COLONIC EPITHELIAL STEM CELLS
The adult colonic epithelium has a well-defined architecture organized into crypts, dynamic structures which are constantly self-renewing[1]. Each crypt unit is maintained by adult multipotent stem cells (SCs), located at the bottom of the structure itself, that are able to simultaneously self-renew and generate a population of transiently amplifying cells which in turn generate more mature cells. Three differentiated cell types mediate the function of colonic epithelium: the colonocytes, also termed absorptive enterocytes, the mucus-secreting goblet cells and the enteroendocrine cells.
Adult SCs are defined by several key functional properties including: self-renewal, potential for multilineage differentiation and tissue regeneration. Two different models have been proposed to localize the intestinal SCs; the “+4 position” model and the “stem cell zone” model[2]. According to the former, the intestinal SCs are located at +4 position relative to the bottom of the small intestine crypt, just above the non-cycling Paneth cells. These cells are actively cycling and, through asymmetric division, give rise to their differentiated progeny. The more recent “stem cell zone” model states that small undifferentiated cycling cells, termed crypt base columnar (CBC) cells, are the true intestinal SCs. These cells are interspersed between the Paneth cells in the small intestine or located at the very bottom of the crypt in the colon.
Despite the fact that colonic crypts have long been known to harbor a functional stem cell compartment, the identification, isolation and characterization of colonic crypt SCs has been hampered by the absence of reliable molecular markers. Over the last 30 years, many studies have been performed to indirectly localize intestinal SCs within the colonic crypts by using techniques such as long-term retention of DNA label[3] or histone-GFP marking[4], both based on the “immortal strand hypothesis” proposed by Cairns[5]. According to this theory, SCs might selectively retain their old DNA strands, while donating the newly synthesized DNA strands to their progeny. However, this hypothesis is currently a subject of controversy due to the demonstrated absence of asymmetric genetic material segregation in hematopoietic stem cells[6]. Recently, several molecules have been proposed as markers of SCs in the intestine including the RNA-binding protein musashi-1 (Msi-1)[7] and Hes-1, a transcriptional repressor transactivated by Msi-1[8]. Hes-1 and Msi-1 were shown to be co-expressed by the putative SCs at the crypt base, but immunoreactivity was also observed in a broader population of cells.
Other putative biomarkers have been evaluated for distinguishing the SC population within the colon, such as members of the integrin superfamily of transmembrane glycoproteins including α2 and β1 subunits[9]. Additionally, Eph-B receptors have been described as important regulators of migration and proliferation in the intestinal epithelium[10]. However, the inhibition of Eph-B2/Eph-B3 signaling has been shown to reduce the number of proliferating cells without altering the stem cell number, suggesting that Eph-B receptors are unlikely to be independent biomarkers of colonic SCs.
Another possible intestinal stem cell marker recently identified is the polycomb protein Bmi-1, known to be involved in the maintenance of hematopoietic and neural stem cells[11]. In the small intestine, this factor is expressed in cells with stem cell features located near the crypt bottom[12].
The colonic epithelium is replaced every five days[13]. This high rate of tissue renewal depends on a complex interplay between processes involving cell proliferation, differentiation, migration, adhesion and cell death that are finely coordinated by a relatively small number of highly evolutionarily conserved signaling pathways including BMP, Sonic hedgehog, Notch and Wnt, the latter playing a critical role in the regulation of epithelial SCs in the intestinal tract.
Wnt signaling is required for self-renewal of gut SCs; it is involved in intestinal embryogenesis and adult intestinal epithelial cell proliferation[14,15]. The unique role played by the Wnt pathway in the physiology of the intestine led to the identification of the Wnt target gene Lgr5 as a biomarker of intestinal SCs in mouse small intestine and colon. The Lgr5 gene encodes a leucine-rich repeat containing G-protein coupled receptor, also known as Gpr49. Lgr5 expression is restricted to cycling CBC cells and it has been demonstrated that Lgr5-expressing cells differentiate into the expected functional lineages of the colonic epithelium[16].
Interestingly, the ability of single Lgr5+ SCs to establish long-term culture and to generate crypt-villus organoid, without requiring a mesenchymal niche, has been also described[17].
Transcriptome analysis of Lgr5+ epithelial cells isolated from the bottom of the small intestinal crypts led to the identification of a gene signature for these Lgr5+ SCs[18]. Not surprisingly, many genes on the list were Wnt target genes whose expression was confirmed to be restricted to cells at the base of the crypts, as revealed by in situ hybridization. By this technique olfactomedin-4 (OLFM4) was also identified as a highly-specific and robust marker for Lgr5+ SCs, even though its expression was not under the control of Wnt. The OLFM-4 gene encodes a secreted molecule with unknown function, originally cloned from human myeloblasts[19], which is enriched in human colon crypts[20]. Due to the very low expression levels of Lgr5, OLFM-4 has been recently proposed as a more faithful SC marker highly expressed in CBC cells in human small intestine and colon[21].
COLORECTAL CANCER STEM CELL IDENTIFICATION
Tumors are composed of a heterogeneous mixture of cancer cells at various levels of differentiation, very similar to the structure of an organ. Recently, the “cancer stem cell” model of tumorigenesis has proposed that within the tumor mass there is a predetermined cell population with a ‘‘stem cell’’ phenotype, able to perpetuate the cancer, while the rest of the tumor cells are incapable of self-renewal. Even though it has long been assumed that mutations within adult colonic stem cells may induce neoplastic transformation, the proof of existence of colorectal cancer stem cells (CRC-SCs) has been hindered in the past years by difficulties in identifying a specific biomarker for this rare cell population. Only recently, new evidence has been provided that supports the existence of CRC-SCs, confirming that the tumorigenic cell population of CRC can be isolated on the basis of the expression of specific cell surface biomarkers. The standard analysis to ascertain the existence of a subpopulation of cancer stem cells (CSCs) is the demonstration that these cells can transfer the tumor in immunocompromised mice and replicate the phenotypic heterogeneity of the parental tumor. Several recent studies have evaluated the functionality of specific CRC-SC biomarkers by using a combination of flow cytometry to identify a “putative” SC population and xenograft models involving immunodeficient mice to determine their tumor initiating potential[22]. In the first two studies, CD133, also known as Prominin-1, was employed to identify the tumorigenic cell population within CRC[23,24]. O’Brien et al[23] isolated CD133+ cells from seven primary colon cancers and ten extracolonic (metastatic) sites. When transplanted into the renal capsule of NOD/SCID mice, CD133+ cells readily developed tumors displaying morphologic features equivalent to those of the parental cancer. Tumor phenotype was further maintained upon serial transplantation[23]. Similarly, in the second study, a population of CD133+ cells, accounting for approximately 2.5% of tumor cells, was isolated from colon cancer specimens and perpetuated in vitro as floating colonies or “tumor spheres”. These tumor spheres, expressing the epithelial adhesion molecule BerEp4, but not differentiation markers such as cytokeratin 20 (CK20), were enriched in a tumorigenic population and could be maintained for serial in vitro passages[24]. Both studies demonstrated the expression of CD133 also in normal colon tissue, although at a lower frequency, reinforcing the hypothesis that CD133+ CRC-initiating cells in cancer samples might result from oncogenic transformation of normal colonic SCs. Subsequently, Dalerba et al[25] proposed CD44 and the epithelial surface antigen (EpCAM) as CRC-SC-specific markers, with further enrichment by CD166. Purified CD44+/EpCAMHIGH cells injected into NOD/SCID mice resulted in high frequency generation of tumor xenograft. In contrast, CD44-/EpCAMLOW cells lack tumor-initiating activity[25]. Further subfractionation of the CD44+/EpCAMHIGH cell population by using the mesenchymal stem cell marker CD166 increased the success of tumor xenograft. Finally, in a more recent study, aldehyde dehydrogenase 1 (ALDH) has been proposed as a promising new marker for normal and malignant human colonic SCs[26]. As few as 25 ALDH1+ cancer cells, isolated by flow cytometry, were able to generate tumor xenografts. Notably, a subsequent isolation of cancer cells using a second marker (CD44 or CD133 serially) produced a modest further enrichment of tumor-initiating ability.
Significant controversies exist over the functional role of these CRC-SC markers. Major questions have been raised regarding CD133. Indeed, Shmelkov et al[27], using a transgenic mouse model in which the CD133 promoter drove LacZ reporter expression, demonstrated that CD133 was expressed by both mature and undifferentiated colonic epithelial cells, suggesting that CD133 is not restricted to the SC compartment[27]. Moreover, using confocal microscopy the authors reported that in primary colon cancer samples from humans and mice, CD133 was expressed in all epithelial, EpCAM+ cells in the malignant tissue and that CD133 expression was excluded from the non-epithelial cell components of the tumor. Thus, they proposed that the inability of CD133- to generate tumors could be simply due to their non-epithelial nature. Furthermore, the same authors demonstrated that both CD133+/EpCAM+ and CD133-/EpCAM+ cell populations, isolated from liver metastasis of colon cancer, were able to generate tumors upon serial transplantation into NOD/SCID mice. Taken together these data appear to be in contrast with those previously published. However, the different techniques employed do not allow a direct comparison between the studies. Indeed, it is possible that primary and metastatic tumors may have a different expression pattern for CD133. Moreover, when Shmelkov et al[27] analysed primary tumors they did not perform the same functional assessment of tumor-initiating activity as was done in previous reports.
Regardless of the ongoing debate as to CD133 as a CRC-SC marker and the lack of evidence for a functional role in tumorigenesis of this marker, clinical data support the significance of this molecule in CRC, particularly as an independent negative prognostic marker[28]. Furthermore, the combined evaluation of CD133 and nuclear β-catenin can identify high risk cases of low stage CRC[29]. A comparison of expression of the three markers CD133, CD44 and CD166 that have been associated with CRC-SCs revealed that the expression of CD133 correlates with that of CD166, whereas both do not correlate with CD44, confirming that CD133 is, alone, the best marker to predict poor patient survival[30]. In a more recent report, Artells et al[31] observed longer relapse-free interval and increased overall survival in patients with lower levels of CD133.
To address the clinical relevance of CD133 for CRC metastasis in patients, Horst et al[32] analyzed CD133 expression in a matched case-control collection of 54 pairs of CRC with and without synchronous liver metastasis. They demonstrated that there is a strong correlation between high CD133 expression and synchronous liver metastasis. However, the authors did not observe any effect, when knocking down CD133, on colon cancer cell line proliferation, migration, invasion and colony formation. Thus, they conclude that CD133 is a marker with high prognostic impact for CRC, even though it seems to have no relevant functional role as a determinant of tumor progression.
Taken together these data confirm the need to identify biomarkers for CRC-SCs in order to improve our understanding of the mechanisms underlying tumor growth and progression. In this regard, Barker et al[33] have shown that deletion of APC in Lgr5+ expressing cells leads to their transformation within days, suggesting that Lgr5 may mark not only normal intestinal stem cells, but also a limited population of CSCs. Simultaneously, using knock-in LacZ reporter mice within the Prominin-1 (Prom1) locus, Zhu et al[34] have shown that Prom1+ cells, located at the base of the crypts in the small intestine, co-express Lgr5, generate the entire intestinal epithelium and are susceptible to neoplastic transformation.
From a clinical point of view, a recent study showed that Lgr5 was markedly over-expressed in the majority of advanced CRCs compared with normal mucosal tissue[35]. As expected, in situ hybridization analysis confirmed the expression of Lgr5 in CBC cells in both small intestine and colon. This Lgr5 expression, which was variable among CRC cases, correlated significantly with lymphatic and vascular invasion, lymph node metastasis and tumor stage, suggesting the involvement of this marker in tumor progression.
A similar correlation has been described for the “stemness” gene Bmi1 by Du et al[36]. Real-time analysis of 98 samples of CRC showed that high expression of Bmi-1 is directly correlated with poor patient survival.
IMPLICATION OF CRC-SC MODEL FOR THERAPY
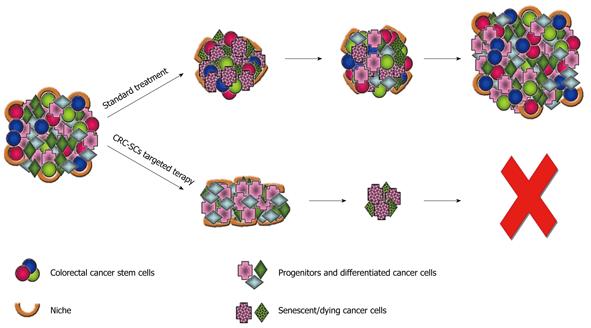
The CSC model has important implications for cancer therapy. Many current cancer therapies target the most rapidly dividing cells, which represent the majority of the tumor cell population. This can result in a remarkable but frequently transitory clinical remission. Failure of conventional treatment options to eliminate the CSC compartment might result in tumor relapse and, more importantly, in the proliferation of therapy-resistant and more aggressive tumor cells, which ultimately reduce patient survival (Figure 1). Several features of CSCs may make them hard to eliminate. CSCs are relatively quiescent and this allows them to escape from chemotherapeutic regimens that typically target actively cycling cells. Moreover, as shown for their normal counterpart, CSCs have been proposed to exhibit high level expression of multidrug transporter family genes, likely resulting in more efficient efflux of chemotherapeutic drugs and innate multidrug resistance[37]. In addition, signaling pathways that regulate self-renewal of normal colonic SC population, such as Wnt, Hedgehog or Notch, are dysregulated in CRC leading to tumor development. The development of an efficient therapeutic approach would therefore require the identification of distinctive molecular pathways active in CSCs and the identification of agents that can either block CSC proliferation or induce CSC differentiation, thus enhancing sensitivity to chemotherapeutic drugs.
Figure 1 Therapeutic implication of cancer stem cells.
The failure of current standard therapies in tumor eradication can be explained by assuming that colorectal cancer stem cells (CRC-SCs) are able to survive treatments leading to an only transitory clinical remission. Therapeutic strategies that specifically target the CRC-SC pool, by eliminating the self-renewing component of the tumor mass, could be more effective in eradicating the tumor and reducing the risk of relapse and metastasis.
Together with resistance to chemotherapy, CSCs are frequently resistant to standard radiotherapy regimens. In this respect, it has been recently demonstrated that resistance to radiation of CD133+ glioblastoma SCs can result from elevated expression of DNA damage response genes[38]. Radiotherapy for glioblastoma is associated with an increase in the proportion of the CD133+ fraction. Similarly, Dylla et al[39] showed that CRC-SCs are enriched in residual tumors following chemotherapy and remain capable of rapidly regenerating tumor from which they were derived. The authors have further demonstrated that resistance is mediated, at least in part, by ALDH1 enzyme activity.
Together with intrinsic factors, the microenvironment, or niche, may influence the ability of CSCs to proliferate, migrate or invade. The niche is an anchoring site for CSCs, and adhesion molecules or microenvironmental soluble molecules, including growth factors and cytokines, can significantly contribute to the refractoriness to therapy.
Todaro et al[40] have recently demonstrated that the upregulation of interleukin-4 (IL-4) in CD133+ CRC-SCs is an important mechanism that protects these tumorigenic cells from apoptosis. CD133+ CRC-SCs produce IL-4 as an autocrine growth factor promoting tumor resistance to chemotherapeutic agents such as 5-fluorouracil and oxaliplatin. Growth inhibition by these agents was significantly increased when cells were first treated with antibodies blocking the IL-4 signal. This phenomenon was confirmed in xenografts in which the administration of anti-IL-4 antibodies significantly reduced tumor growth after chemotherapy.
When human chemotherapy-resistant CRC cell lines (HT29/5FU-R and HT29/OxR) were developed following exposure of HT29 to increasing doses of 5-fluorouracil and oxaliplatin, there was a marked enrichment of CD133+ and CD44+ double positive cells. Phosphorylated and total insulin-like growth factor receptor (IGF-IR) levels were also increased in the resistant cell lines and their derived-tumors showed significantly greater growth inhibition in response to an IGF-IR mAb than parental cells, demonstrating that IGF-IR activation provided for enhanced sensitivity of CRC-SCs[41]. Other potential targets include specific targeting of symmetric stem cell division. To study new approaches to develop drugs that target CSCs, Boman et al[42] used computer modelling. They demonstrated that exponential increase in both SC and non-SC populations in CRC development involves an enhanced symmetric SC division. This finding suggests that any systemic therapy designed to effectively treat CRC and other cancers must act to control or eliminate symmetrical CSC division in tumors, while minimally affecting normal SCs[42].
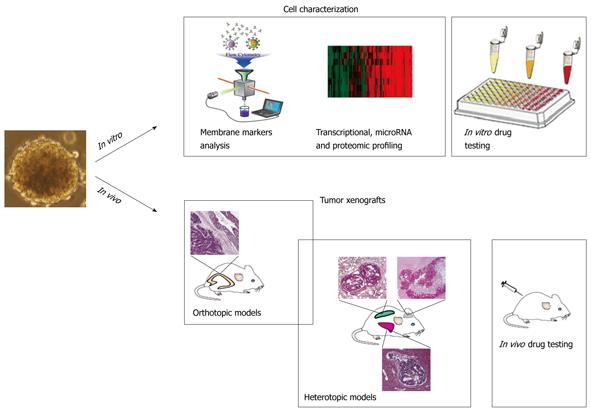
Another important aspect to be considered with regard to the designing of novel and more effective therapies against CRC is that CRC-SCs represent an excellent tool for the preclinical evaluation of new anticancer therapies both in vitro and in vivo, where they generate xenografts that phenocopy the human tumor of origin (Figure 2). Currently available mouse models of CRC are based on chemically-induced tumors, genetically engineered animals and tumor implants in immunocompromised mice, and none of them faithfully replicates all the aspects of human tumor development. Reliable mouse models of human CRC are essential to understand the mechanisms underlying tumor development and pathogenesis. In this respect CSCs represent an innovative and powerful tool in cancer research by conjugating the advantages of in vitro amplification, high grafting efficiency and, more importantly, the ability to closely reproduce the original tumors in terms of histology and drug sensitivity.
Figure 2 Colorectal cancer stem cells as a tool for drug discovery.
In vitro, colorectal cancer stem cells (CRC-SCs), isolated from the tumor specimen, are propagated as “tumor spheres”. Membrane marker analysis together with transcriptional, microRNA and proteomic profiling lead to the identification of molecular targets that are expressed by this cell population. These findings can be used to evaluate the cytotoxic ability of new compounds. In vivo, CRC-SCs can be orthotopically and heterotopically injected into immunocompromised mice generating tumors that mimic the cytoarchitecture of the parental tumors. The use of such mouse models of CRC allows for drug testing analyses in order to eradicate the primary tumor and avoid the formation of incurable metastases.
Thus, a systematic approach to identify and challenge the CSC survival machinery appears to be mandatory in order to develop novel and more efficient stem cell-based therapies. Genome-wide analyses of cancer have revealed the existence of a great genetic variation among individual tumors, which makes extremely complex the use of an exclusively genomic approach to cancer biology. At the same time, it is increasingly clear that tumors share common features in terms of protein pathway level, suggesting that a pathway-orientated perspective would represent the most effective approach to drug discovery and therapy. In a recent study, Fang et al[43] generated CD133+ tumor sphere cultures from several colon cancer specimens and performed mass-spectrometry-based quantitative proteomics in order to identify cell surface proteins enriched on culture tumor cells. These cells retain the expression of cell surface markers such as CD133, CD166, CD44 and EpCAM as well as other stem cell-associated proteins including nestin, Bmi1 and Msi-1, thus confirming the value of this in vitro model for biological analysis of CSC populations as well as for drug screening experiments. Therefore, integrated strategies based on high-throughput proteomic, drug screening and gene expression-based approaches will allow in the near future the identification and targeting of survival signaling pathways in CSCs.
CONCLUSION
Increasing evidence shows that CSCs may play a critical role in tumor development and progression. CSC resistance to conventional therapies may explain why it is difficult to completely eradicate cancer and why recurrence is often inevitable. Thus, the identification and molecular characterization of CSCs is critical to develop therapeutic strategies that specifically target this rare population of cells and that are likely to be effective in eradicating tumors and in reducing the risk of relapse and metastasis.
Peer reviewers: Yu-Yuan Li, Professor, Department of Gastroenterology, First Municipal People’s Hospital of Guangzhou, 1 Panfu Road, Guangzhou 510180, Guangdong Province, China; Wendy Wilhelmina Johanna de Leng, PhD, Department of Pathology, Internal address H04.312, Heidelberglaan 100, Postbox 85500, Utrecht, 3508 GA, The Netherlands
S- Editor Wang JL L- Editor Logan S E- Editor Ma WH