Published online Aug 21, 2010. doi: 10.3748/wjg.v16.i31.3950
Revised: June 17, 2010
Accepted: June 24, 2010
Published online: August 21, 2010
AIM: To study the mechanism underlying carbon tetrachloride (CCl4)-induced alterations of protein synthesis in liver.
METHODS: Male Sprague-Dawley rats were given CCl4 (1 mL/100 g body weight) and 3H-leucine incorporation. Malondialdehyde (MDA) level in the liver, in vitro response of hepatocyte nuclei nucleotide triphosphatase (NTPase) to free radicals, and nuclear export of total mRNA with 3’-poly A+ were measured respectively. Survival response of HepG2 cells to CCl4 treatment was assessed by methyl thiazolyl tetrazolium. Km and Vmax values of nuclear envelope NTPase activity in liver of rats treated with CCl4 were assayed by a double-reciprocal plot.
RESULTS: The protein synthesis was inhibited while the MDA level was significantly increased in liver of rats treated with CCl4. In addition, CCl4 decreased the NTPase binding capacity of nuclear envelope (Km value) in cultured HepG2 cells. Moreover, in vitro ferrous radicals from Fenton’s system suppressed the NTPase activity of liver nuclear envelope in a dose-dependent manner. Down-regulation of the nuclear envelope NTPase activity indicated a lower energy provision for nucleocytoplasmic transport of mRNA molecules, an evidence in CCl4-treated HepG2 cells correspondingly supported by the nuclear sequestration of poly (A)+ mRNA molecules in morphological hybridization research.
CONCLUSION: Inhibition of mRNA transport, suggestive of decreased NTPase activity of the nuclear envelope, may be involved in carbon tetrachloride-inhibited protein synthesis in liver.
- Citation: Li XW, Zhu R, Li B, Zhou M, Sheng QJ, Yang YP, Han NY, Li ZQ. Mechanism underlying carbon tetrachloride-inhibited protein synthesis in liver. World J Gastroenterol 2010; 16(31): 3950-3956
- URL: https://www.wjgnet.com/1007-9327/full/v16/i31/3950.htm
- DOI: https://dx.doi.org/10.3748/wjg.v16.i31.3950
Carbon tetrachloride (CCl4) is an organic solvent widely used in chemical industry. Its hepatotoxicity includes fatty liver and hepatic necrosis[1,2]. Moreover, CCl4 has also an ability to inhibit protein synthesis in liver with early hepatotoxic damage[3]. Early studies showed that free radicals, such as trichloromethyl (· CCl3) and oxygen-centered lipid radicals (LO· or LOO, or both), are generated during CCl4 metabolism by hepatic cellular cytochrome P450. These radicals can cause hepatic injury resulting from CCl4 exposure. Protein synthesis is a cytoplasmic event in eukaryotic cells, in which mRNA templates in the form of a genetic code are employed to guide protein synthesis. A nuclear envelope (NE) of eukaryotic cells causes spatial separation between mRNA and protein synthesis. Thus, mRNA templates in nuclei have to be transported into cytoplasm, a limiting process of protein synthesis. Nuclear membrane-associated nucleotide triphosphatase (NTPase), a specific energy driver of mRNA nucleocytoplasmic export, enables mRNA cytoplasmic accumulation[4,5]. Our previous work demonstrated that reduced NTPase activity in the NE correspondingly decreases mRNA transport[6]. In this report, we studied the role and mechanism of NTPase activity in CCl4-induced inhibition of protein synthesis in liver.
Trypsin, 4,6-diamidino-2-phenylindole dihydrochloride (DAPI), dimethyl sulfoxide (DMSO), DL-dithiothreitol (DTT), adenosine triphosphate (ATP), guanine triphosphate (GTP), cytosine triphosphate (CTP), thymine triphosphate (TTP), and methyl thiazolyl tetrazolium (MTT) were purchased from Sigma Company (St. Louis, MO, USA). DMEM medium was purchased from Invitrogen (Cat.12100-046, Carlsbad, CA, USA), and fetal bovine serum (FBS) was purchased from Beijing New Probe Biotechnology Co., Ltd. (Beijing, China). Trichloride acetic acid (TCA, analytic purity grade), a precipitating agent for protein, and CCl4 (analytic purity grade) were obtained from Beijing Chemical Reagent Factory (Beijing, China). L-[3, 4-3H]-leucine was purchased from Beijing Atom High Tech., Ltd. (Beijing, China).
HepG2 cells (Cat. HB-8065, human hepatic carcinoma cell line) were obtained from American Type Culture Collection (Manassas, VA, USA). HepG2 cells were grown in a DMEM supplemented with 10% newborn calf serum and maintained at 37°C in a humidified atmosphere containing 5% CO2. The 80% confluent-cultured HepG2 cells were treated for 2 h with fresh DMEM with or without CCl4, the former containing a final concentration at 4 mmol/L CCl4 (CCl4 group), and the latter with the same volume of DMSO (control group).
Sprague-Dawley rats were supplied by the Experimental Animal Center, Peking University Health Science Center. The following procedure was approved by the Medical Animal Ethics Committee of the Health Science Center, Peking University. Male rats weighing 180-220 g were fasted for 14 h prior to the experiment, and then had free access to standard animal food and tap water. The animals were housed in plastic cages and maintained at 23 ± 1°C under a natural light-dark cycle in a well-ventilated room.
Rats (n = 6) in the CCl4 group were given CCl4 (dissolved in an equal part of vegetable oil) at 1 mL/100 g of body weight under anesthesia. Rats in the control group (n = 6) received the same volume of vegetable oil. Two weeks following CCl4 treatment, 50 μCi 3H-leucine (100 g of body weight) was injected into each animal via the tail vein. Rats were dispatched by cervical dislocation 1 h later with their livers quickly isolated and perfused in ice-chilled phosphate buffer saline (PBS). Liver tissue samples, free of blood, were divided into two parts and stored at -80°C. To measure the total liver radioactivity, one aliquot of liver tissue was baked at 60°C until its weight remained constant. The liver tissue samples were then ground into powder and digested at 70°C in 5 mL 70:30 mixture of perchloric acid/H2O2 for 30 min. To measure the radioactivity retained at the TCA-insoluble portion of the liver protein, another aliquot of liver tissue was homogenized in ice-chilled PBS using a Teflon glass homogenizer. Homogenates were precipitated with 10% TCA, and TCA-insoluble sediments were digested at 70°C in the same volume of perchloride acid/H2O2 mixture for 30 min. Protein level in TCA-insoluble sediments was measured using the Bradford method. Radioactivity remaining either in the entire liver or in TCA-insoluble portion was determined with a Beckman liquid scintillation counter. 3H-leucine incorporations in the entire liver or in the TCA-insoluble protein were expressed as count per minute (CPM) over mg of dry liver tissue or TCA-insoluble protein portion[7].
Twenty male rats were divided into control group, CCl4-treatment group, taurine group, and CCl4/taurine group, 6 rats in each group. The rats were treated with or without CCl4 (dissolved in vegetable oil) via subcutaneous injection, and oral administration of taurine if necessary. Two weeks after treatment, the rats were sacrificed under anesthesia with urethane (l g/kg, ip). Their livers were perfused in situ at 37°C with PBS via the left ventricle. Liver tissue samples were homogenized in approximately 10 times the volume of saline to tissue weight. MDA level was measured by assaying rat liver homogenates with a MDA kit (Bio-lab Materials Institute, Beijing) following its manufacturer’s instructions. Results were calibrated to their protein content and expressed as nmol MDA/mg protein.
Cytotoxicity of HepG2 cells to CCl4 was detected by MTT metabolic viability assay as previously described[8]. Briefly, HepG2 cells were seeded into a 96-well plate (1 × 105 cells/well), incubated for 12 h, and then switched to a DMEM containing CCl4 at the indicated doses (0, 2, 10, and 50 mmol/L). After incubated for another 4 h at 37°C and further incubated with 1 mg/mL MTT for 2 h, the activity of cellular mitochondrial succinic dehydrogenase was measured. Absorbance at 570 nm wavelength with a reference wavelength of 655 nm reflected the number of surviving cells.
Twenty-four hours after incubation, HepG2 cells were harvested to isolate their nuclei as previously described[5]. Suspended cells were homogenized in Teflon (10 strokes) and centrifuged at 800 g for 10 min. The nuclei were suspended in DS/PMSF buffer, layered over cushions in the same buffer, and spun down at 70 000 g for 60 min. Isolated nuclei were resuspended in STM/PMSF buffer, relayered over cushions of DS/PMSF buffer, and centrifuged at 70 000 g for 30 min. The final pellet was resolved with STM/PMSF to obtain a final protein concentration of 1 mg/mL and stored at -70°C. The marker enzyme activities specific to nuclei or cytoplasm were determined for monitoring the purity of isolated NE preparations.
NE NTPase activity was assayed as described by Ramjiawan et al[9] with some modifications. In the assay, 40 μL nuclear extracts (containing 40 μg protein) of HepG2 cells with 2 mmol/L CCl4 or DMSO treatment was preincubated in 200 μL buffer for 10 min at 30°C. After different levels of ATP or GTP substrates were added into the nuclear extracts to initiate NTPase reaction and incubated at 30°C for 10 min, reactions were terminated by adding 10% SDS and test tubes were immediately placed in ice. Inorganic phosphates produced in these reactions were detected according to the method of Tiffany[10]. The two characteristic constants, i.e. maximum velocity (Vmax) and Michaelis constant (Km) for NTPase, were obtained using the Line weaver-Burke equation, and expressed as nmol Pi/mg, Pr/min and nmol/L, respectively, for ATP or GTP. Preliminary experiments showed a linear relationship between NTPase activity and incubation time.
Double staining of HepG2 cells with a fluorescein isothiocyanate (FITC) probe conjugated with oligo-(dT)15 specific for mature mRNA with 3’-poly A+ tailing and 4, 6-diamidino-2-phenylindole (DAPI) for localization of nuclei was carried out as previously described[11]. Briefly, after formaldehyde fixation, HepG2 cells cultured on cover slips were permeabilized with PBS/0.5% Triton X-100 for 10 min at room temperature. Cover slips were equilibrated in 2 × SSC for 10 min, and incubated with a FITC-labeled oligo-(dT)15 probe (showing green fluorescence) for 30 min. The cells on cover slips were washed three times with PBS, and incubated for another 30 min at room temperature with a DAPI solution (showing blue fluorescence). The cells were then washed once with PBS/0.2% Triton X-100 and twice with PBS. To ensure that the FITC-hybridization of mRNA was not resulted from DNA binding, RNase A was introduced into the sample to digest all remaining RNA. Total mature mRNA with 3’-poly A+ tailing was clearly observed either in nuclei or in cytoplasm with FITC-oligo (dT)15 labeling under a fluorescence microscope.
Hepatocyte nuclei from rats were prepared and characterized using the method described in section “Isolation and characterization of nuclei”. Different levels of hydroxyl radicals from in vitro Fenton reaction[12] system were freshly prepared by mixing FeSO4 and H2O2 in the ratio of 0/0, 0.1/0.5, 0.5/2.5, 1/5, and 5/25 [(μmol/L)/(μmol/L)], respectively. The mixture was immediately added to the hepatocyte nuclei. After hepatocyte nuclei were incubated with different levels of hydroxyl radicals for 30 min at 30°C, NE NTPase activity for the ATP or GTP was determined according to the method described in section “Kinetics assay of NE NTPase activity”.
All data were expressed as mean ± SE (n = 4-6). Statistical analysis was performed by one-way analysis of variance followed by Student-Newman-Keuls test. P < 0.05 was considered statistically significant.
Electron micrographs showed an early and widespread dislocation of ribonucleoprotein particles from membranes of the rough endoplasmic reticulum in hepatocytes of rats 3 h after CCl4 administration. Furthermore, amino acid incorporations into two liver-produced proteins, albumin and fibrinogen, were decreased in the same CCl4-treated rats. These findings suggest that protein synthesis in liver is influenced by CCl4 treatment[3].
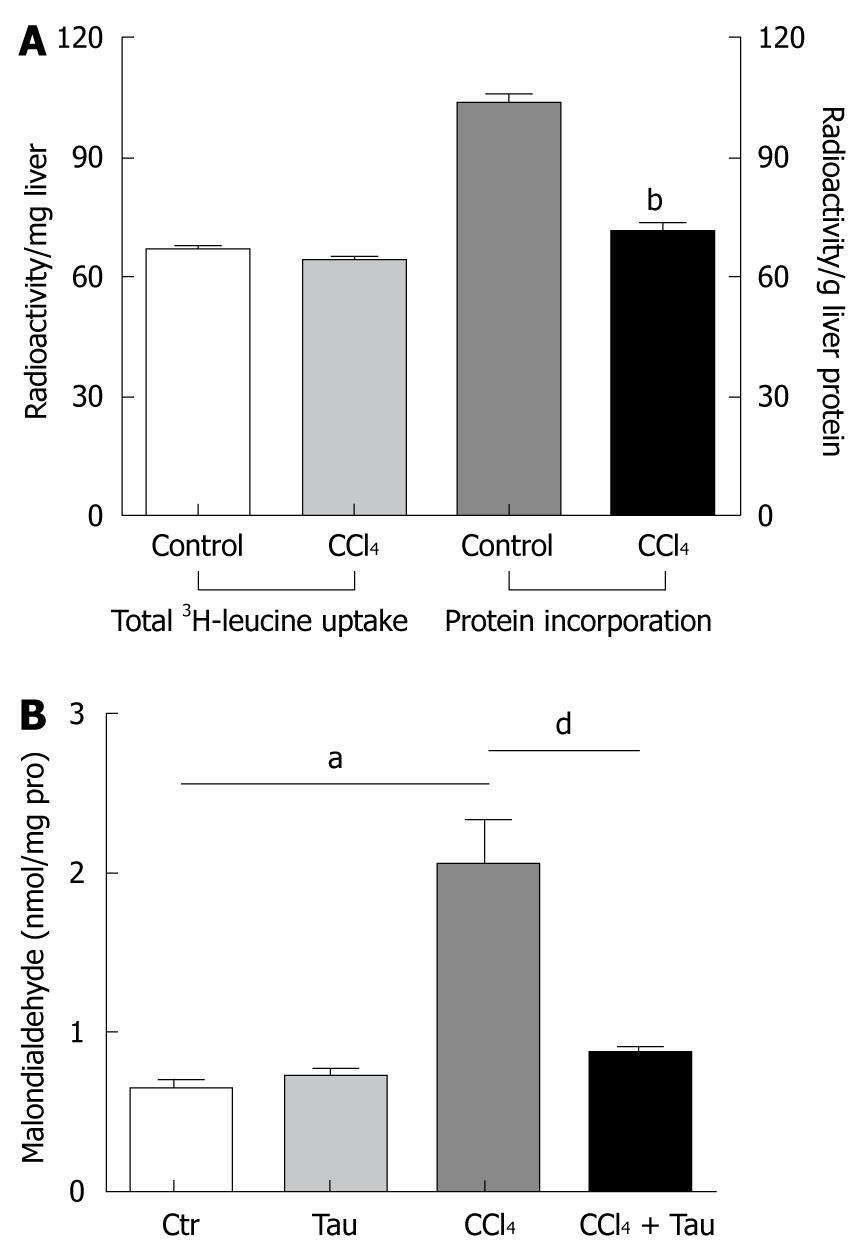
To confirm that the incorporation of amino acid into liver protein is impaired, 50 μCi 3H-leucine (100 g of body weight) was administered to the rats through the tail vein 2 h after CCl4 treatment. Trichloride acetic acid (TCA), a general protein-precipitating agent, was used in tissue homogenates to separate total protein from rat liver tissues. The rates of 3H-leucine incorporation into the liver homogenate and TCA-precipitated liver protein are shown in Figure 1A. Treatment with CCl4 significantly decreased 3H-leucine incorporation into TCA-precipitated liver protein (103.7 ± 5.4 cpm/mg in control group vs 71.8 ± 3.8 cpm/mg in CCl4 treatment group).
No significant difference was observed in total radioactivity in the entire liver between CCl4 treatment and control groups (66.7 ± 2.4 cpm/g vs 64.2 ± 1.8 cpm/g), indicating that the decreased total protein synthesis in CCl4 treatment group is not due to the difference in leucine uptake between the two groups.
· CCl3 radicals produced in reactions by liver microsomes of animals exposed to CCl4 were assumed to attack the membrane lipid in hepatocyte endoplasmic reticulum. When these free radicals attacked the membrane lipid in hepatocyte endoplasmic reticulum, malondialdehyde was brought out quickly, suggesting that MDA formation is recognized as an indicator of lipid peroxidation initiated by reactive radicals. Taurine has been well-known to be an effective scavenger of reactive radicals in vivo and in vitro through an unknown mechanism[5], which ameliorates MDA production in oxidative damaged liver. In the current work, the MDA level was 220% higher in liver of rats treated with CCl4 than in liver of rats not treated with CCl4. Meanwhile, the MDA level was 42% lower in liver of rats treated with combined CCl4 and taurine than in liver of rats treated with CCl4 alone (Figure 1B), indicating that reactive radicals have formed in liver of rats during CCl4 metabolism.
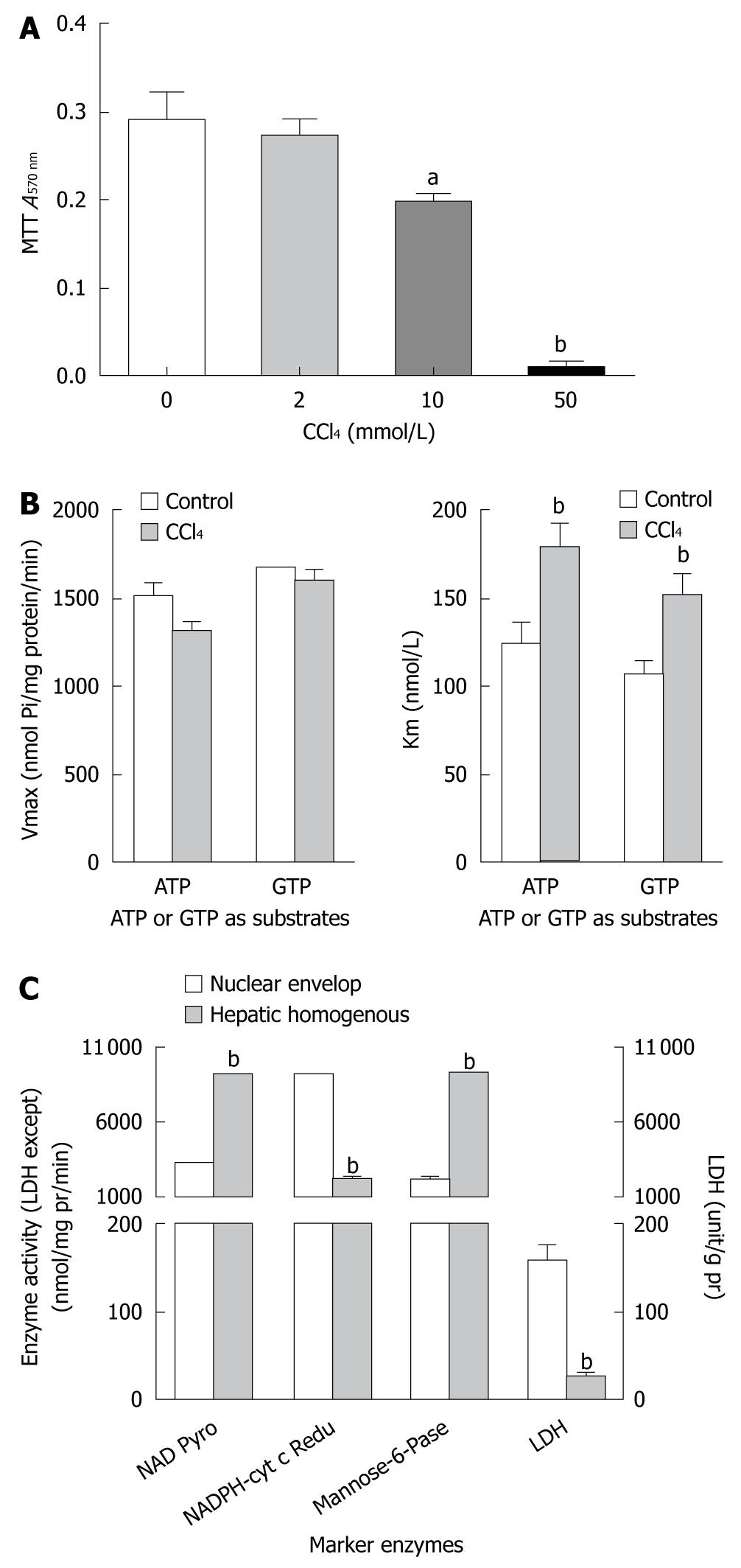
To further investigate the mechanism underlying CCl4-inhibied protein synthesis in liver, the CCl4 dose acting on cultured HepG2 cells was maximized by measuring the hepatotoxicity of CCl4 in a MTT cell survival assay. CCl4 at the dose of 10 mmol/L and 50 mmol/L decreased the survival rate of HepG2 cells to 32% (0.290 ± 0.08 vs 0.196 ± 0.04) and 97% (0.290 ± 0.08 vs 0.009 ± 0.01), respectively (P < 0.01, Figure 2A). Therefore, CCl4 at a dose of lower than 10 mmol/L was used to explore the mechanism underlying CCl4-inhibited protein synthesis in liver of rats in subsequent experiments.
Protein synthesis in liver, regulated by the richness of cytoplasmic template of mRNA, is a key event in cell survival and metabolism. Down-regulation of nuclear export of mature mRNA into cytoplasm is related to its depressed NE NTPase activity in aged rats (data not shown). NTPase, embedded in eukaryotic nuclear envelope, provides nucleocytoplasmic transport energy and promotes shuttling of macromolecules between nuclei and cytoplasm via its hydrolysis action on NTP substrates, such as ATP, GTP, CTP, and TTP. In this study, nuclear envelopes of HepG2 cells were isolated to study the effect of CCl4 on mRNA export into cytoplasm through NE NTPase action in HepG2 cells.
To characterize the purity of nuclei, several marker enzymes for different organelles were used to assess the quality of isolated nuclear envelopes with reference to their hepatic homogenous counterparts (Figure 2C). The activity of NAD pyrophosphorylase was 6.9-fold higher in the prepared nuclear envelope, previously assumed to be exclusively located in nuclei, than in homogenates of HepG2 cells. The activity of NADPH cytochrome-C reductase (as a marker enzyme in microsome) was only 22% higher in the prepared nuclear envelope than in homogenates of HepG2 cells. The activities of other marker enzymes, such as mannose-6-phosphatase (an indicator of both nuclei and cytoplasm) and lactose dehydrogenase (an indicator of cytoplasm) had a similar changing tendency as NAD pyrophosphorylase and NADPH cytochrome-C reductase, showing that these nuclear envelopes, prepared from HepG2 cells in this experiment, have less cytoplasmic contamination and are comparable with the prepared nuclear envelopes, and can be used in analysis of NTPase activity.
In the present study, CCl4 significantly depressed the NTPase activity of hepatic nuclei, regardless of whether the ATP or GTP acted as a substrate, in terms of Km but not Vmax value. An estimation using the double-reciprocal plot method (Figure 2B) showed that CCl4 interfered with the HepG2 NE NTPase Km but not the Vmax, in ATP or GTP acting as a substrate, suggesting that the Km value of liver nuclear membrane NTPase contributes to a lower hydrolysis efficiency on NTP molecules in the CCl4 treatment group than in the control group, and decreases energy provision for nucleocytoplasmic transport.
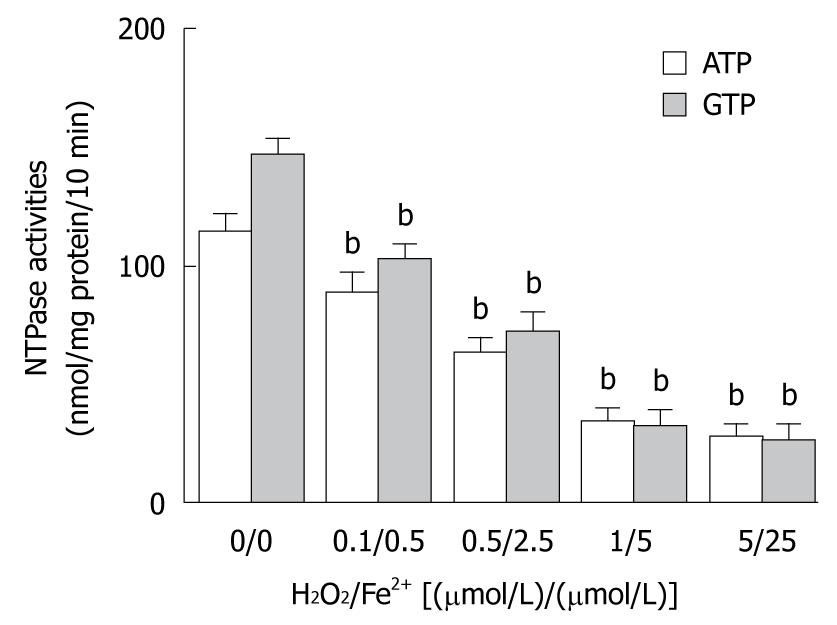
The elevated MDA level in liver tissue of Sprague-Dawley rats treated with CCl4 was significantly correlated with membrane lipid peroxidation initiated by reactive radicals. To observe the involvement of reactive radicals produced by CCl4 metabolism in hepatic nuclear NTPase activity, Fenton reaction system, an in vitro mimic system, was used. In brief, ferrous sulfate and hydroperoxide were simultaneously mixed in different ratios [0.1/0.5, 0.5/2.5, 1/5, and 5/25, (μmol/L)/(μmol/L)] and placed into cultured media to produce unstable reactive radicals. HepG2 nuclear envelope fractions were incubated with an equal volume of in vitro radicals at different concentrations. The results showed that the in vitro radicals decreased the nuclear NTPase activity in a dose-dependent manner, regardless of whether ATP or GTP was used as a substrate (Figure 3). After nuclei of HepG2 cells were incubated with reactive radicals at a mixing ratio of (5 μmol/L)/(25 μmol/L) of Fe2+/H2O2, the NTPase activity on the NE was decreased by 75.4% (for ATP) and 82.2% (for GTP), respectively (P < 0.01), indicating that the decreased nuclear NTPase activity in HepG2 cells treated with CCl4 may be attributed to hepatic microsome CCl4 metabolites through the action of its reactive radicals.
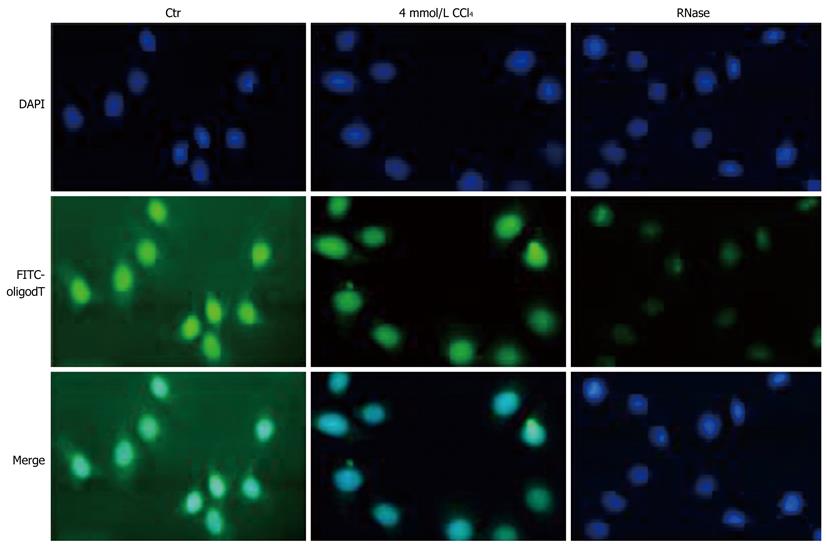
As described above, the nuclear NTPase activity in HepG2 cells was sharply suppressed by CCl4 treatment, the exporting process of mRNA templates into cytoplasm was thus assumed to be influenced in CCl4-treated HepG2 cells. To investigate whether the concurrent export inhibition of mRNA occurs in CCl4-treated HepG2 cells, the poly (A)+ RNA molecular (i.e. mRNA) shuttling between nuclei and cytoplasm in cultured HepG2 cells treated or not treated with CCl4 was morphologically observed by fluorescence in situ hybridization with a FITC-labeled oligo-(dT)15 probe, a specific targeting probe for endogenous 3’-poly(A)+ tailing maturing mRNA (Figure 4). The FITC-labeled oligo-(dT)15 hybridization in HepG2 cells not treated with CCl4 was almost evenly distributed throughout the whole cells. Moreover, a weaker hybridization was observed in cytoplasm of HepG2 cells treated with CCl4. Pretreatment with RNase A nearly abolished the observed results mainly from the specific hybridization to poly(A)+ mRNA with FITC-labeled oligo-(dT)15 staining, demonstrating that CCl4 treatment inhibits the export of poly (A)+ mRNA molecules from the nuclear site by inhibiting nuclear NTPase activity.
In 1987, Enzan[13] conducted a study to observe the protein synthesis of Ito cells in acute liver injury induced by CCl4. He used small liver tissue blocks from normal and CCl4-treated mice that was incubated in cultured medium supplemented with 3H-leucine for one hour, and found that a few Ito cells in normal mouse liver tissues were labeled with 3H-leucine, while no labeling over the Ito cells was observed in CCl4-treated mouse liver without any cellular degeneration, revealing that protein synthesis is blocked in Ito cells after CCl4 treatment[13]. In this study, 3H-leucine incorporation into liver protein was decreased 30% in CCl4-treated rats, which is consistent with the findings of Enzan[13].
Both protein synthesis and degradation occur actively in liver in order to maintain the normal metabolism through appropriate steady-state levels of liver proteins. Among the liver protein homeostasis, mature 3’-poly (A)+ mRNA acting as a template appears to be an important determinant regulator for protein synthesis. Thus, all eukaryotic cytoplasmic mature 3’-poly (A)+ mRNA molecules have to be transported from their original nuclei by traversing through the nuclear pore, an active process in which the nuclear membrane-associated NTPase can provide mechanic energy by catalyzing the hydrolysis of nucleoside tri-phosphates[4,10]. Moreover, the effect of changes in nuclear membrane phosphatidylcholine composition by phospholipase A2 (PLA2) on mRNA transport is related to the changes in NTPase activity responded to PLA2[6]. Based on this assumption, we further explored whether protein synthesis in liver inhibited by CCl4 is related to alterations in nuclear NTPase activity. In brief, the Km value of NE NTPase in CCl4-treated HepG2 cells increased to 44% (ATP as a substrate) and 42% (GTP as a substrate), respectively (Figure 2B), indicating that NE NTPase in CCl4-treated HepG2 cells has a lower substrate binding affinity and a lower energy provision for the export process.
Although the nuclear NTPase activity was inhibited by CCl4 treatment, whether inhibition of protein synthesis in liver inhibited by CCl4 is also brought out by the decreased the nucleocytoplasmic transport of total mRNA remains unknown. A fluorescent FITC-probe conjugated with 5’-oligomerization with 15 consequent thymine bases was designed to test this hypothesis (Figure 4). The probe can specifically bind to mature mRNA with 3’-poly A+. Human HepG2 cells treated with CCl4 decreased the fluorescence strength in cytoplasm, suggesting that CCl4 inhibits mature mRNA nuclear export and that decreased NE NTPase activity is involved in CCl4-induced inhibition of total mRNA nucleocytoplasmic transport and protein synthesis in liver.
In this study, the MDA level was 220% higher in CCl4 treatment group than in control group. However, taurine, a potent scavenger of reactive oxidative radicals, clearly reduced the MDA level in liver of rats treated with CCl4 (Figure 1B), displaying that CCl4-induced liver oxidative damage is caused by reactive radicals via liver cytochrome P450 metabolism of CCl4. To further study the effect of free radical levels on alteration in nuclear NTPase activity, the nuclear extracts were co-incubated with Fenton’s radicals at different concentrations for 30 min. The nuclear NTPase activity was inhibited by Fenton’s radicals in a dose-dependent manner (Figure 3), suggesting that metabolism of CCl4 radicals is an intermediate step in inhibiting nuclear NTPase activity, mRNA nucleocytoplasmic transport, and protein synthesis.
The cellular concentration of a protein is determined by a delicate balance of at least seven processes, each one under several potential points of regulation. It has been shown that gene expression is down-regulated by CCl4 and other chemicals. However, the mechanical mechanism underlying inhibition of protein synthesis in liver induced by CCl4 is correlated to the decreasing nucleocytoplasmic transport of mature mRNA. Advances in modern chemical industry demand to evaluate the safety of various chemical substances. The comprehensive evaluation of these chemical substances has to be further studied in hepatotoxicity.
Many proteins produced in liver, such as albumin and prothrombin, are involved in coordination in the body’s metabolism. The importance of protein function in pathogenesis of liver disease has been increasingly recognized. Carbon tetrachloride (CCl4) is an organic solvent widely used in chemical industry. Its hepatotoxicity includes fatty liver and hepatic necrosis, and evidence also shows that CCl4 inhibits protein synthesis in liver with early hepatotoxic damage. The mechanism underlying alterations in hepatic protein synthesis induced by CCl4 treatment, however, is largely unknown.
Among the steady-state levels of liver protein, mature mRNA acting as a template is an important determinant regulator for protein synthesis. The mRNA nucleocytoplasmic transport has to be promoted by mechanic energy from the nuclear membrane-associated nucleotide triphosphatase (NTPase), which can catalyze the hydrolysis of nucleoside triphosphate. Inhibition of protein synthesis in liver is correlated with down-regulation of nucleocytoplasmic transport in total mRNA during hepatotoxicity induced by CCl4.
This study focused on the effect of mRNA nucleocytoplasmic transport on protein synthesis in liver. The results showed that the decreased NTPase activity of hepatic nuclear envelope inhibits mRNA nucleocytoplasmic transport during protein synthesis in liver inhibited by CCl4.
This study highlighted the importance of nucleocytoplasmic mRNA transport in regulation of protein synthesis in liver. Nucleocytoplasmic mRNA transport and gene transcription are deserved in assessment of hepatotoxicity of prospective chemical substances.
A nuclear envelope NTPase is a key enzyme involved in nucleocytoplasmic transport by catalyzing the hydrolysis of nucleoside triphosphate. When the activity of injured NTPase in nuclear envelope is decreased, the number of mRNA templates in cytoplasm is influenced.
The manuscript is well written and the hypothesis contributes to our understanding of the mechanism underlying CCl4-induced liver damage.
| 1. | Masuda Y. [Learning toxicology from carbon tetrachloride-induced hepatotoxicity]. Yakugaku Zasshi. 2006;126:885-899. |
| 2. | Quan J, Yin X, Xu H. Boschniakia rossica prevents the carbon tetrachloride-induced hepatotoxicity in rat. Exp Toxicol Pathol. 2009;Epub ahead of print. |
| 3. | Smuckler EA, Iseri OA, Benditt EP. An intracellular defect in protein synthesis induced by carbon tetrachloride. J Exp Med. 1962;116:55-72. |
| 4. | Agutter PS, McArdle HJ, McCaldin B. Evidence for involvement of nuclear envelope nucleoside triphosphatase in nucleocytoplasmic translocation of ribonucleoprotein. Nature. 1976;263:165-167. |
| 5. | Li JX, Pang YZ, Tang CS, Li ZQ. Protective effect of taurine on hypochlorous acid toxicity to nuclear nucleoside triphosphatase in isolated nuclei from rat liver. World J Gastroenterol. 2004;10:694-698. |
| 6. | Li JX, Li ZQ, Pang YZ, Tang CS. Phospholipase A2 inhibits nuclear nucleoside triphosphatase activity and mRNA export in isolated nuclei from rat liver. Life Sci. 2003;73:969-980. |
| 7. | Livne E, Weiss A. In vitro effect of hormones and growth factors on the incorporation of [3H]leucine, [35S]sulfate and [3H]proline by chondrocytes of aging mice. Mech Ageing Dev. 1993;72:213-229. |
| 8. | Zhang LJ, Li ZQ, Yang YP, Li XW, Ji JF. Tunicamycin suppresses cisplatin-induced HepG2 cell apoptosis via enhancing p53 protein nuclear export. Mol Cell Biochem. 2009;327:171-182. |
| 9. | Ramjiawan B, Czubryt MP, Massaeli H, Gilchrist JS, Pierce GN. Oxidation of nuclear membrane cholesterol inhibits nucleoside triphosphatase activity. Free Radic Biol Med. 1997;23:556-562. |
| 10. | Tiffany BR, White BC, Krause GS. Nuclear-envelope nucleoside triphosphatase kinetics and mRNA transport following brain ischemia and reperfusion. Ann Emerg Med. 1995;25:809-817. |
| 11. | Watanabe M, Fukuda M, Yoshida M, Yanagida M, Nishida E. Involvement of CRM1, a nuclear export receptor, in mRNA export in mammalian cells and fission yeast. Genes Cells. 1999;4:291-297. |
| 12. | Rachmilovich-Calis S, Masarwa A, Meyerstein N, Meyerstein D, van Eldik R. New mechanistic aspects of the Fenton reaction. Chemistry. 2009;15:8303-8309. |
| 13. | Enzan H. Protein synthesis in Ito cells (fat-storing cells) of cultured liver tissues from CCL4-treated mice. A light and electron microscopic autoradiographic study. Acta Pathol Jpn. 1987;37:225-230. |
Peer reviewers: Jordi Camps, PhD, Centre de Recerca Biomèdica, Hospital Universitari de Sant Joan, C. Sant Joan s/n, 43201 Reus, Catalunya, Spain; Islam Khan, PhD, Professor, Department of Biochemistry, Faculty of Medicine, Kuwait University, PO box 24923, 13110 Safat, Kuwait
S- Editor Wang YR L- Editor Wang XL E- Editor Zheng XM