Published online May 7, 2010. doi: 10.3748/wjg.v16.i17.2176
Revised: February 20, 2010
Accepted: February 27, 2010
Published online: May 7, 2010
AIM: To investigate the aberrant expression of nuclear matrix proteins in human gastric cancer cells before and after hexamethylene bisacetamide (HMBA) treatment.
METHODS: Proteomics analysis of differential nuclear matrix proteins was performed by two dimensional electrophoresis polyacrylamide gel electrophoresis and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. The expression levels of three nuclear matrix proteins were further confirmed by Western blotting and their locations in nuclear matrix filament were observed by quantum dots-based immunofluorescence.
RESULTS: Proteomics analysis showed that 43 protein spots were significantly changed due to HMBA treatment. Fifteen proteins were identified in the HMBA-induced differentiation of gastric tumor cells. Eight proteins spots were down-regulated while seven were up-regulated. Among these proteins, prohibitin, nucleophosmin and hnRNP A2/B1 were significantly decreased in HMBA-treated human gastric cancer cells, and their locations in nuclear matrix were altered by HMBA. Our results proved the alteration of specific nuclear matrix proteins during the differentiation of human gastric cancer cells. And the aberrant expressions of nuclear matrix proteins were of significance in revealing the regulatory mechanism of tumor cell proliferation and differentiation.
CONCLUSION: The aberrant expressions and intracellular redistributions of nuclear matrix proteins before and after HMBA treatment indicated that nuclear matrix proteins play a pivotal role in the differentiation of gastric cancer cells.
- Citation: Jing GJ, Xu DH, Shi SL, Li QF, Wang SY, Wu FY, Kong HY. Aberrant expression of nuclear matrix proteins during HMBA-induced differentiation of gastric cancer cells. World J Gastroenterol 2010; 16(17): 2176-2182
- URL: https://www.wjgnet.com/1007-9327/full/v16/i17/2176.htm
- DOI: https://dx.doi.org/10.3748/wjg.v16.i17.2176
Gastric cancer is the most common malignant gastrointestinal cancer, which accounts for 25% of cancer deaths[1]. Researches on gastric cancer cell differentiation will not only contribute to the diagnosis of gastric cancer, but also the regulatory mechanism of tumor cell differentiation. Studies have been carried out on the mechanism of induced differentiation of gastric cancer cells. But reports about the proteins related to the differentiation and proliferation of gastric cancer are rare. It is important to elucidate the regulatory mechanism of gastric cancer cell differentiation. Nuclear matrix is a filamentous protein framework for eukaryotic cell chromatin. Nuclear matrix proteins closely relate to DNA duplication and transcription[2]. Some regulatory proteins or enzymes are nuclear matrix proteins or nuclear matrix-binding proteins[3], and they regulate cell differentiation. Protein components of nuclear matrix are dynamic throughout cell proliferation and differentiation. Protein composition of nuclear matrix in tumor cells differ from normal tissue-derived cells[4]. In our previous study we found that the protein compositions of nuclear matrix were changed during the induced-differentiation of various tumor cells[5,6], suggesting that analysis on the aberrant expression of nuclear matrix proteins was of great importance for an in-depth investigation in the mechanism of cell carcinogenesis and reversal. Hexamethylene bisacetamide (HMBA) is a hybrid bipolar compound and originally developed as a potent inducer of cell differentiation[7]. Hereby, based on the induced effects of HMBA on the differentiation of human gastric cancer cells[8,9], we extended our study to determine the aberrant expression of nuclear matrix proteins in HMBA-treated human gastric adenocarcinoma BGC-823 cells. We aimed to identify specific nuclear matrix proteins related to gastric cancer cells and provide further scientific evidences for the mechanism of gastric cancer cell proliferation and differentiation.
BGC-823 cells, obtained from China Center for Type Culture Collection, were routinely cultured in RPMI 1640 (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Hyclone) and 100 U/mL penicillin, 100 μg/mL streptomycin and 50 μg/mL kanamycin at 37°C in a humidified atmosphere containing 5% CO2.
To prepare the whole cell extracts, cells were washed in phosphate buffered saline (PBS) and then lysed in ice-cold lysis buffer {7 mol/L urea, 2 mol/L thiourea, 4% 3-[(3-Cholanidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS), 1.5% Triton X-100, 1% pharmalyte (pH 3-10 Bio-rad), 65 mmol/L DL-Dithiothreitol (DTT), 40 mmol/L Tris, 5 mg/mL aprotinin, 1 mg/mL leupeptin, 1 mg/mL pepstatin, 2 mmol/L phenylmethanesulfonyl fluoride (PMSF), and 5 mmol/L ethylenediaminetetraacetic acid (EDTA)}. The suspension was then sonicated for 20 min at 0°C and centrifuged at 8000 ×g for 30 min.
Nuclear matrix proteins were prepared using a method described by Michishita et al[10]. After washed in ice-cold PBS twice, BGC-823 cells were suspended in cytoskeleton (CSK) buffer [100 mmol/L KCl, 3 mmol/L MgCl2, 5 mmol/L ethylene glycol tetraacetic acid, 10 mmol/L piperazine-N,N’-bis(2-ethanesulfonic acid), 300 mmol/L sucrose, 0.5% Triton X-100, and 2 mmol/L PMSF, pH 6.8] for 10 min at 0°C. After being centrifuged at 1000 ×g for 5 min, the pellet was resuspended in digestion buffer (identical to CSK buffer except for 50 mmol/L NaCl instead of KCl) containing 400 mg/mL DNase I for 30 min at room temperature and centrifuged at 1000 ×g twice. Cold ammonium sulfate at a final concentration of 0.25 mol/L was used to precipitate proteins. After centrifugation, the pellet was dissolved in lysis buffer [7 mol/L urea, 2 mol/L thiourea, 4% CHAPS, 1.5% Triton X-100, 1% Pharmalyte (pH 3-10 Bio-rad), 65 mmol/L DTT, 40 mmol/L Tris, 5 mg/mL aprotinin, 1 mg/mL leupeptin, 1 mg/mL pepstatin, 2 mmol/L PMSF, and 5 mmol/L EDTA] and then sonicated at 0°C for 20 min. Finally, the suspension was centrifuged at 10 000 ×g at 4°C for 30 min and the supernatants were used as nuclear matrix extracts.
Protein concentrations were determined by BCA assay.
2-D polyacrylamide gel electrophoresis (PAGE) was performed as follows. Protein lysates were diluted in sample buffer with 2% Immobiline™ DryStrip gel (IPG) buffer, pH 3-10, nonlinear (GE Heathcare). The samples were applied to 18 cm, immobilized pH gradient strips of pH 3-10 (IPG Drystrips, GE Healthcare). After isoelectric focusing was completed, the strips were equilibrated and the second dimensional electrophoresis was carried out overnight at 3 W/gel at 20°C. The triplicate sets of silver-stained gels were scanned using a UMAX POWER LOOK III photometer and analyzed with the PD Quest 8.0 software (Bio-rad). The digitalized two dimensional electrophoresis (2-DE) gel images were compared by matching method. Differentially expressed spots were analyzed and annotated.
The spots were cut and digested using 12.5 ng/μL trypsin (Promega, Madison, WI, USA) in 50 mmol/L ammounium bicarbonate (pH 8.0, Sigma). After that the samples were eluted with 2 μL of matrix solution containing 10 mg/mL α cyano-4-hydroxy cinnamic acid (CHCA, Sigma) and were submitted to Bruker III matrix-assisted laser desorption/ionization time-of-flight mass spectrometer. The spectra were internally calibrated using the trypsin autolysis products [842.51 (M + H) and 2211.11 (M + H)] by Flex Analysis software and searched against Swiss-prot and NCBI database using the Mascot tool from matrix science. All the searches were analyzed with a 50 ppm mass tolerance.
For Western blotting experiments, 20 μg cell lysates were loaded and separated on polyacrylamide gles and then transferred to positively-charged nylon membranes (Millipore, Bedford, MA) according to standard protocol. These blots were blocked for 1 h at room temperature in 5% skim milk. The target proteins were probed with primary antibodies and horseradish peroxidase-labeled secondary antibodies (Santa Cruz). β-actin was used as an indicator for equality of lane loading. Antibody positive bands were visualized using ECL Western blotting detection reagents (Pierce). The X-ray film was scanned and the band density was calculated using the ImageJ software[11].
The NM-IF system was prepared according the methods described by Liang et al[12]. After being prefixed with 4% paraformaldehyde at room temperature for 10 min, the cover slips were blocked with 3% bovine serum albumin for 1 h and then incubated with nucleophosmin (NPM), prohibitin (PHB) and hnRNP A2/B1 primary antibodies at 37°C for 1 h. After washing, the cover slips were rinsed in biotin-labeled secondary antibodies for 45 min at 37°C, washed with tris-buffered saline Tween-20, and incubated with streptavidin-conjugated quantum dots (size scale, 605 nm) for 1 h at 37°C. After that, the cover slips were enveloped with 90% glycerol and observed under fluorescence microscope.
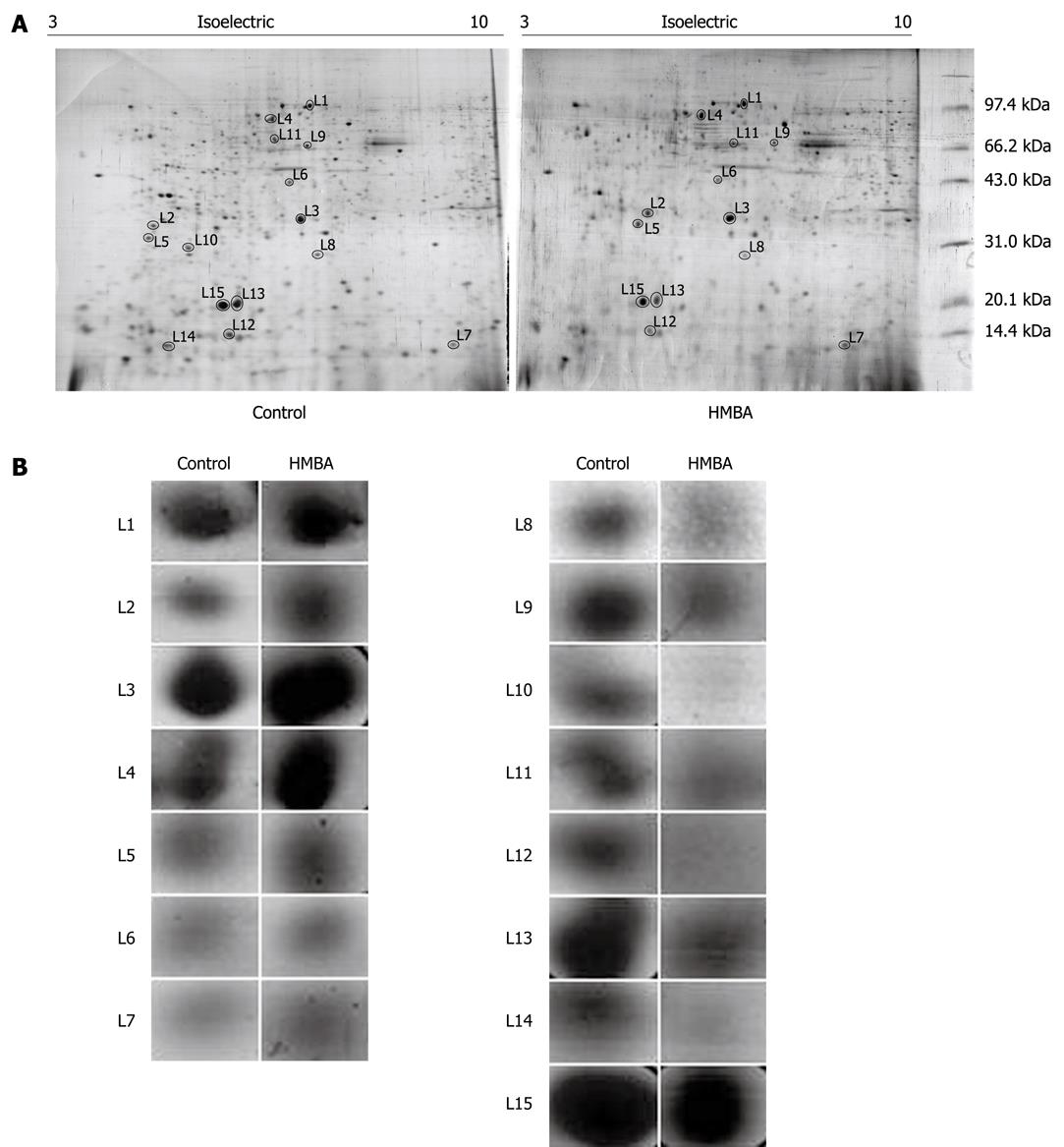
For proteomics analysis, cell lysates from control and HMBA-treated cells were submitted to 2D-PAGE followed by silver-staining of the gel. The procedures were independently repeated three times and the representative gel images are shown in Figure 1A. And Figure 1B displayed the enlarged maps of changed expression of nuclear matrix proteins from BGC-823 cells. PD Quest 8.0 software (Bio-rad) detected about 342 ± 10 proteins spots on the silver-stained gels of control group while 280 ± 13 protein spots of HMBA-treated group, suggesting that the result of 2-DE PAGE had a high repeatability. Forty-three changed spots were excised and digested with trypsin, and were identified by mass spectrometry. Fifteen proteins were identified in the HMBA-induced differentiation of gastric tumor cells. The identified proteins are listed in Table 1. The expressions of Zinc finger protein 268, calcium-binding protein CaBP5, MAGUK p55 subfamily member 3, vacuolar protein sorting 33B, and Ras-related protein Rab-30, Vimentin, Calnexin (Precursor) were up-regulated while peroxiredoxin 2, transcriptional repressor CTCFL, PHB, Keratin 1 type II cytoskeletal, PACAP protein, hnRNP A2/B1, C-type lectin and NPM were down-regulated. Another 28 protein spots were not identified for their low abundances. We next focused on PHB, NPM and hnRNP A2/B1, which were also identified as aberrantly expressed proteins in our previously studies[12-15].
| Spot number | Protein name | Accession number | Mol. mass calc (Da) | pI (calc) | Score |
| Up-regulated proteins | |||||
| L1 | Zinc finger protein 268 | Q14587 | 108 373 | 9.14 | 57 |
| L2 | Calcium-binding protein CaBP5 | Q9NP86 | 19 812 | 4.46 | 61 |
| L3 | MAGUK p55 subfamily member 3 | Q13368 | 66 168 | 6.27 | 45 |
| L4 | Vacuolar protein sorting 33B | Q9H267 | 70 570 | 6.29 | 54 |
| L5 | Ras-related protein Rab-30 | Q15771 | 23 058 | 4.91 | 64 |
| L6 | Vimentin | 57471646 | 49 623 | 5.19 | 48 |
| L7 | Calnexin (Precursor) | 4262127 | 10 638 | 5.30 | 89 |
| Down-regulated protein | |||||
| L8 | Peroxiredoxin 2 | P32119 | 21 892 | 5.66 | 72 |
| L9 | Transcriptional repressor CTCFL | Q8NI51 | 75 668 | 8.58 | 67 |
| L10 | Prohibitin | P35232 | 29 859 | 5.57 | 79 |
| L11 | Keratin 1, type II, cytoskeletal | 7428712 | 65 455 | 6.03 | 86 |
| L12 | PACAP protein | 18204192 | 20 681 | 5.37 | 161 |
| L13 | hnRNP A2/B1 | P22626 | 36 600 | 8.67 | 97 |
| L14 | C-type lectin | Q2K157 | 22 130 | 5.47 | 68 |
| L15 | Nucleophosmin | Q96EA5 | 32 726 | 4.64 | 76 |
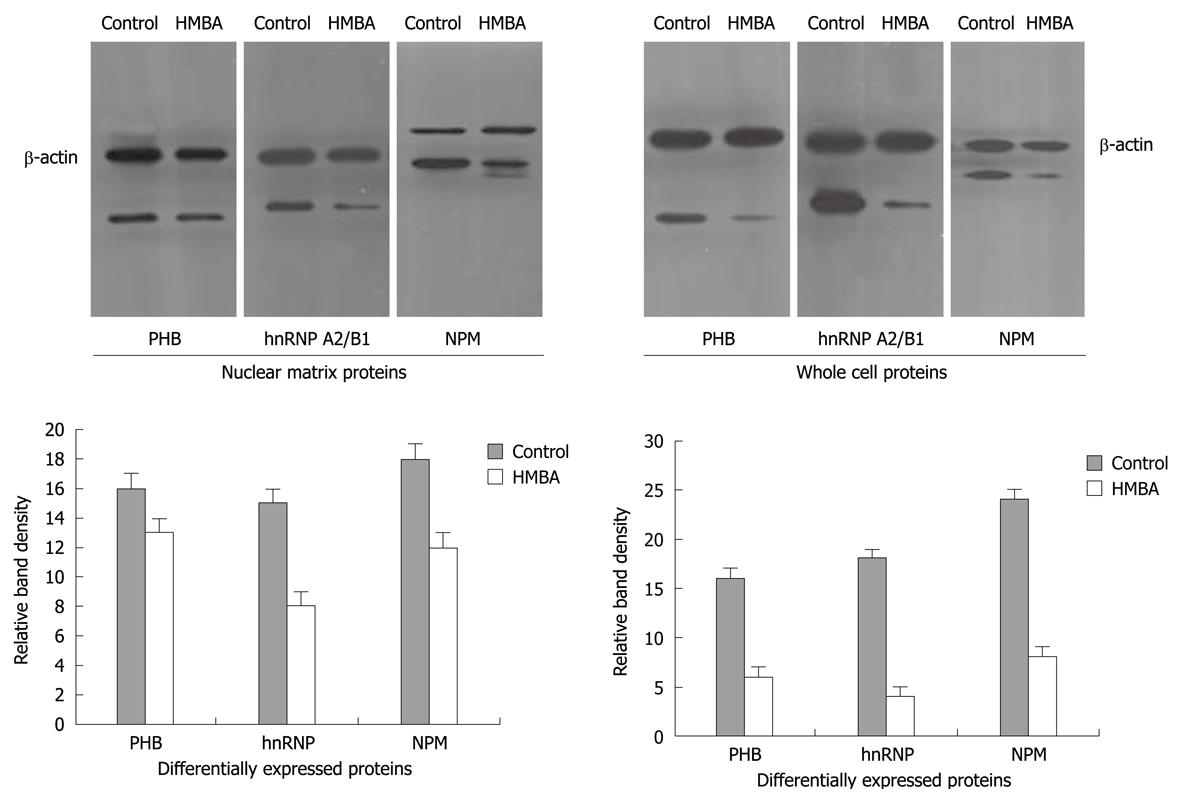
To further verify the aberrant changes of PHB, NPM and hnRNP A2/B1, Western immunoblotting was employed to confirm the expression levels of these proteins in cells before and after HMBA treatment, and the intensities of protein bands were densitometrically quantified as described by Sheffield JB[11]. Results showed that signals of PHB, NPM and hnRNP A2/B1 in HMBA-treated cells were much lower than that in control cells, suggesting that the expressions of these three proteins at 7 d of treatment were significantly inhibited by HMBA (Figure 2). All the Western blotting results were consistent with the proteomic analysis.
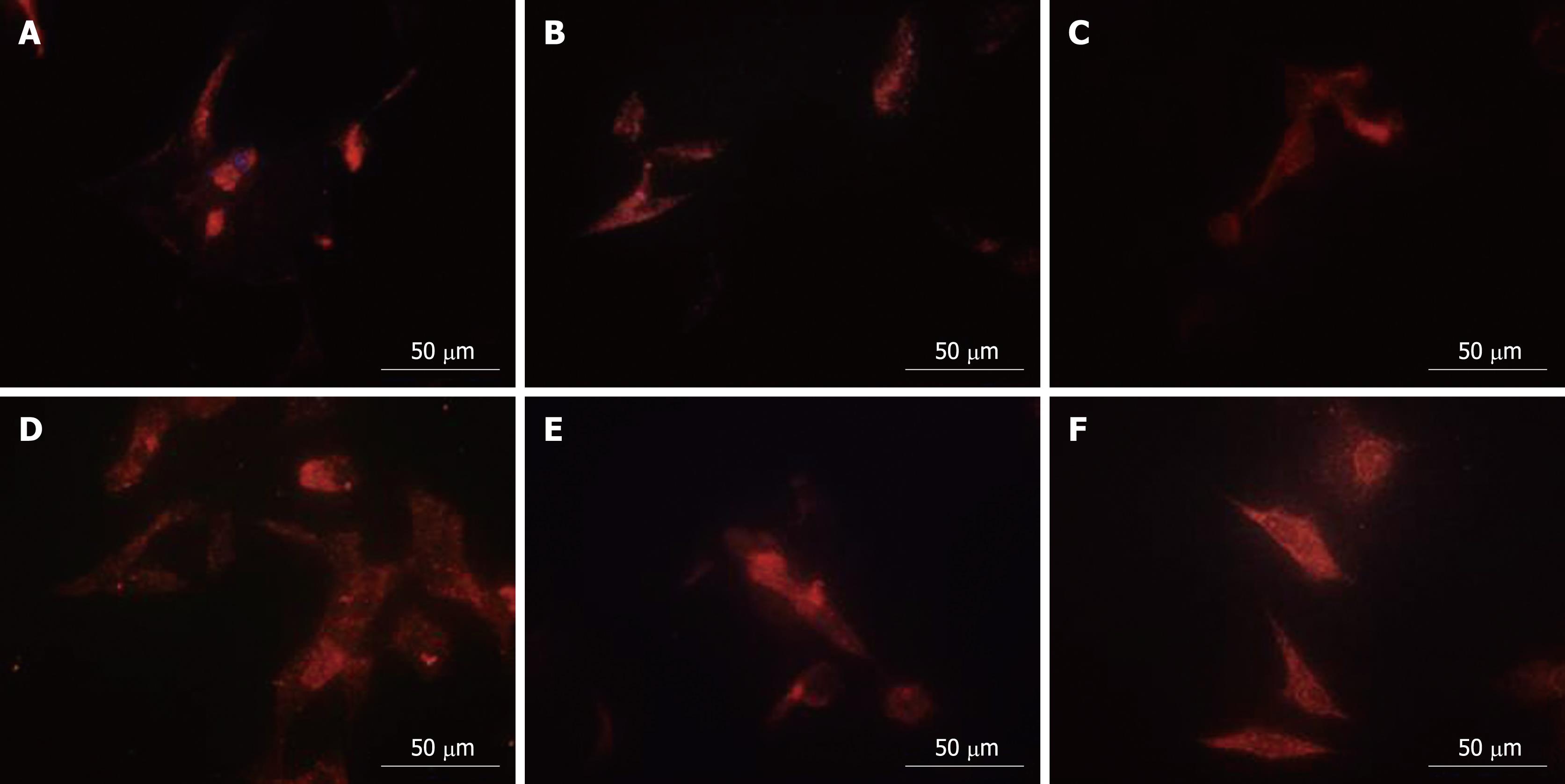
Proteomics analysis showed that PHB, NPM and hnRNP A2/B1 were nuclear matrix proteins. QDs-based location experiments were conducted to corroborate the alteration of the physical distribution of these three proteins in nuclear matrix-intermediate system. Clearly altered distribution patterns for NPM and hnRNP A2/B1 were observed in the nucleus while PHB in at the cytoplasm. The red fluorescence representing PHB mainly accumulated at the nucleus periphery in control cells (Figure 3C), while PHB was uniformly distributed in the whole cytoplasm after HMBA treatment, suggesting that HMBA obviously altered the location of PHB (Figure 3D). Highly intensified NPM fluorescence mainly localized in the residue of nucleoli in correspondence with the fact that NPM was an established marker for the granular components[16], with very subtle expression in the cytoplasm (Figure 3A). HMBA treatment resulted in the low intensity of fluorescence in nucleolus and faint fluorescence in cytoplasm derived from NPM, suggesting that NPM had a tendency of translocation from nucleoli regions to nuclear matrix (Figure 3B). When referred to hnRNP A2/B1, a similar distribution pattern to NPM was observed. In control cells, the fluorescence representing hnRNP A2/B1 was mainly detected in nuclear region with subtle signals in cytoplasm region (Figure 3E), whereas after HMBA treatment, a clear translocation appeared from nuclear to cytoplasm region, and hnRNP A2/B1was uniformly distributed in the cytoplasm but not in the nucleus (Figure 3F).
The alteration of nuclear matrix components is closely related to cell carcinogenesis. Configuration and protein composition of nuclear matrix in tumor cells differ from normal tissue-derived cells. For further investigating the effects of HMBA on the protein components of nuclear matrix of human gastric cancer cells, proteomics analysis was employed to screen the aberrant nuclear matrix proteins of human gastric cancer cells before and after HMBA treatment. And 15 differentially expressed proteins were identified, seven of which, including Zinc finger protein 268, nebulin, etc. were up-regulated while the other eight proteins, including PHB, KeHMBAtin 1 type II, cytoskeletal, hnRNP A2/B1, NPM, etc. were down-regulated. Getzenberg et al[17] reported that nuclear matrix profiles of bladder cancer, liver cancer and colon cancer were significantly altered in comparison with that of the normal tissues; moreover, differential nuclear matrix proteins were tumor-specific[17-19]. In our previous studies, we found variational nuclear matrix profiles of tumor cells[5,6]. HMBA could change protein compositions of gastric cancer cell nuclear matrix. The alteration of nuclear matrix protein expressions during the differentiation of gastric cancer cells was closely related to their functional transformation.
In this article we focused on the aberrant expression of nuclear matrix proteins in the induced-differentiation of gastric cancer cells. Among all the differentially expressed proteins, the down-regulated proteins, PHB, NPM and hnRNP A2/B1, were also identified in our previous studies on the induced differentiation of gastric cancer MGC80-3 cells, suggesting that PHB, NPM and hnRNP A2/B1 were common aberrantly expressed nuclear matrix-specific proteins in different tumor cell lines[8,12-15]. Proteomics analysis showed that expressions of PHB, NPM and hnRNP A2/B1 were down-regulated both in the whole cell lysates and nuclear matrix lysates, and the expression changes were further confirmed by Western blotting. Quantum dots-based immunofluorescent assay was performed to investigate the localization of specific nuclear matrix proteins in nuclear matrix-intermediate filament system. And results displayed that PHB, NPM and hnRNP A2/B1 were localized in nuclear matrix fibers, and their locations were altered by HMBA treatment. Our data further proved that PHB, NPM and hnRNP A2/B1 were common differential nuclear matrix proteins. And the aberrant expression and relocation of these three proteins were closely related to their regulatory roles in the process of tumor cell differentiation.
The importance of differentially expressed nuclear matrix proteins was embodied not only by their locations in nuclear matrix fibers but also their regulations on cell differentiation and apoptosis. For example, PHB presented anti-proliferation activities by inhibiting the activity of nuclear estrogen receptors, and it was usually over-expressed in tumor tissues[20]. PHB was also a biomarker protein for differentiating benign prostate hyperplasia from prostate cancer[21]. NPM was an abundant nucleolar phosphoprotein, and it could cause the suppression of cell differentiation and anti-apoptosis in human carcinogenesis. Inhibiting the activity of NPM led to the over-expression of P53[22]. Cell proliferation could be inhibited by blocking the shuttle of NPM from nuclear to cytoplasm[23]. hnRNP A2/B1 played a pivotal role in DNA damage repair[24]. The expression and location of PHB, NPM and hnRNP A2/B1 were changed in gastric tumor cells before and after HMBA treatment, suggesting their pivotal biological functions in the differentiation of gastric tumor cells. Continued investigation on specific nuclear matrix proteins will contribute to better understanding the regulatory mechanisms of nuclear matrix proteins in gastric carcinogenesis and reversal.
Gastric cancer is the most common malignant gastrointestinal cancer and accounts for 25% of cancer deaths. Nuclear matrix proteins are closely related to DNA duplication and transcription. Researches on nuclear matrix proteins of gastric tumor cells will contribute to the diagnosis and prevention of gastric cancer.
It is important to identify specific nuclear matrix proteins related to gastric cancer cells and provide further scientific evidences for the mechanism of gastric cancer cell proliferation and differentiation.
Forty-three protein expressions were significantly changed in hexamethylene bisacetamide (HMBA)-treated human gastric cancer cells. Moreover, the locations of three proteins in nuclear matrix fibers were obviously altered after HMBA treatment.
The differentially expressed nuclear matrix proteins might be intracellular target proteins of HMBA and the tumor markers for gastric cancer.
This study examined the effects of HMBA on nuclear matrix protein expression in gastric cancer cell BGC-823. Forty-three proteins were affected significantly by the treatment, among them 15 were identified including 7 up-regulated and 8 down-regulated ones. The study is neatly designed and the figures are presented with good quality. The manuscript is highly interesting, well-written and valuable.
| 1. | Parkin DM, Pisani P, Ferlay J. Estimates of the worldwide incidence of 25 major cancers in 1990. Int J Cancer. 1999;80:827-841. |
| 2. | Brünagel G, Schoen RE, Bauer AJ, Vietmeier BN, Getzenberg RH. Nuclear matrix protein alterations associated with colon cancer metastasis to the liver. Clin Cancer Res. 2002;8:3039-3045. |
| 3. | Marchisio M, Santavenere E, Paludi M, Gaspari AR, Lanuti P, Bascelli A, Ercolino E, Di Baldassarre A, Miscia S. Erythroid cell differentiation is characterized by nuclear matrix localization and phosphorylation of protein kinases C (PKC) alpha, delta, and zeta. J Cell Physiol. 2005;205:32-36. |
| 4. | Coffey DS. Nuclear matrix proteins as proteomic markers of preneoplastic and cancer lesions : commentary re: G. Brunagel et al., nuclear matrix protein alterations associated with colon cancer metastasis to the liver. Clin. Cancer Res., 8: 3039-3045, 2002. Clin Cancer Res. 2002;8:3031-3033. |
| 5. | Shi SL, Wang YY, Liang Y, Li QF. Effects of tachyplesin and n-sodium butyrate on proliferation and gene expression of human gastric adenocarcinoma cell line BGC-823. World J Gastroenterol. 2006;12:1694-1698. |
| 6. | Zhao CH, Li QF, Zhao Y, Niu JW, Li ZX, Chen JA. Changes of nuclear matrix proteins following the differentiation of human osteosarcoma MG-63 cells. Genomics Proteomics Bioinformatics. 2006;4:10-17. |
| 7. | Contreras X, Barboric M, Lenasi T, Peterlin BM. HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription. PLoS Pathog. 2007;3:1459-1469. |
| 8. | Zhao CH, Li QF. Altered profiles of nuclear matrix proteins during the differentiation of human gastric mucous adenocarcinoma MGc80-3 cells. World J Gastroenterol. 2005;11:4628-4633. |
| 9. | Zhang G, Wang G, Wang S, Li Q, Ouyang G, Peng X. Applying proteomic methodologies to analyze the effect of hexamethylene bisacetamide (HMBA) on proliferation and differentiation of human gastric carcinoma BGC-823 cells. Int J Biochem Cell Biol. 2004;36:1613-1623. |
| 10. | Michishita E, Kurahashi T, Suzuki T, Fukuda M, Fujii M, Hirano H, Ayusawa D. Changes in nuclear matrix proteins during the senescence-like phenomenon induced by 5-chlorodeoxyuridine in HeLa cells. Exp Gerontol. 2002;37:885-890. |
| 11. | Sheffield JB. ImageJ, A useful tool for biological image processing and analysis. Microsc Microanal. 2007;13 Suppl 2:200-201. |
| 12. | Liang Y, Li QF, Zhang XY, Shi SL, Jing GJ. Differential expression of nuclear matrix proteins during the differentiation of human neuroblastoma SK-N-SH cells induced by retinoic acid. J Cell Biochem. 2009;106:849-857. |
| 13. | Li QF, Shi SL, Liu QR, Tang J, Song J, Liang Y. Anticancer effects of ginsenoside Rg1, cinnamic acid, and tanshinone IIA in osteosarcoma MG-63 cells: nuclear matrix downregulation and cytoplasmic trafficking of nucleophosmin. Int J Biochem Cell Biol. 2008;40:1918-1929. |
| 14. | Xu DH, Tang J, Li QF, Shi SL, Chen XF, Liang Y. Positional and expressive alteration of prohibitin during the induced differentiation of human hepatocarcinoma SMMC-7721 cells. World J Gastroenterol. 2008;14:5008-5014. |
| 15. | Tang J, Niu JW, Xu DH, Li ZX, Li QF, Chen JA. Alteration of nuclear matrix-intermediate filament system and differential expression of nuclear matrix proteins during human hepatocarcinoma cell differentiation. World J Gastroenterol. 2007;13:2791-2797. |
| 16. | Krüger T, Zentgraf H, Scheer U. Intranucleolar sites of ribosome biogenesis defined by the localization of early binding ribosomal proteins. J Cell Biol. 2007;177:573-578. |
| 17. | Getzenberg RH, Konety BR, Oeler TA, Quigley MM, Hakam A, Becich MJ, Bahnson RR. Bladder cancer-associated nuclear matrix proteins. Cancer Res. 1996;56:1690-1694. |
| 18. | Brünagel G, Vietmeier BN, Bauer AJ, Schoen RE, Getzenberg RH. Identification of nuclear matrix protein alterations associated with human colon cancer. Cancer Res. 2002;62:2437-2442. |
| 19. | Konety BR, Nguyen TS, Dhir R, Day RS, Becich MJ, Stadler WM, Getzenberg RH. Detection of bladder cancer using a novel nuclear matrix protein, BLCA-4. Clin Cancer Res. 2000;6:2618-2625. |
| 20. | Gamble SC, Chotai D, Odontiadis M, Dart DA, Brooke GN, Powell SM, Reebye V, Varela-Carver A, Kawano Y, Waxman J. Prohibitin, a protein downregulated by androgens, represses androgen receptor activity. Oncogene. 2007;26:1757-1768. |
| 21. | Ummanni R, Junker H, Zimmermann U, Venz S, Teller S, Giebel J, Scharf C, Woenckhaus C, Dombrowski F, Walther R. Prohibitin identified by proteomic analysis of prostate biopsies distinguishes hyperplasia and cancer. Cancer Lett. 2008;266:171-185. |
| 22. | Qi W, Shakalya K, Stejskal A, Goldman A, Beeck S, Cooke L, Mahadevan D. NSC348884, a nucleophosmin inhibitor disrupts oligomer formation and induces apoptosis in human cancer cells. Oncogene. 2008;27:4210-4220. |
| 23. | Maggi LB Jr, Kuchenruether M, Dadey DY, Schwope RM, Grisendi S, Townsend RR, Pandolfi PP, Weber JD. Nucleophosmin serves as a rate-limiting nuclear export chaperone for the Mammalian ribosome. Mol Cell Biol. 2008;28:7050-7065. |
| 24. | Zech VF, Dlaska M, Tzankov A, Hilbe W. Prognostic and diagnostic relevance of hnRNP A2/B1, hnRNP B1 and S100 A2 in non-small cell lung cancer. Cancer Detect Prev. 2006;30:395-402. |
Peer reviewers: Dr. Jianyuan Chai, PhD, MS, BS, Assistant Professor, Research (09-151), VA Long Beach Healthcare System, 5901 E. 7th St, Long Beach, CA 90822, United States; Dr. Mukaddes Esrefoglu, Professor, Department of Histology and Embryology, Inonu University, 44280, Malatya, Turkey
S- Editor Wang JL L- Editor Ma JY E- Editor Zheng XM