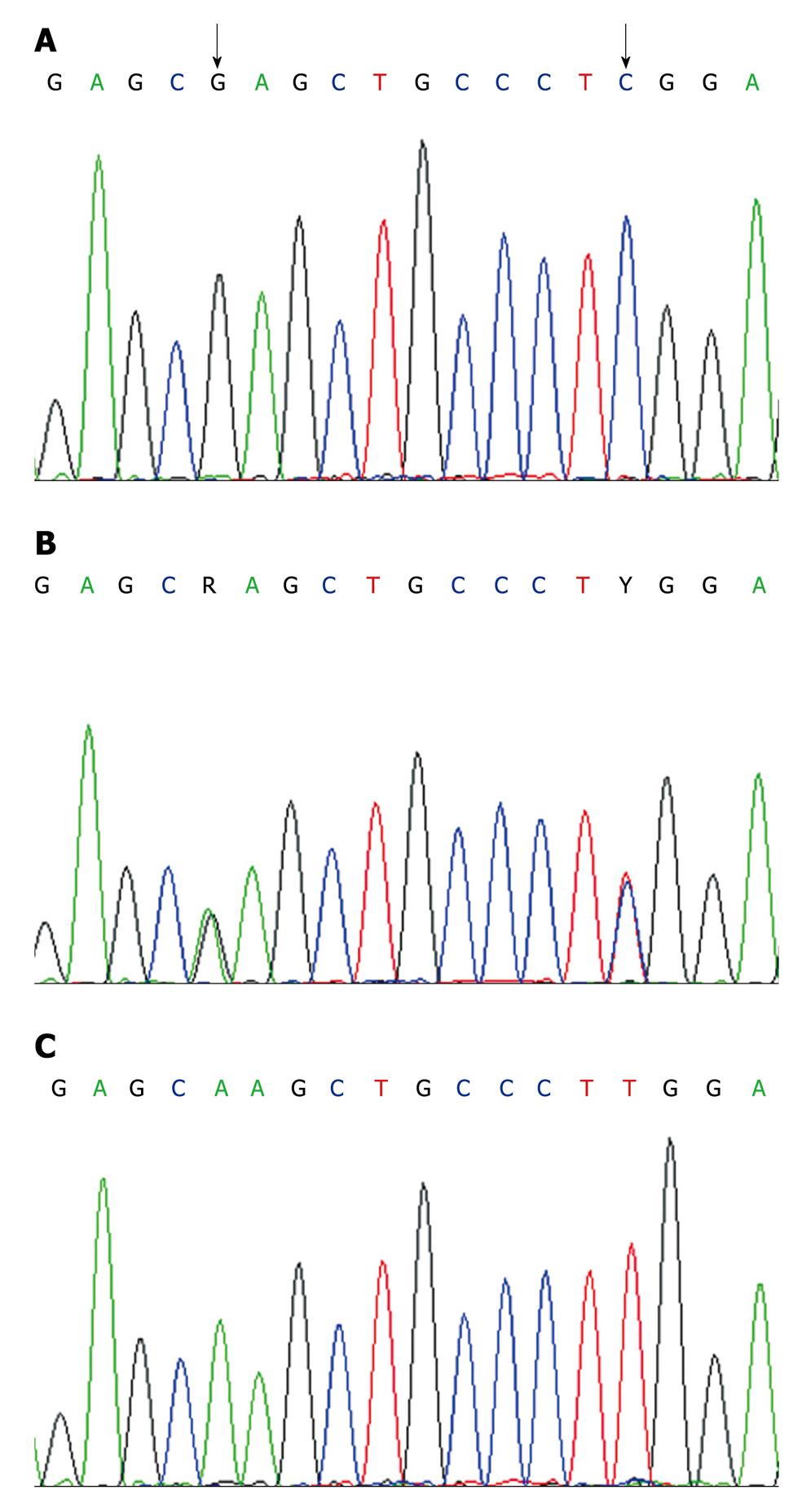
DNA amplification and genotyping of the p73 GC/AT polymorphism were performed by the method of the polymerase chain reaction with confronting two-pair primers (PCR-CTPP). DNA was amplified with the primers: 5'CCACGGATGGGTCTGATCC (F1) and 5'GGCCTCCAAGGGCAGCTT (R1) for the A allele, and 5'CCTTCCTTCCTGCAGAGCG (F2) and 5'TTAGCCCAGCGAAGGTGG (R2) for the G allele. Allele specific bases are underlined. PCR was performed in a volume of 20 μL containing 100 ng of DNA template, 0.2 mmol/L each deoxynucleotide triphosphate, 1 × PCR buffer (50 mmol/L KCl, 10 mmol/L Tris HCl, and 0.1% Triton X-100), 1.5 mmol/L MgCl2, 1 U of Taq polymerase (Sigma-Aldrich Biotechnology) and 10 pmol of each of four primers. For amplification, an initial denaturation step at 95°C for 10 min was followed by 35 cycles of 95°C for 1 min, 62°C for 45 seconds, and 72°C for 1 min, and a final extension step at 72°C for 5 min. The amplified DNA was visualized on a 2% agarose gel with ethidium bromide staining. The p73 G4A polymorphism was genotyped as a 193 base pair band for the G allele, a 270 base pair band for the A allele, and a 428 base pair common band (GenBank: AL 136528, Figure 1). The genotyping by PCR-CTPP analysis was confirmed by DNA sequencing analysis; the results of PCR-CTPP genotyping and sequencing analysis were completely concordant (Figure 2). The genotype distributions of controls were in agreement with Hardy-Weinberg equilibrium (P = 0.71).
Immunohistochemical study was performed for p73 protein expression with tissue microarray (TMA) slides prepared by the avidin-biotin-peroxidase complex method. Deparaffinization of all the sections was performed through a series of xylene baths, and rehydration was performed with a series of graded alcohol solutions. To enhance immunoreactivity, microwave antigen retrieval was performed at 750 W for 30 min in Tris-EDTA buffer (pH 9.0). After blocking endogenous peroxidase activity with 5% hydrogen peroxidase for 10 min, primary antibody incubation was performed for 1 h at room temperature. The primary antibody was a mouse monoclonal antibody directed against p73 (abcam, Cambridge, UK) used in a 1:100 dilution. An Envision Chem Kit (DakoCytomation, Carpinteria, CA, USA) was used for the secondary antibody at room temperature for 30 min. After washing the tissue samples in Tris-buffered saline for 10 min, 3,3’-diaminobenzidine was used as a chromogen, and Mayer’s hematoxylin counterstain was applied.
All the slides were evaluated without knowledge of any of the clinicopathological data. p73 immunoreactivity was defined by a nuclear staining pattern of the lesional tissue with a minimal background. The percentage of immunoreactive tumor cells was categorized into four groups: 0 (0%), 1 (1%-10%), 2 (11%-50%), and 3 (> 50%). The staining intensity was also categorized into four groups: 0 (negative), 1 (weak), 2 (moderate), and 3 (strong). A final score was obtained for each case by multiplying the percentage and the intensity score. Finally, tumors with multiplied scores exceeding 4 (i.e. tumors with a moderate and strong intensity of > 10% of the tumor cells) were recorded as positive immunoreactivity to p73; all the other scores were considered negative.
Colorectal cancer risks were estimated as the odds ratios (OR) and 95% confidence intervals (CI) calculated by an unconditional logistic regression model adjusted for age and gender. Chi-squared (χ2) tests were used to assess the relationship between the immunoreactivity of p73 protein and the clinicopathological characteristics of colorectal cancer patients and p73 genotypes. In survival analysis, survival curves were computed according to the Kaplan-Meier method. All statistical analysis was performed with the Statistical Package Service Solution software (SPSS for Windows, Standard version 14.0, Chicago, IL, USA).
Due to frequency matching, the median ages were similar between cases (60.4 ± 11.6 years) and controls (60.2 ± 6.2 years) (data not shown).
The genotype frequencies of the p73 G4C14 to A4T14 polymorphism among the controls and cases are shown in Table 1. The frequency of the p73 genotypes in the colorectal cancer patients (GC/GC 47.8%, GC/AT 44.6%, AT/AT 7.6%) was significantly different from that in the control groups (GC/GC 57.8%, GC/AT 36.9%, AT/AT 5.3%). When the GC/GC genotype was used as the reference, the GC/AT and the AT/AT genotypes were significantly associated with the risk for colorectal cancer (OR = 1.46, 95% CI: 1.10-1.94; and OR = 1.72, 95% CI: 0.98-3.03, respectively; Ptrend = 0.01). In a dominant model, the combined variant genotype (GC/AT or AT/AT) increased the risk by 1.5-fold compared with the GC/GC genotype (OR = 1.50, 95% CI: 1.14-1.96). Stratified analysis showed that the association was restricted to patients less than 60 years of age (GC/AT or AT/AT vs GC/GC: 2.22, 1.39-3.55) and male (GC/AT or AT/AT vs GC/GC: 1.91, 1.31-2.77) (Table 2).
| p73 genotype | Cases | Controls | OR (95% CI)1 |
| (n = 383) | (n = 469) | ||
| GC/GC | 183 (47.8) | 271 (57.8) | 1.00 (reference) |
| GC/AT | 171 (44.6) | 173 (36.9) | 1.46 (1.10-1.94) |
| AT/AT | 29 (7.6) | 25 (5.3) | 1.72 (0.98-3.03) |
| GC/AT or AT/AT | 200 (52.2) | 198 (42.2) | 1.50 (1.14-1.96) |
| Ptrend | 0.01 |
| Variables | p73 genotype | Odds ratio of GC/AT + AT/AT, OR (95% CI)1 | |
| GC/GC (case/control) | GC/AT + AT/AT (case/control) | ||
| Age (yr) | |||
| < 60 | 70/81 | 96/50 | 2.22 (1.39-3.55) |
| ≥ 60 | 113/190 | 104/148 | 1.18 (0.84-1.66) |
| Gender | |||
| Male | 88/153 | 114/104 | 1.91 (1.31-2.77) |
| Female | 95/118 | 86/94 | 1.14 (0.76-1.69) |
When various clinicopathological characteristics of the colorectal cancer patients were compared by the p73 GC/AT genotypes, p73 G4C14 to A4T14 polymorphism was correlated with age (P = 0.003), but there were no significant associations of p73 polymorphism with gender, histological type, invasion depth, lymph node metastasis, and Dukes’ stage (each P > 0.05) (data not shown).
p73 immunoreactivity was detected in 252 (65.8%) out of the 383 colorectal cancer tissues (Table 3). The proportion of positive p73 expression was higher among those with GC/AT or AT/AT genotypes (54.0%) than that among those with GC/GC genotype (46.0%) (P = 0.40). Although there was no statistical significance, AT/AT genotype (6.0%) showed a tendency towards lower expression of the p73 protein expression than those with GC/GC genotype (46.0%) and GC/AT genotype (48.0%) when stratified by three genotypes (P = 0.09).
| Genotype | Cases | p73 protein expression | P | |
| Positive | Negative | |||
| (n = 252) | (n = 131) | |||
| GC/GC | 183 | 116 (46.0) | 67 (51.1) | 0.09 |
| GC/AT | 171 | 121 (48.0) | 50 (38.2) | |
| AT/AT | 29 | 15 (6.0) | 14 (10.7) | |
| Ptrend | 0.40 | |||
| GC/GC | 183 | 116 (46.0) | 67 (51.1) | |
| GC/AT+AT/AT | 200 | 136 (54.0) | 64 (48.9) | |
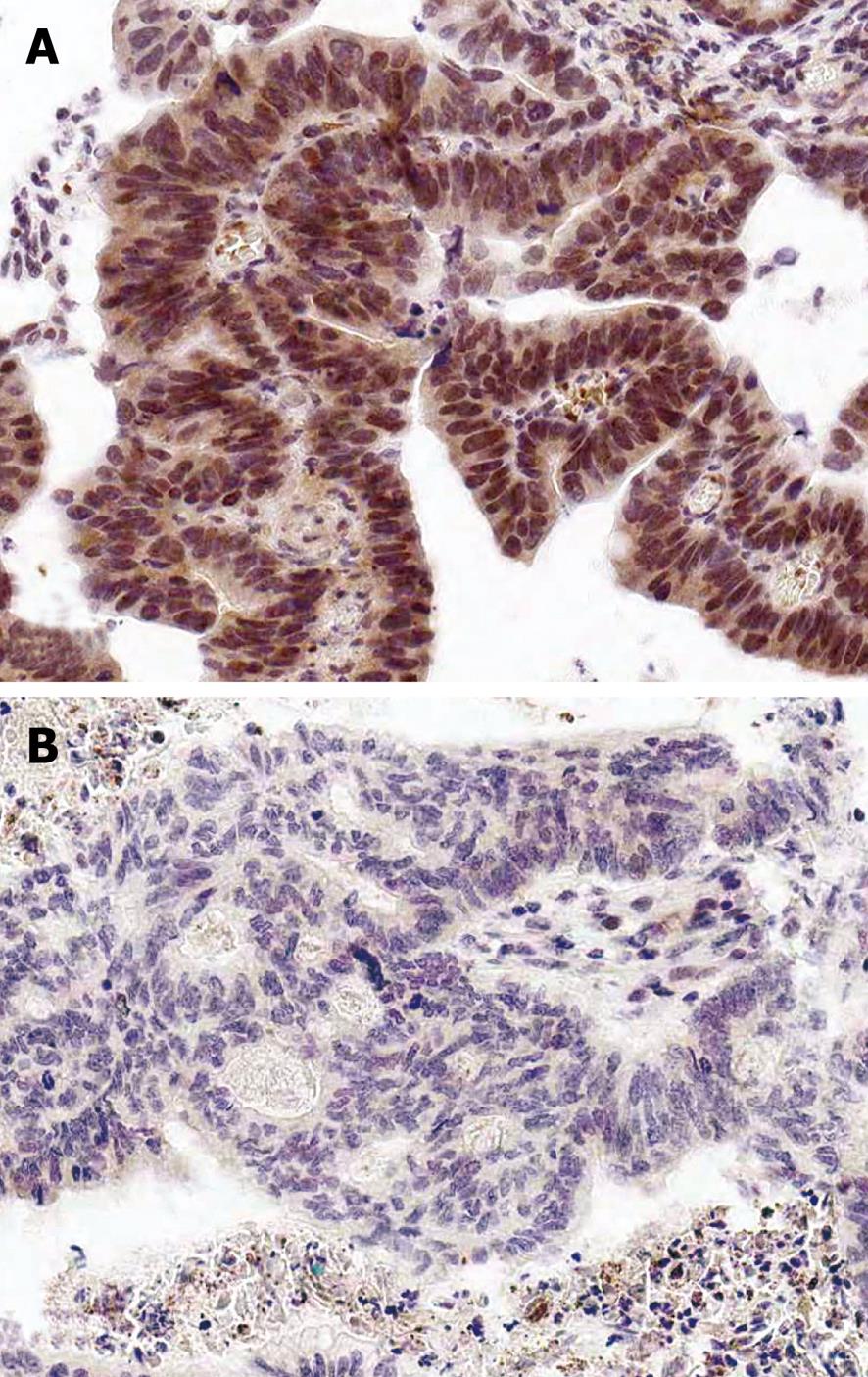
Among various clinicopathological characteristics, invasion depth (P = 0.003) and advanced Duke’s stage (P = 0.06) of colorectal cancer were correlated with p73 expression. There were no significant associations of p73 protein expression with age, gender, histological type, and lymph node metastasis (each P > 0.05, Table 4, Figure 3).
| Clinicopathological characteristics | No. of cases | p73 protein expression | P | |
| Positive | Negative | |||
| (n = 252) | (n = 131) | |||
| Age (yr) | 0.45 | |||
| < 60 | 166 | 113 (68.1) | 53 (31.9) | |
| ≥ 60 | 217 | 139 (64.1) | 78 (35.9) | |
| Gender | 1.00 | |||
| Male | 203 | 134 (66.0) | 69 (34.0) | |
| Female | 180 | 118 (65.6) | 62 (34.4) | |
| Histological type | 0.19 | |||
| Well | 256 | 169 (66.0) | 87 (34.0) | |
| Moderate | 102 | 64 (62.7) | 38 (37.3) | |
| Poorly | 12 | 7 (58.3) | 5 (41.7) | |
| Mucinous | 13 | 12 (92.3) | 1 (7.7) | |
| Invasion depth | 0.003 | |||
| Mucosa | 3 | 1 (33.3) | 2 (66.7) | |
| Submucosa | 9 | 9 (100) | 0 (0) | |
| Muscle | 47 | 39 (83.0) | 8 (17.0) | |
| Subserosa | 324 | 203 (62.7) | 121 (37.3) | |
| Lymph node metastasis | 0.67 | |||
| Positive | 175 | 113 (64.6) | 62 (35.4) | |
| Negative | 208 | 139 (66.8) | 69 (33.2) | |
| Dukes’ stage | 0.06 | |||
| A | 3 | 1 (33.3) | 2 (66.7) | |
| B1 | 43 | 36 (83.7) | 7 (16.3) | |
| B2 | 162 | 102 (63.0) | 60 (37.0) | |
| C1 | 13 | 10 (76.9) | 3 (23.1) | |
| C2 | 148 | 92 (62.2) | 56 (37.8) | |
| D | 14 | 11 (78.6) | 3 (21.4) | |
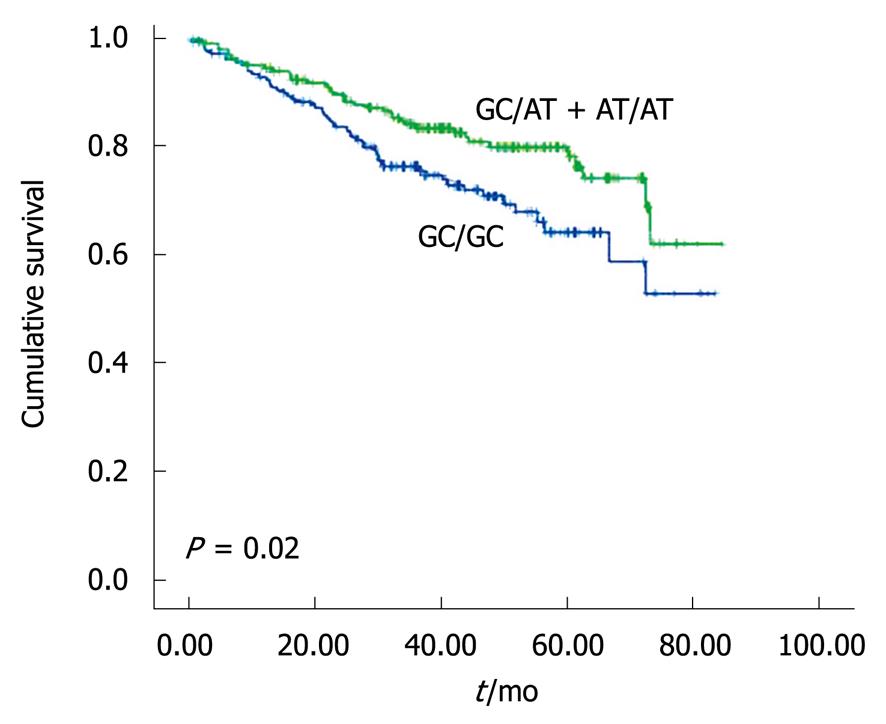
The patients with GC/GC genotype were associated with worse survival compared with those with the other genotypes (P = 0.02, Figure 4). The 5-year overall survival (OS) was 64.1% among those with GC/GC genotype and 76.3% among those with GC/AT or AT/AT genotypes.
In our study, the p73 GC/AT polymorphism was associated with an increased risk of colorectal cancer. When stratified by age and gender, male subjects less than 60 years of age with p73 GC/AT polymorphisms were at an increased risk of colorectal cancer. We suggest that the p73 GC/AT polymorphisms, which is associated with an early onset of less than 60, might play a more important role in the development of colorectal cancer among younger patients.
The association of the p73 GC/AT polymorphism with cancer risk has been investigated in a variety of cancers[11-13,19,20]. There are several studies indicating that subjects with the p73 GC/AT polymorphism might have an increased risk for head and neck cancer[12] and lung cancer in an American population[13]. Pfeifer et al[11] found a relationship of the AT/AT genotype with high risk for colorectal cancer in a Swedish population, which is consistent with our results. Other studies, however, reported that the p73 GC/AT polymorphism was associated with a significantly decreased risk for lung cancer in a Chinese population[19] and esophageal cancer in an Irish population[20]. A few studies even failed to find any correlation between the p73 GC/AT polymorphism and risk for lung cancers in Korean and Japanese populations[21,22]. These inconsistent findings suggest that the p73 polymorphism might have different roles in various cancer types and ethnic populations.
The mechanism whereby the p73 polymorphism influences colorectal cancer development remains to be elucidated. One study has shown that the GC to AT change might lead to formation of a stem-loop structure, which could influence p73 translation efficiency[1]. Another study suggested that the p73 GC/AT polymorphism has a critical role in the development or progression of early cancerous lesions, possibly due to alteration in expression of the p73 protein, perhaps due to alternative splicing[20]. Another study has reported that the GC/AT polymorphism is in linkage disequilibrium with other functional polymorphisms that affect either the expression or activity levels of proteins or enzymes involved in tumorigenesis[19]. However, we note that no significant relationship between the p73G4C14-to-A4T14 polymorphism and p73 protein expression was found in our study. The exact relationship between p73 polymorphism and p73 protein expression might not have been detected due to limitations of this study such as small number of AT/AT groups, relatively small and heterogeneous histological and stage subgroups, and the retrospective study design.
In our study, the expression of p73 was associated with invasion depth and advanced Duke’s stage of colorectal cancer. These results suggest that p73 protein has an important role in colorectal cancer progression. p73 overexpression has been found in various types of cancer, including breast[23], lung[7,8], colorectal[24], and liver[25]. These reports indicated that p73 might be involved in the development of cancers. p73 was initially suggested to be a tumor suppressor gene[1]. However, the result that p73 knockout mice do not develop spontaneous tumors[26] and the p73 gene is rarely mutated in a human cancers[16], suggesting that the activation of a silent allele or overexpression of p73, rather than an impairment of tumor suppressor function, might contribute to tumor development.
To explain the role and mechanism of p73 protein expression, this study investigated the associations between p73 polymorphism and p73 protein expression. There was no statistical significance, when the positive p73 expression in cases with GC/GC genotype (46.0%), GC/AT genotype (48.0%), and AT/AT genotype (6.0%) were stratified by three genotypes. Sun[14] found that overexpression of p73 protein was a valuable prognostic factor to predict poor outcome for patients with colorectal cancer. Tannapfel et al[15] found that patients with p73 positive tumors had a poorer prognosis than those with p73 negative tumors in hepatocellular carcinoma patients. In this study, the patients with GC/GC genotype were associated with worse survival compared with those with the other genotypes.
In conclusion, this study provides the first indication that the p73 G4C14 to A4T14 polymorphism is significantly associated with an increased colorectal cancer risk in a Korean population. Moreover, our study suggests an association between the p73 GC/AT polymorphisms and men with early-onset colorectal cancer. There was a significant association of p73 protein expression with invasion depth, which suggests that p73 immunoreactivity may be a useful pathological marker for colorectal cancer progression. However, patients with the GC/GC genotype were significantly associated with worse survival than those with GC/AT polymorphism. Future molecular epidemiologic studies are needed to elucidate the relationship between p73 polymorphism, its related genes, such as members of the p53 family, and p73 protein expression in colorectal cancer development, and to define whether the association of p73 protein expression may independently occurs of p73 polymorphism or not.
Colorectal cancer is a multigenetic disease whereby biologically relevant single nucleotide polymorphisms might play role as a genetic susceptibility factors. As one of the p53 tumor suppressor family, p73 is known to inhibit cell growth in a p53-like manner, by inducing apoptosis or cell cycle arrest. However, overexpression of wild-type p73 might have some p53-independent functions either as an oncogene or as a tumor suppressor gene in human tumorigenesis.
There are several studies indicating that subjects with the p73 GC/AT polymorphism might have an increased risk for certain types of cancers, including colorectal cancer. However, the genotype-phenotype relationship between p73 polymorphism and p73 protein expression in colorectal cancer has not yet been studied. In this study, patients with the GC/GC genotype were associated with worse survival compared with those with the other genotypes. However, no significant relationship was observed between the p73 G4C14-to-A4T14 polymorphism and p73 protein expression in colorectal cancer.
This study provides the first evidence that the p73 G4C14 to A4T14 polymorphism is significantly associated with an increased colorectal cancer risk in the Korean population. Moreover, our study suggests an association between the p73 GC/AT polymorphisms and men with early-onset colorectal cancer. There was a significant association of p73 protein expression with invasion depth, which suggests that p73 protein has a certain important role in colorectal cancer progression.
To define whether the association of p73 protein expression occurs independently of the p73 polymorphism or not, further study is required. This study provides a valuable prognostic factor to predict poor outcome for patients with colorectal cancer.
The author examined the p73 G4C14 to A4T14 polymorphism and p73 protein expression. The p73 GC/AT polymorphism is associated with an increased colorectal cancer risk and survival in the Korean population. The expression of p73 was associated with invasion depth and advanced Duke’s stage of colorectal cancer. The results are interesting and might provide a useful pathological marker for colorectal cancer progression.
| 1. | Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A, Minty A, Chalon P, Lelias JM, Dumont X. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809-819. |
| 2. | Jost CA, Marin MC, Kaelin WG Jr. p73 is a simian [correction of human] p53-related protein that can induce apoptosis. Nature. 1997;389:191-194. |
| 3. | Flores ER, Tsai KY, Crowley D, Sengupta S, Yang A, McKeon F, Jacks T. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature. 2002;416:560-564. |
| 4. | Yokomizo A, Mai M, Tindall DJ, Cheng L, Bostwick DG, Naito S, Smith DI, Liu W. Overexpression of the wild type p73 gene in human bladder cancer. Oncogene. 1999;18:1629-1633. |
| 5. | Nimura Y, Mihara M, Ichimiya S, Sakiyama S, Seki N, Ohira M, Nomura N, Fujimori M, Adachi W, Amano J. p73, a gene related to p53, is not mutated in esophageal carcinomas. Int J Cancer. 1998;78:437-440. |
| 6. | Choi HR, Batsakis JG, Zhan F, Sturgis E, Luna MA, El-Naggar AK. Differential expression of p53 gene family members p63 and p73 in head and neck squamous tumorigenesis. Hum Pathol. 2002;33:158-164. |
| 7. | Mai M, Yokomizo A, Qian C, Yang P, Tindall DJ, Smith DI, Liu W. Activation of p73 silent allele in lung cancer. Cancer Res. 1998;58:2347-2349. |
| 8. | Tokuchi Y, Hashimoto T, Kobayashi Y, Hayashi M, Nishida K, Hayashi S, Imai K, Nakachi K, Ishikawa Y, Nakagawa K. The expression of p73 is increased in lung cancer, independent of p53 gene alteration. Br J Cancer. 1999;80:1623-1629. |
| 9. | Peters MA, Janer M, Kolb S, Jarvik GP, Ostrander EA, Stanford JL. Germline mutations in the p73 gene do not predispose to familial prostate-brain cancer. Prostate. 2001;48:292-296. |
| 10. | Li Q, Athan ES, Wei M, Yuan E, Rice SL, Vonsattel JP, Mayeux RP, Tycko B. TP73 allelic expression in human brain and allele frequencies in Alzheimer's disease. BMC Med Genet. 2004;5:14. |
| 11. | Pfeifer D, Arbman G, Sun XF. Polymorphism of the p73 gene in relation to colorectal cancer risk and survival. Carcinogenesis. 2005;26:103-107. |
| 12. | Li G, Sturgis EM, Wang LE, Chamberlain RM, Amos CI, Spitz MR, El-Naggar AK, Hong WK, Wei Q. Association of a p73 exon 2 G4C14-to-A4T14 polymorphism with risk of squamous cell carcinoma of the head and neck. Carcinogenesis. 2004;25:1911-1916. |
| 13. | Li G, Wang LE, Chamberlain RM, Amos CI, Spitz MR, Wei Q. p73 G4C14-to-A4T14 polymorphism and risk of lung cancer. Cancer Res. 2004;64:6863-6866. |
| 14. | Sun XF. p73 overexpression is a prognostic factor in patients with colorectal adenocarcinoma. Clin Cancer Res. 2002;8:165-170. |
| 15. | Tannapfel A, Wasner M, Krause K, Geissler F, Katalinic A, Hauss J, Mössner J, Engeland K, Wittekind C. Expression of p73 and its relation to histopathology and prognosis in hepatocellular carcinoma. J Natl Cancer Inst. 1999;91:1154-1158. |
| 16. | Bénard J, Douc-Rasy S, Ahomadegbe JC. TP53 family members and human cancers. Hum Mutat. 2003;21:182-191. |
| 17. | Hamilton SR, Alatonen LA. World Health Organization Classification of Tumours. Pathology and genetics of tumors of the digestive system. Lyon: IARC Press 2004; . |
| 18. | Astler VB, Coller FA. The prognostic significance of direct extension of carcinoma of the colon and rectum. Ann Surg. 1954;139:846-852. |
| 19. | Hu Z, Miao X, Ma H, Tan W, Wang X, Lu D, Wei Q, Lin D, Shen H. Dinucleotide polymorphism of p73 gene is associated with a reduced risk of lung cancer in a Chinese population. Int J Cancer. 2005;114:455-460. |
| 20. | Ryan BM, McManus R, Daly JS, Carton E, Keeling PW, Reynolds JV, Kelleher D. A common p73 polymorphism is associated with a reduced incidence of oesophageal carcinoma. Br J Cancer. 2001;85:1499-1503. |
| 21. | Hiraki A, Matsuo K, Hamajima N, Ito H, Hatooka S, Suyama M, Mitsudomi T, Tajima K. Different risk relations with smoking for non-small-cell lung cancer: comparison of TP53 and TP73 genotypes. Asian Pac J Cancer Prev. 2003;4:107-112. |
| 22. | Choi JE, Kang HG, Chae MH, Kim EJ, Lee WK, Cha SI, Kim CH, Jung TH, Park JY. No association between p73 G4C14-to-A4T14 polymorphism and the risk of lung cancer in a Korean population. Biochem Genet. 2006;44:543-550. |
| 23. | Zaika AI, Kovalev S, Marchenko ND, Moll UM. Overexpression of the wild type p73 gene in breast cancer tissues and cell lines. Cancer Res. 1999;59:3257-3263. |
| 24. | Sunahara M, Ichimiya S, Nimura Y, Takada N, Sakiyama S, Sato Y, Todo S, Adachi W, Amano J, Nakagawara A. Mutational analysis of the p73 gene localized at chromosome 1p36.3 in colorectal carcinomas. Int J Oncol. 1998;13:319-323. |
| 25. | Herath NI, Kew MC, Whitehall VL, Walsh MD, Jass JR, Khanna KK, Young J, Powell LW, Leggett BA, Macdonald GA. p73 is up-regulated in a subset of hepatocellular carcinomas. Hepatology. 2000;31:601-605. |
Peer reviewer: Dr. Yutao Yan, Medicine Department, Emory University, 615 Michael ST, Whitehead Building/265, Atlanta 30322, United States
S- Editor Tian L L- Editor Stewart GJ E- Editor Ma WH