Published online Jan 21, 2010. doi: 10.3748/wjg.v16.i3.320
Revised: November 11, 2009
Accepted: November 18, 2009
Published online: January 21, 2010
AIM: To evaluate how widely Helicobacter pylori (H. pylori) HopE and HopV porins are expressed among Chilean isolates and how seroprevalent they are among infected patients in Chile.
METHODS: H. pylori hopE and hopV genes derived from strain CHCTX-1 were cloned by polymerase chain reaction (PCR), sequenced and expressed in Escherichia coli AD494 (DE3). Gel-purified porins were used to prepare polyclonal antibodies. The presence of both genes was tested by PCR in a collection of H. pylori clinical isolates and their expression was detected in lysates by immunoblotting. Immune responses against HopE, HopV and other H. pylori antigens in sera from infected and non-infected patients were tested by Western blotting using these sera as first antibody on recombinant H. pylori antigens.
RESULTS: PCR and Western blotting assays revealed that 60 and 82 out of 130 Chilean isolates carried hopE and hopV genes, respectively, but only 16 and 9, respectively, expressed these porins. IgG serum immunoreactivity evaluation of 69 H. pylori-infected patients revealed that HopE and HopV were infrequently recognized (8.7% and 10.1% respectively) compared to H. pylori VacA (68.1%) and CagA (59.5%) antigens. Similar values were detected for IgA serum immunoreactivity against HopE (11.6%) and HopV (10.5%) although lower values for VacA (42%) and CagA (17.4%) were obtained when compared to the IgG response.
CONCLUSION: A scarce expression of HopE and HopV among Chilean isolates was found, in agreement with the infrequent seroconversion against these antigens when tested in infected Chilean patients.
-
Citation: Lienlaf M, Morales JP, Díaz MI, Díaz R, Bruce E, Siegel F, León G, Harris PR, Venegas A.
Helicobacter pylori HopE and HopV porins present scarce expression among clinical isolates. World J Gastroenterol 2010; 16(3): 320-329 - URL: https://www.wjgnet.com/1007-9327/full/v16/i3/320.htm
- DOI: https://dx.doi.org/10.3748/wjg.v16.i3.320
Helicobacter pylori (H. pylori) are Gram-negative, microaerophilic, spiral-shaped bacteria isolated from human gastric biopsies in 1983[1]. In order to survive in this aggressive environment, H. pylori are able to neutralize their close surrounding space by production of urease, which catalyzes the conversion of urea into ammonium and CO2, raising pH close to neutral. In addition, to colonize the epithelium, this bacterium is able to bind to the epithelial cell surface, partially avoiding its removal by natural peristalsis or mucus renewal. These characteristics allow H. pylori to persist for decades.
H. pylori infection affects one half of the world population, roughly 73% in Chile[2], with higher prevalence as age increases. After several years of chronic gastric infection, approximately 10%-15% of infected patients develop severe gastrointestinal diseases such as chronic gastritis, peptic ulcer and gastric carcinoma[3,4]. In Chile, 5% of the infected population develops gastric cancer[2] and this malignancy is the second cause of death by cancer in the country.
H. pylori carries various virulence factors, and some may have potential as vaccine antigens. These factors may be grouped as: (1) colonization factors, which allow bacterial residence; (2) persistence factors which enable bacteria to accomplish an effective and lasting survival; and (3) disease inducing factors which cause adverse pathological effects on the gastric mucosa[5].
Based on a bioinformatics analysis of the H. pylori genome, a family of outer membrane proteins (OMPs) composed of 33 members has been identified[6]. These proteins are assembled into the outer membrane exposing, on the bacterial surface, small peptide loops which may act as epitopes to induce an immune response. This feature may be useful when selecting appropriate antigens for vaccine design. All these members contain an N-terminal signal peptide (processed by signal peptidase type I or II) that allows these proteins to cross the inner membrane on their way towards the outer membrane. The H. pylori OMPs form 2 families: the Hop members (21 proteins) and the Hor members (12 proteins). Hor proteins lack a characteristic N-terminal Hop motif[7]. Hop proteins have structural homology with the Escherichia coli (E. coli) outer membrane protein F (OmpF) porin[8]. Currently, 5 H. pylori Hop members (HopA, HopB, HopC, HopD and HopE) from strain 26 695 have been characterized as porins using planar bilayer techniques[7,9] and some also behave as adhesins[10,11]. These properties make them attractive candidates as vaccine antigens. In fact, other bacterial porins from Salmonella, Pseudomonas, Chlamydia and Neisseria, have been found to be strong immunogens[12-15]. However, in the case of H. pylori, it has been suggested that not all the genes encoding OMPs may be functional at a given time. Some of these genes are under a control mechanism that operates by strand slippage during DNA replication or DNA repair. DNA polymerase slippage may easily add or remove nucleotides when DNA synthesis occurs in front of a homopolymeric tract or dinucleotide repeats at the template strand (i.e. polyG or polyCA gene segments) causing mutations either at the promoter or at the coding region. This type of mutation may turn off or on some hop genes that may include these polynucleotide features. For instance, the hopC gene has been reported to carry a polyT tract (13 Ts in length) near the 5’ end but hopA, hopB and hopE do not have such long polyT tracts either at their coding regions or 5’ upstream at the promoter regions. Gene switching will produce a change in the antigenic bacterial surface, a strategy that will distract the host immune system. For this reason, whether any H. pylori OMP would be considered as a vaccine antigen, omp genes containing long homopolymeric tracts or dinucleotide repeats should be avoided.
Regarding HopV and HopW, genetic heterogeneity in orthologous members of the Hop family among H. pylori strains has been described[16]. These new members were defined as part of the HopA/HopE family, because of their homology at the N-terminal sequence and the presence of 7 homologous domains in the C-terminus region. Regarding functional aspects, HopV and HopW have pore sizes similar to that of the E. coli OmpF porin[16] and HopE has been defined as the homolog to the E. coli OmpF porin[17].
Since the use of porins as antigens has been reported as successful[12-15], we decided to evaluate members of the H. pylori Hop family as putative antigen candidates for vaccine development by determining how widely they are expressed among Chilean H. pylori isolates and how often Chilean patients develop antibodies against them. A brief bioinformatic survey indicated that some genes of the Hop family had homopolymeric tracts or dinucleotide repeats in their coding sequences and promoter regions, with potential capability to promote strand slippage which may affect stability of gene expression[18]. Considering this aspect, only porin genes having single homopolymeric tracts or dinucleotide repeats no longer than 6 bases in their coding sequences were chosen as source of putative useful antigens for a vaccine. For this reason, among several OMP genes, only hopE and hopV sequences were selected for the present study.
E. coli DH5α was used for polymerase chain reaction (PCR) cloning, and E. coli BL21 (DE3), JM109 (DE3) and AD494(DE3)pLysS as host for porin expression assays. For cloning of PCR fragments plasmid pGEM-T Easy from Promega was used. For expression studies pET21a and pET21d (Novagen) were selected. H. pylori Chilean strain CHCTX-1 was used as DNA source for gene amplifications[19]. A collection of 130 H. pylori strains isolated from infected patients living in different Chilean cities was already available[20].
E. coli cells were grown overnight in LB media at 37°C with shaking. H. pylori strains were grown under 50 mL/L CO2 and 80% humidity in Brucella agar plates enriched with 5% horse blood cells and grown at 37°C for 2-3 d. E. coli strains were kept for short periods in LB plates at 4°C. Bacterial strains containing 14% glycerol were stored frozen for longer periods at -70°C.
Plasmids were usually detected by the “one step” method[21], and purified by alkaline lysis method[22]. Restriction digestions, DNA ligations and plasmid dephosphorylations were done according to standard procedures[22]. Electroporation in 0.2 cm electrode separation cuvettes was performed as previously described[23], in a Gene Pulser™ apparatus. Electrocompetent cells were prepared according to described protocols[23] with a yield of 1 × 109 to 1 × 1010 cells/mL.
Primers corresponding to the 5’ and 3’ ends as well as the internal sequences of the hopE and hopV genes (Table 1) were designed based on H. pylori 26 695 GenBank sequences. As templates, chromosomal DNA from the CHCTX-1 strain[19] and from clinical isolates was prepared according to described procedures[24]. PCR reactions were carried out in a BioRad Mastercycler II thermocycler, with Pfu polymerase (Stratagene, CA, USA) or Taq polymerase (Promega, Madison WI, USA). Assays were done in 25 μL final volume following the manufacturer’s instructions. Gene amplicons were detected by 1% agarose gel electrophoresis. Other conditions were as previously described[25]. VacA alleles were determined by using primers and assay conditions described by Atherton et al[26].
| Name | 5'→3' sequence | Restriction sites1 |
| HopV12 | GGGCCCATATGCTCAATTTTATGACAAAGAAGAAAAATAGAATGC | NdeI |
| HopV22 | GGATCCCATGGTTAAAAATCCCTCAAGTAACTGATTTG | BamHI |
| HopE12 | GGCGCCATGGAATTTATGAAAAAGTTTGTAGCTTTAGG | NcoI |
| HopE22 | CGCGAAGCTTTTAAAAAGTGTAGTTATACCCTAAATAAAG | HindIII |
| HopE113 | GCAAGTGGTTTGGTTTTAGAG | - |
| HopE223 | ACCATATCCAACTGGATTTT | - |
| HopV113 | GGCGTGGGGTTAGATACGCTG | - |
| HopV223 | ACCATGTTTTCTTTATTCAC | - |
| HopVint3 | ATGCGTTATTATGGGTTTTTTGACT | - |
| pETT7d4 | TAATACGACTCACTATAGGG | - |
| pETT7r4 | GCTAGTTATTGCTCAGCGG | - |
According to standard procedures[27], anti-HopE and anti-HopV rabbit antibodies were prepared. Proteins were obtained from E. coli clones expressing the H. pylori porins after separation by SDS-PAGE and purification from gel slices by electroelution as previously described[28]. Pathogen-free New Zealand adult female rabbits (approximately 1400 g in weight) were immunized with 250 μg of each porin dissolved in 2 mL of Tris-glycine buffer mixed (1:1) with complete Freund’s adjuvant. Two animals were used for each porin inoculation and 100 μL aliquots were applied subcutaneously in the back. This was followed by 3 boosters every 15 d.
Lysates from clones expressing HopE and HopV were separated by polyacrylamide gels (12% or 15%) and run in minichambers according to Laemmli[29]. Western blotting were done as previously described[30]. As first antibody, patient serum (1:100 dilution) or rabbit anti-porin antibodies against HopE or HopV (1:1000 dilution) were used. As a second antibody for patient assays, goat anti-human IgA or anti-human IgG conjugated to peroxidase, were incubated (1:1000) overnight at 4°C. For anti-porin rabbit sera, a goat anti-rabbit peroxidase-conjugated antibody (1:1000) second antibody was used. To reduce cross reactions, rabbit antisera and human antisera were adsorbed with sonicated lysates from E. coli AD494(DE3)pLysS/pET21d and BL21 (DE3), respectively. Human sera immunoblotting were done with lysates expressing HopE or HopV porins and products derived from expression of cagA and vacA gene fragments cloned from strain CHCTX-1[19].
A sera panel from 69 infected patients (63 with gastritis, 6 with ulcers) recruited from the Universidad Católica de Chile Medical Center in Santiago, with signed consent, was available. Each patient’s infected condition was defined by endoscopy, positive urease rapid test and detection of hematoxylin/eosin-stained curved bacteria on gastric tissue biopsies. Also, 8 non-infected patients were included in this study. The local ethics committee approved the protocol for this study.
In order to obtain a cleaner IgA reaction in Western blotting assays using patient serum antibodies, protein G-plus-Agarose (Santa Cruz Biotechnology, catalogue #sc-2002) was utilized to first remove IgG from serum by immunoprecipitation. One hundred microliters of each serum without pre-adsorption treatment were incubated overnight with 200 μL of protein G-plus-agarose at 4°C with mild shaking. After sedimentation for 5 min at 2500 r/min and 4°C, the supernatant of each sample was used as a source of IgG-free serum.
DNA samples were previously purified by a commercial kit, and sequenced at our Sequencing Core Facility. T7 and internal primers for DNA sequencing are listed in Table 1.
Selection for cloning and expression studies of the hopE and hopV genes were based on known gene sequences from strain H. pylori 26 695. Since our study was focused on antigens obtained from local strains, H. pylori CHCTX-1 strain, a clinical isolate obtained from a Chilean patient[19] was selected as the DNA source for gene cloning in this study.
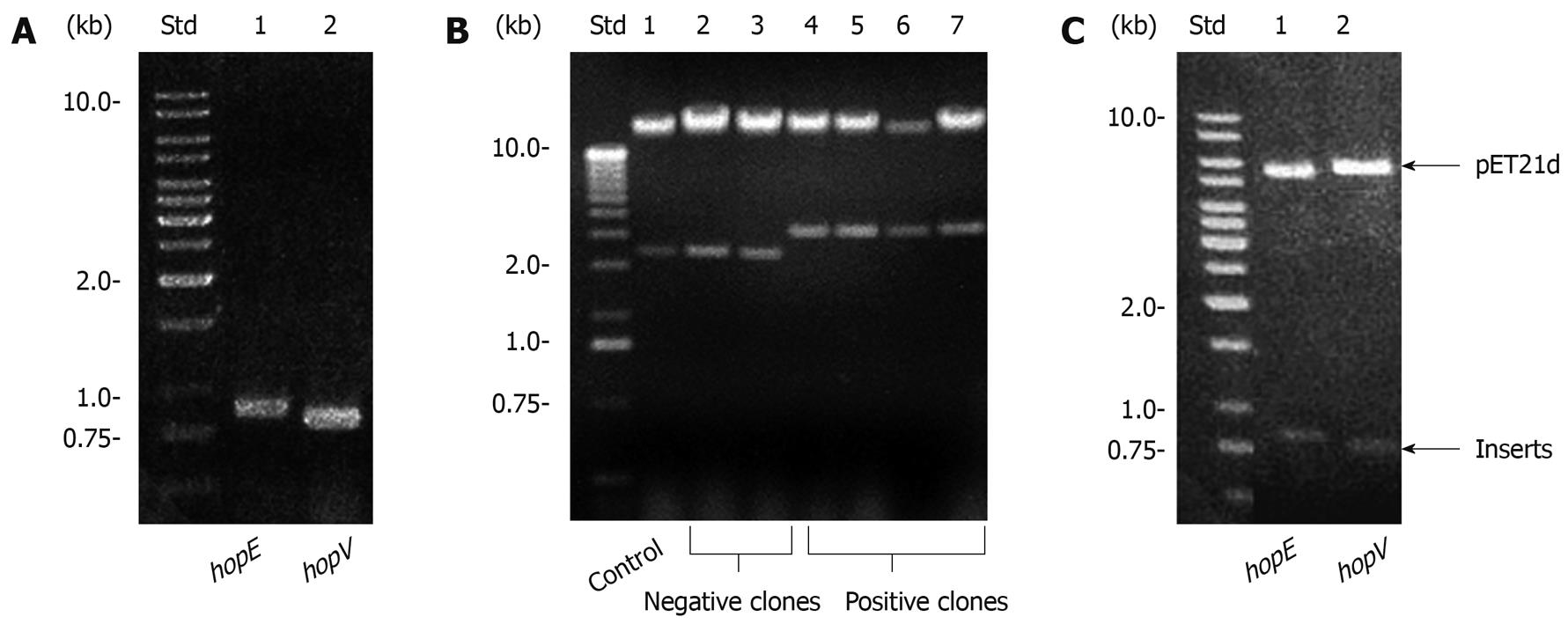
Cloning of HopE and HopV porin genes, including their signal peptide regions, was done by PCR. Primers and assay conditions are described in Table 1 and Methods, respectively. Amplicons from hopE, and hopV genes were separated in a 1% agarose gel (Figure 1A), purified and treated with Taq polymerase and dATP to be ligated to pGEM-T. Recombinant plasmids were detected by insert release after EcoRI digestion and separation in 1% agarose gel electrophoresis. The hopE and hopV cloned inserts were subjected to NcoI-HindIII and NdeI-BamHI double digestions and ligated to pET21d and pET21a, respectively. As expected, fragments with sizes corresponding to these genes were observed. For expression purposes, plasmids were transferred to E. coli AD494(DE3)pLysS cells and visualized by the “one step method”[21]. Some clones containing plasmids with the hopE gene are displayed in Figure 1B. Purified plasmids were used for restriction digestions and also as DNA templates for PCR gene detection. NcoI-HindIII and NdeI-BamHI double digestions of plasmid DNA isolated from single clones were analyzed by agarose gel electrophoresis, and released inserts with sizes close to the expected ones (819 bp for hopE and 735 bp for hopV) were observed (Figure 1C).
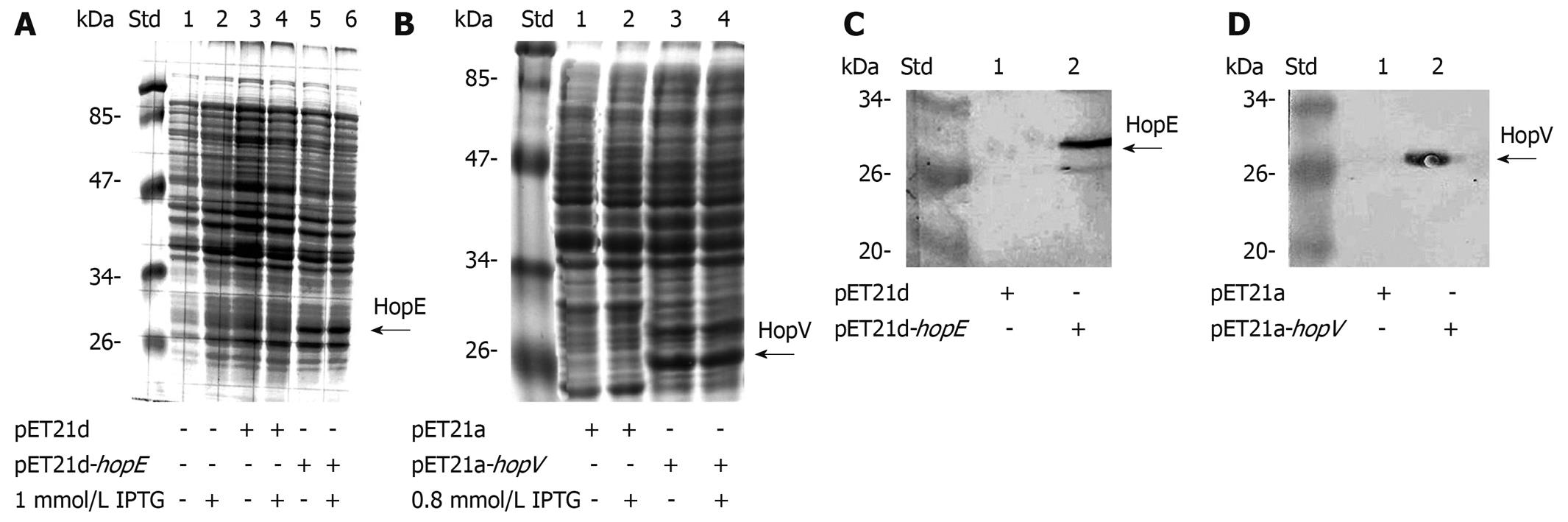
E. coli AD494(DE3)pLysS was able to express detectable amounts of HopE and HopV porins, as seen after SDS-PAGE and Coomassie blue staining (Figure 2A and B) and Western blotting assays (Figure 2C and D).
Expression conditions were optimized by 5 h induction with 1 mmol/L isopropyl β-D-thiogalactoside (IPTG) for hopE and 0.8 mmol/L for hopV on previously saturated cultures. Protein sizes of 32 kDa for HopE and 28 kDa for HopV were observed. These porins displayed a certain amount of expression without IPTG induction, partially explained by the fact that the induction procedure was done on saturated cultures.
The hopE (clone 1) and hopV (clone 13) gene sequences from the CHCTX-1 strain were obtained using external (T7 promoter and T7 terminator) as well as internal primers (Table 1) as described in Methods. Both hopE and hopV sequences were deposited at GenBank (accession numbers #EF635415 and #EF635416, respectively). As expected, these genes did not contain nonsense or frameshift mutations at their coding regions. Also, neither homopolymeric nor dinucleotide tracts longer than 6 nucleotides were found.
From a collection of 240 clinical strains previously isolated[20], we selected 130 colonies (1 to 5 isolates per patient) as representatives from 6 Chilean cities: Iquique (IQ) in the North, Valparaiso (VA) and Santiago (SA) in the central region, Los Angeles (LA) and Valdivia (VL) in the South, and Punta Arenas (PA), the Southernmost city, to evaluate the distribution of strains carrying hopE and hopV genes and their expression throughout the country.
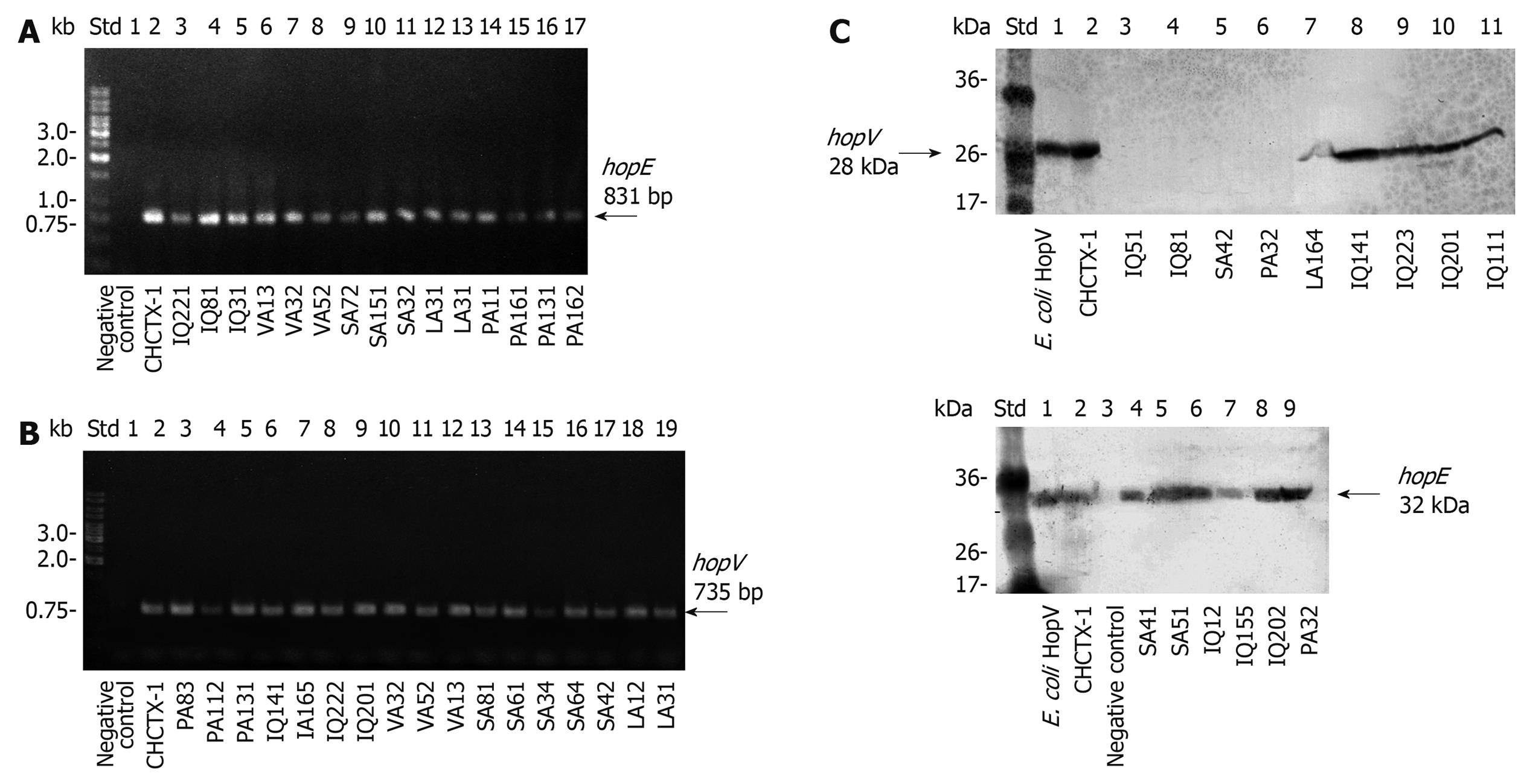
Detection of the genes was done by standard PCR. The amplicons were almost identical in size to those expected for hopE and hopV genes from strain 26 695. Representative groups of isolates carrying hopE and hopV genes are shown in Figure 3A and B, respectively.
The hopE and hopV genes were detected in 46.9% (61 out of 130 strains) and 63.1% (82 out of 130) of the studied strains, respectively, and 40% (52 out of 130) of the strains revealed the presence of both genes simultaneously (Table 2). Among different cities, hopE and hopV gene contents varied between 30% and 69%. Curiously, hopV was frequently detected (69.2%) in strains from infected patients living in PA, the southernmost city. Patients from VL (mostly descendants from ancient aborigines) carried strains with a lower content (42.8%) of hopV gene. All PCR reactions were done at least twice using a pair of primers which bound to the gene ends. For those cases with negative amplification, additional assays using 2 primer combinations, including in each pair of primers one of the internal primers (Table 1), were performed. In most cases negative PCR reactions were confirmed and just a few strains showed positive PCR amplification only with pairs including internal primers, indicating that our initial estimation about the reduced presence of these genes in Chilean isolates was valid.
The positive results of HopE and HopV Western blotting expression assays in these isolates revealed no protein size variation, and selected results are displayed in Figure 3C.
Regarding porin expression, results showed that only 13.1% of the 130 strains expressed HopE and 6.9% expressed HopV. Altogether, 83.8% (109 out of 130 strains) did not express HopE or HopV porins either because of a lack of these genes, random inactivating gene mutations or gene silencing by the strand slippage mechanism (Table 2).
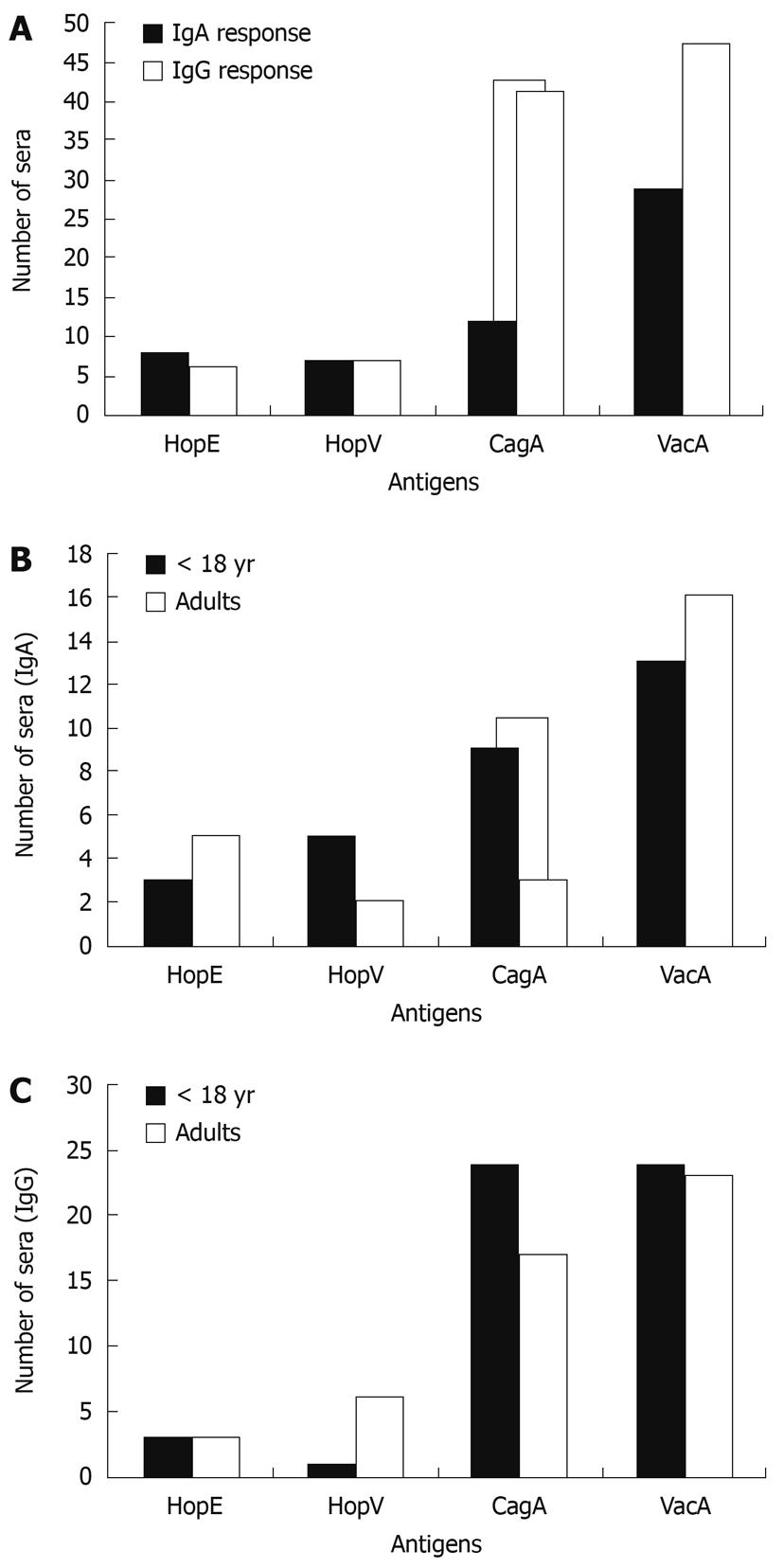
In order to evaluate the capability of sera from Chilean H. pylori-infected patients to recognize recombinant H. pylori HopE and HopV porins, sera from 69 infected and 8 non-infected patients were tested. IgG and IgA serum antibodies against HopE and HopV antigens expressed as recombinant proteins in E. coli clones were tested using Western blotting assays on these bacterial lysates. VacA and CagA expressed similarly were used as immunodominant controls. The number of infected patient sera able to recognize these antigens are shown in Figure 4A. It was found that, as expected, IgG human antibodies more frequently recognized VacA (68.1% or 47 out of 69) and CagA (59.4%), but rarely recognized HopE (8.7%) and HopV (10.1%) porins. A similar distribution for HopE (11.6%) and HopV (10.5%), but lower distribution values for VacA (42%) and CagA (17.4%) were found for IgA antibodies. The lower number of anti-HopE and anti-HopV reactive sera can be explained by the low proportion of H. pylori strains able to express these genes, being 13.1% (17/130) for HopE and 6.9% (9/130) for HopV (Table 2). Taken together, these results strongly suggest that H. pylori possesses a mechanism to switch on/off these OMP genes as a strategy to evade the host immune response.
In addition, considering that the immune response in children[31,32] could be quite different from that in adults[33], the IgA (Figure 4B) and IgG (Figure 4C) immune responses of the infected patients were plotted for 2 age groups: under 18 years of age and adults. It was noted that the number of sera with IgA and IgG responses against CagA antigen was significantly higher in patients under 18 than in adults. In contrast, IgA responses against HopE and VacA and the IgG response against HopV seemed to be more frequent in adults than in children.
H. pylori colonizes the human gastric epithelium in half of the world’s population and induces strong serological and inflammatory responses in the host which persist during the entire life of the subject, rendering the host unable to eradicate infection. Knowledge of the most frequently recognized antigens in the infected population may contribute to an understanding of the bacterium survival strategy. In addition, this could also help to select appropriate antigens for vaccine design. The most extensively studied H. pylori virulence factors as potential vaccine antigens are urease subunits[34,35], VacA and CagA[36], H. pylori adhesin A[37] and neutrophil-activating protein[38,39]. However, results indicating reduction in colonization after oral vaccination of human subjects have been rather modest[40,41]. As new antigens are needed, and there are few studies comparing porin genes among different H. pylori strains, we have looked for H. pylori porins suitable for vaccine development. The present report provides new information, particularly about Chilean strains, regarding porin expression among clinical isolates.
Folded porins have exposed loops at the bacterial surface which may induce a strong immune response. Porins have been proposed for the design of oral vaccines to eradicate H. pylori[6]. This pathogen has an extensive collection of OMPs with defined pore characteristics, such as the Hop group. However, some may display a unique on/off mechanism affecting gene expression, based on DNA strand slippage during replication. The instability of expression status makes antigen selection a very important issue. Antigens that will not be permanently expressed in most of the strains should be avoided, since they will provide a limited protection.
PCR assays carried out on 130 Chilean isolates detected hopE and hopV genes in 46.9% and 63.1%, respectively. However porin expression was infrequently detected in these strains (HopE = 13.1%, HopV = 6.9%), concurring with low seroprevalence of these porins among sera of infected patients, suggesting that these genes may be under DNA strand slippage control.
Infrequent detection of expression cannot be explained by low immunoreactivity of the rabbit antisera resulting from amino acid sequence diversity among strains, since these 2 porin sequences seem to be conserved. HopE and HopV amino acid sequence identities among those from strain CHCTX-1 and the corresponding sequences from fully sequenced H. pylori genomes, were in the range of 98%-99% and 94%-96%, respectively.
As a complementary approach, 69 patient sera were tested by Western blotting on E. coli clones expressing CagA, VacA, HopE or HopV. It was revealed that HopE and HopV porins were not often recognized within the analyzed sample. Only 6 sera (8.7%) showed IgG-type immune reaction against HopE-containing lysates and 7 (10.1%) against HopV. Similar results were obtained analyzing the IgA response. These results agree with the fact that HopE and HopV porins are sporadically expressed. In contrast, CagA (its gene is present in about 50% of the strains) and VacA (its gene is present in almost 100% of the strains) reacted with 59.4% and 68.1% of the IgG patient sera, respectively.
Regarding nucleotide sequence features, dinucleotide repeats in hopE and hopV sequences from the CHCTX-1 strain barely reached 5 nucleotides in length. However, they contained CCCCCC and TTTTTT tracts after codons 58 and 66, respectively. These findings, taken together with the low number of strains expressing these porins, and their low seroprevalence among Chilean patients, suggest that hopE and hopV may be under strand slippage gene control. Confirmation should be done by sequencing strains carrying silenced genes.
In H. pylori, at least 3 porin genes from the Hop family (hopZ, hopP and hopO) may be subjected to this on/off switching[11,42]. Another study[43] showed a similar case: 3 different H. pylori strains re-isolated after Maccacus rhesus infection lost expression of BabA adhesin which binds Lewis b antigen. These observations support the idea that H. pylori can modulate expression of some OMP genes. This feature provides an adaptive mechanism to avoid induction of a strong host immune response. This is supported by the large repertory of OMPs genes in the H. pylori genome. Functional redundancy of porins may explain emergence of mutations in these genes without affecting bacteria viability. It has been proposed that the role of such redundancy of outer membrane proteins is to sustain antigenic variation to support pathogen survival by evasion of the host immune response[44]. The strand slippage mechanism is not normally operating in E. coli, therefore, in most cases, lack of heterologous expression of H. pylori genes in E. coli should be due to mutations that previously affected the H. pylori genome.
In spite of the low content of homopolymeric and dinucleotide repeats found in CHCTX-1 hopE and hopV genes, some strains may have switched these genes off but, in a few cases, expression could be restored by the same mechanism. This may restrict the use of these H. pylori OMPs as a single source of antigens for vaccine design. However, in order to provide a wider and stronger immune response, vaccines based on a mixture of H. pylori antigens with the inclusion of HopE and HopV should be considered.
Few studies have been done on HopE and HopV porins from Helicobacter pylori (H. pylori). These proteins have been described as part of a large family of outer membrane proteins having similar functions, mainly as gating for influx of nutrients. Compared to other enterobacteria, such as Escherichia coli (E. coli) with only 3 major porins, H. pylori have shown a remarkable redundancy in this kind of outer membrane protein. The explanation for such redundancy has not arisen. A study looking at how widely these porin genes are distributed and what proportion is actually active among clinical isolates should provide some answers.
Switching on/off in outer membrane genes in a few bacteria has been described as a mechanism to distract the immune system during infection by changing the proteins displayed on the surface. The authors found that H. pylori HopE and HopV porin genes seemed to be absent in some isolates, and about half of those who carried them did not express them. In addition, sera from infected patients do not frequently recognize these antigens. This feature may contribute to the ability of these bacteria to avoid the host immune response allowing their persistence in humans for an extended period of time.
Recent reports have shown that some outer membrane protein genes from H. pylori could be turned on/off by random nucleotide insertions or deletions either at the promoter or within the coding region, through a mechanism called “strand slippage” during DNA replication. This is the first report proposing that this switching may also occur in the hopE and hopV genes, explaining why around 70%-90% of these genes are shut down in Chilean clinical isolates.
Determining whether a protein is subjected to on/off switching during its expression at the bacterial surface, together with the knowledge of its immunoreactivity will be useful to select potential antigen candidates to be used in the design and development of vaccines.
H. pylori HopE and HopV proteins are part of a large family of outer membrane proteins and are located at the bacterial surface. They are defined as porins because they form a pore structure to allow the influx of small size nutrients and other compounds. They may constitute a target for the immune system. The “strand slippage” mechanism to control gene expression is a result of random errors during strand DNA replication consisting of nucleotide insertions or deletions that alter the genetic code of the protein or the functionality of elements (i.e. promoter) that control gene expression.
The manuscript by Lienlaf et al assesses the patterns of 2 porin genes in H. pylori. H. pylori remains a significant problem in developing countries around the world. Although much is now known about the pathogenesis of this bacterium and about host responses to infection, the organism remains a clinical problem. Various investigators have focused upon the establishment of a vaccine for this pathogen. An appropriate selection of a conserved and widely expressed antigen of H. pylori clinical isolates will assure a suitable design of a protective vaccine. This work is promising.
| 1. | Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;1:1311-1315. |
| 2. | Ferreccio C, Rollán A, Harris PR, Serrano C, Gederlini A, Margozzini P, Gonzalez C, Aguilera X, Venegas A, Jara A. Gastric cancer is related to early Helicobacter pylori infection in a high-prevalence country. Cancer Epidemiol Biomarkers Prev. 2007;16:662-667. |
| 3. | Sepulveda AR, Graham DY. Role of Helicobacter pylori in gastric carcinogenesis. Gastroenterol Clin North Am. 2002;31:517-535, x. |
| 4. | Suerbaum S, Josenhans C. Helicobacter pylori evolution and phenotypic diversification in a changing host. Nat Rev Microbiol. 2007;5:441-452. |
| 5. | Peek RM Jr, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer. 2002;2:28-37. |
| 6. | Alm RA, Bina J, Andrews BM, Doig P, Hancock RE, Trust TJ. Comparative genomics of Helicobacter pylori: analysis of the outer membrane protein families. Infect Immun. 2000;68:4155-4168. |
| 7. | Exner MM, Doig P, Trust TJ, Hancock RE. Isolation and characterization of a family of porin proteins from Helicobacter pylori. Infect Immun. 1995;63:1567-1572. |
| 8. | Bond PJ, Sansom MS. The simulation approach to bacterial outer membrane proteins. Mol Membr Biol. 2004;21:151-161. |
| 9. | Doig P, Exner MM, Hancock RE, Trust TJ. Isolation and characterization of a conserved porin protein from Helicobacter pylori. J Bacteriol. 1995;177:5447-5452. |
| 10. | Ilver D, Arnqvist A, Ogren J, Frick IM, Kersulyte D, Incecik ET, Berg DE, Covacci A, Engstrand L, Borén T. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science. 1998;279:373-377. |
| 11. | Peck B, Ortkamp M, Diehl KD, Hundt E, Knapp B. Conservation, localization and expression of HopZ, a protein involved in adhesion of Helicobacter pylori. Nucleic Acids Res. 1999;27:3325-3333. |
| 12. | Secundino I, López-Macías C, Cervantes-Barragán L, Gil-Cruz C, Ríos-Sarabia N, Pastelin-Palacios R, Villasis-Keever MA, Becker I, Puente JL, Calva E. Salmonella porins induce a sustained, lifelong specific bactericidal antibody memory response. Immunology. 2006;117:59-70. |
| 13. | Worgall S, Krause A, Rivara M, Hee KK, Vintayen EV, Hackett NR, Roelvink PW, Bruder JT, Wickham TJ, Kovesdi I. Protection against P. aeruginosa with an adenovirus vector containing an OprF epitope in the capsid. J Clin Invest. 2005;115:1281-1289. |
| 14. | Holland MJ, Faal N, Sarr I, Joof H, Laye M, Cameron E, Pemberton-Pigott F, Dockrell HM, Bailey RL, Mabey DC. The frequency of Chlamydia trachomatis major outer membrane protein-specific CD8+ T lymphocytes in active trachoma is associated with current ocular infection. Infect Immun. 2006;74:1565-1572. |
| 15. | Toropainen M, Saarinen L, Vidarsson G, Käyhty H. Protection by meningococcal outer membrane protein PorA-specific antibodies and a serogroup B capsular polysaccharide-specific antibody in complement-sufficient and C6-deficient infant rats. Infect Immun. 2006;74:2803-2808. |
| 16. | Peck B, Ortkamp M, Nau U, Niederweis M, Hundt E, Knapp B. Characterization of four members of a multigene family encoding outer membrane proteins of Helicobacter pylori and their potential for vaccination. Microbes Infect. 2001;3:171-179. |
| 17. | Bina J, Bains M, Hancock RE. Functional expression in Escherichia coli and membrane topology of porin HopE, a member of a large family of conserved proteins in Helicobacter pylori. J Bacteriol. 2000;182:2370-2375. |
| 18. | Salaün L, Linz B, Suerbaum S, Saunders NJ. The diversity within an expanded and redefined repertoire of phase-variable genes in Helicobacter pylori. Microbiology. 2004;150:817-830. |
| 19. | Müller I, Medina-Selby A, Palacios JL, Martinez P, Opazo P, Bruce E, Mancilla M, Valenzuela P, Yudelevich A, Venegas A. Cloning and comparison of ten gene sequences of a Chilean H. pylori strain with other H. pylori strains revealed higher variability for VacA and CagA virulence factors. Biol Res. 2002;35:67-84. |
| 20. | Diaz MI, Valdivia A, Martinez P, Palacios JL, Harris P, Novales J, Garrido E, Valderrama D, Shilling C, Kirberg A. Helicobacter pylori vacA s1a and s1b alleles from clinical isolates from different regions of Chile show a distinct geographic distribution. World J Gastroenterol. 2005;11:6366-6372. |
| 21. | Beuken E, Vink C, Bruggeman CA. One-step procedure for screening recombinant plasmids by size. Biotechniques. 1998;24:748-750. |
| 22. | Sambrook J, Fritsch E, Maniatis T. Molecular Cloning. A laboratory manual. 2nd ed. Cold Spring Harbor: Cold Spring Harbor Laboratory Press 1989; . |
| 23. | Miller JF. Bacterial transformation by electroporation. Methods Enzymol. 1994;235:375-385. |
| 24. | Owen R, Bickley J. Isolation of H. pylori genomic DNA and restriction analysis. Helicobacter pylori protocols. Methods in Molecular Medicine. Totowa, New Jersey: Humana Press 1997; 81-88. |
| 25. | Serrano C, Diaz MI, Valdivia A, Godoy A, Peña A, Rollan A, Kirberg A, Hebel E, Fierro J, Klapp G. Relationship between Helicobacter pylori virulence factors and regulatory cytokines as predictors of clinical outcome. Microbes Infect. 2007;9:428-434. |
| 26. | Atherton JC, Cao P, Peek RM Jr, Tummuru MK, Blaser MJ, Cover TL. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771-17777. |
| 27. | Cooper HM, Paterson Y. Production of Antibodies. Current protocols in immunology. New York, NY: John Wiley and Sons Inc 1995; 2.4.1-2.4.9. |
| 28. | Every D, Green RS. Purification of individual proteinase isozymes from Bacteroides nodosus by use of polyacrylamide gel electrophoresis, a fluorogenic substrate detection system, and a simple electroelution apparatus. Anal Biochem. 1982;119:82-85. |
| 29. | Laemmli UK, Beguin F, Gujer-Kellenberger G. A factor preventing the major head protein of bacteriophage T4 from random aggregation. J Mol Biol. 1970;47:69-85. |
| 30. | Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350-4354. |
| 31. | Campbell DI, Pearce MS, Parker L, Thomas JE. IgG subclass responses in childhood Helicobacter pylori duodenal ulcer: evidence of T-helper cell type 2 responses. Helicobacter. 2004;9:289-292. |
| 32. | Campbell DI, Pearce MS, Parker L, Thomas JE, Sullivan PB, Dale A. Immunoglobulin G subclass responses to Helicobacter pylori vary with age in populations with different levels of risk of gastric carcinoma. Clin Diagn Lab Immunol. 2004;11:631-633. |
| 33. | D’Elios MM, Manghetti M, De Carli M, Costa F, Baldari CT, Burroni D, Telford JL, Romagnani S, Del Prete G. T helper 1 effector cells specific for Helicobacter pylori in the gastric antrum of patients with peptic ulcer disease. J Immunol. 1997;158:962-967. |
| 34. | Kreiss C, Buclin T, Cosma M, Corthésy-Theulaz I, Michetti P. Safety of oral immunisation with recombinant urease in patients with Helicobacter pylori infection. Lancet. 1996;347:1630-1631. |
| 35. | Michetti P, Kreiss C, Kotloff KL, Porta N, Blanco JL, Bachmann D, Herranz M, Saldinger PF, Corthésy-Theulaz I, Losonsky G. Oral immunization with urease and Escherichia coli heat-labile enterotoxin is safe and immunogenic in Helicobacter pylori-infected adults. Gastroenterology. 1999;116:804-812. |
| 36. | Yan J, Mao YF, Shao ZX. Frequencies of the expression of main protein antigens from Helicobacter pylori isolates and production of specific serum antibodies in infected patients. World J Gastroenterol. 2005;11:421-425. |
| 37. | Nyström J, Svennerholm AM. Oral immunization with HpaA affords therapeutic protective immunity against H. pylori that is reflected by specific mucosal immune responses. Vaccine. 2007;25:2591-2598. |
| 38. | Malfertheiner P, Schultze V, Rosenkranz B, Kaufmann SH, Ulrichs T, Novicki D, Norelli F, Contorni M, Peppoloni S, Berti D. Safety and immunogenicity of an intramuscular Helicobacter pylori vaccine in noninfected volunteers: a phase I study. Gastroenterology. 2008;135:787-795. |
| 39. | D’Elios MM, Andersen LP. Inflammation, immunity, and vaccines for Helicobacter pylori. Helicobacter. 2009;14 Suppl 1:21-28. |
| 40. | Lee CK. Vaccination against Helicobacter pylori in non-human primate models and humans. Scand J Immunol. 2001;53:437-442. |
| 41. | Bumann D, Metzger WG, Mansouri E, Palme O, Wendland M, Hurwitz R, Haas G, Aebischer T, von Specht BU, Meyer TF. Safety and immunogenicity of live recombinant Salmonella enterica serovar Typhi Ty21a expressing urease A and B from Helicobacter pylori in human volunteers. Vaccine. 2001;20:845-852. |
| 42. | Yamaoka Y, Kita M, Kodama T, Imamura S, Ohno T, Sawai N, Ishimaru A, Imanishi J, Graham DY. Helicobacter pylori infection in mice: Role of outer membrane proteins in colonization and inflammation. Gastroenterology. 2002;123:1992-2004. |
| 43. | Solnick JV, Hansen LM, Salama NR, Boonjakuakul JK, Syvanen M. Modification of Helicobacter pylori outer membrane protein expression during experimental infection of rhesus macaques. Proc Natl Acad Sci USA. 2004;101:2106-2111. |
| 44. | Deitsch KW, Moxon ER, Wellems TE. Shared themes of antigenic variation and virulence in bacterial, protozoal, and fungal infections. Microbiol Mol Biol Rev. 1997;61:281-293. |
Peer reviewers: Mario M D’Elios, Professor, University of Florence, viale Morgagni 85, Florence 50134, Italy; Andrew Day, Assistant Professor, University of Otago, Christchurch Hospital, Christchurch 8140, New Zealand
S- Editor Wang JL L- Editor Cant MR E- Editor Ma WH