Published online Mar 21, 2010. doi: 10.3748/wjg.v16.i11.1385
Revised: December 14, 2009
Accepted: December 21, 2009
Published online: March 21, 2010
AIM: To extend the knowledge of the dynamic interaction between Helicobacter pylori (H. pylori) and host mucosa.
METHODS: A time-series cDNA microarray was performed in order to detect the temporal gene expression profiles of human gastric epithelial adenocarcinoma cells infected with H. pylori. Six time points were selected to observe the changes in the model. A differential expression profile at each time point was obtained by comparing the microarray signal value with that of 0 h. Real-time polymerase chain reaction was subsequently performed to evaluate the data quality.
RESULTS: We found a diversity of gene expression patterns at different time points and identified a group of genes whose expression levels were significantly correlated with several important immune response and tumor related pathways.
CONCLUSION: Early infection may trigger some important pathways and may impact the outcome of the infection.
-
Citation: You YH, Song YY, Meng FL, He LH, Zhang MJ, Yan XM, Zhang JZ. Time-series gene expression profiles in AGS cells stimulated with
Helicobacter pylori . World J Gastroenterol 2010; 16(11): 1385-1396 - URL: https://www.wjgnet.com/1007-9327/full/v16/i11/1385.htm
- DOI: https://dx.doi.org/10.3748/wjg.v16.i11.1385
Helicobacter pylori (H. pylori) have been shown to be the principal cause of acute and chronic gastritis and a major risk factor in gastric cancer development. A chronic inflammatory process induced by the pathogen is thought to be the cause of tumor development. It is well known that H. pylori binding to epithelial cells can induce tyrosine phosphorylation of host cell proteins and rearrangement of the cytoskeleton, which may contribute to inflammation and oncogenic transformation[1]. H. pylori colonization to the mucosa may also induce a systemic immune response and be susceptible to Ab-dependent complement-mediated phagocytosis and killing. Infected epithelial cells may also induce a mucosal inflammation under a mechanism of autoantibody-mediated destruction[2]. Some host factors like interleukin (IL)-1β, tumor necrosis factor (TNF)-α, and IL-10 may influence the disease outcome. One investigation on nuclear factor (NF)-κB signaling pathway and iNOS suggests that NF-κB activation may play an important role in protecting mucosol cells from apoptosis through upregulating iNOS[3]. Many previous studies have performed expression profiling to investigate host changes induced by H. pylori infection. These studies have provided some useful and significant information and shed some light for exploring the potential mechanism of H. pylori infection and host immunity[4-10]. However, none of them is designed based on a time-series scheme, the global and sequential profile of H. pylori infection that may be involved in the pathogenetic mechanism by which H. pylori infects and contributes to gastric carcinogenesis remains poorly understood. In this study, human gastric epithelial adenocarcinoma cells (AGS) co-cultured with an H. pylori 26695 strain at different time points were separated and analyzed by a whole genome Illumina microarray. Computer-assisted bioinformatics analysis was conducted to analyze the differential gene expression pattern.
H. pylori strain 26695 was routinely cultured for 24 h on Columbia agar plates (Oxoid) containing 5% goat blood under microaerophilic conditions at 37°C, following a wash in sterile PBS and estimation of the quantity of bacteria by OD600. The human gastric epithelial adenocarcinoma cell line AGS (ATCC CRL 1739) was cultured in RPMI 1640 without antibiotic or antifungal agents, and supplemented with 4 mmol/L L-glutamine and 10% fetal calf serum (Gibco) at 37°C in a humidified atmosphere of 5% CO2. A monolayer of AGS cells grown to 80% confluence was co-cultured with H. pylori at a multiplicity of infection of 300:1 in culture media for 0.5, 1, 2, 4, and 6 h.
Co-culture was stopped at each time point and followed by washing three times with PBS. Total RNA was isolated using Trizol extraction (Gibco/BRL). The quality of the RNA was verified by 1% agarose gel containing ethidium bromide.
Illumina Human-6 v2 BeadChips used for this study contains probes for well characterized genes, gene candidates and splice variants for a total number of 48 000 features. The “Detection Score > 0.99” was used to determine the expression. It was a statistical measure in the BeadStudio software, which was computed based on the Z-value of a gene relative to that of the negative controls. The data were normalized using a cubic spline method, which was generally used as a normalization algorithm in BeadStudio. The differentially expressed genes in different time point were identified using the Illumina custom error model implemented in BeadStudio. DiffScore, the expression difference score, takes into account background noise and sample variability[11]. The formula for the calculation of the DiffScore is: DiffScore = 10 sgn(μcond - μref)log10 (p). The differentially expressed genes with a |Diffscore| > 13 were selected for further analysis. The genes with a fold change > 1.5 were integrated and hierarchically clustered using Mev_4_0 (Multiple Experiment Viewer, TIGR). Gene enrichment in KEGG pathways (Kyoto Encyclopedia of Genes and Genomes) and Gene Ontology (GO) were accomplished with Onto-Tool (Pathway Express, OE2GO)[12,13], and co-expression gene clustering by short time-series expression miner (STEM, Carnegie Mellon University)[14] with a maximum number of model profiles set as 245, and a maximum unit change in model profiles between time points set at 2. Four interesting co-expression profiles were selected for further analyses. To obtain an optimized GO distribution, we also took all differentially expressed genes including those with a fold change < 1.5 as input for STEM analysis, and chose four profiles for GO enrichment using OE2GO. For pathway level analysis, those genes with a fold change > 1.5 were imported into Pathway-Express to obtain the significantly perturbed pathway list and gene mapping. This program was based on an impact analysis that included the classical statistics but also considered other crucial factors such as the magnitude of each gene’s expression change, their type and position in the given pathways, their interactions, etc. The IF of a pathway is calculated as the sum of the following two terms:
Then a simplified network construction was completed based on the genes enriched and mapped to KEGG pathways using STRING (version 8.2)[15], which is a known Predicted Protein-Protein Interactions Database (http://string.embl.de/).
Real-time reverse-transcriptase polymerase chain reaction (Q-RT-PCR) validation of microarray results was carried out for the GFPT2 gene at the five time points which were significantly altered according to the microarray data. RNA samples of different time points were prepared as previously described in RNA isolation. Briefly, 2 g total RNA of each sample was used for cDNA synthesis. Real time PCR was performed on the Rotor-Gene RG-3000 Real-Time Thermal Cycler with the SYBR Premix Ex Taq™ (TakaRa) and GAPDH was used as an internal control. The relative quantification of mRNA expression at each time point was calculated and compared with that of the untreated AGS cells as control. The primers of selected gene for RT-PCR were: (1) GFPT2 forward primer (5'-GACAAGCAGATGCCCGTCAT-3') and reverse primer (5'-AACTTGGAACTTTCAGTATCGTCCTT-3'); and (2) GAPDH forward primer (5'-AGAAGGCTGGGGCTCATTTG-3') reverse primer (5'-AGGGGCCATCCACAGTCTTC-3').
Microarray hybridization results showed that about 3577 genes in total (P < 0.05, DiffScore > 13, named dataset1 in this study) expressed differentially compared with 0 h group. This dataset was generated by taking an integration and alignment for the gene list of different time points using Microsoft Excel software, and the repeated genes were thus excluded. Rows were gene names and columns were differential expression values in different time points. Those genes without fold changes in some time points were set as a value equal to 0. The gene numbers at each time point for the 808 genes (P < 0.05, a fold change > 1.5, named dataset2 in this study) are listed in Table 1 and were selected for further emphatically analysis.
| Time point (h) | Up-regulation (n) | Down-regulation (n) | Total |
| 0.5 | 109 | 209 | 318 |
| 1 | 140 | 242 | 382 |
| 2 | 151 | 203 | 354 |
| 4 | 126 | 291 | 417 |
| 6 | 198 | 156 | 354 |
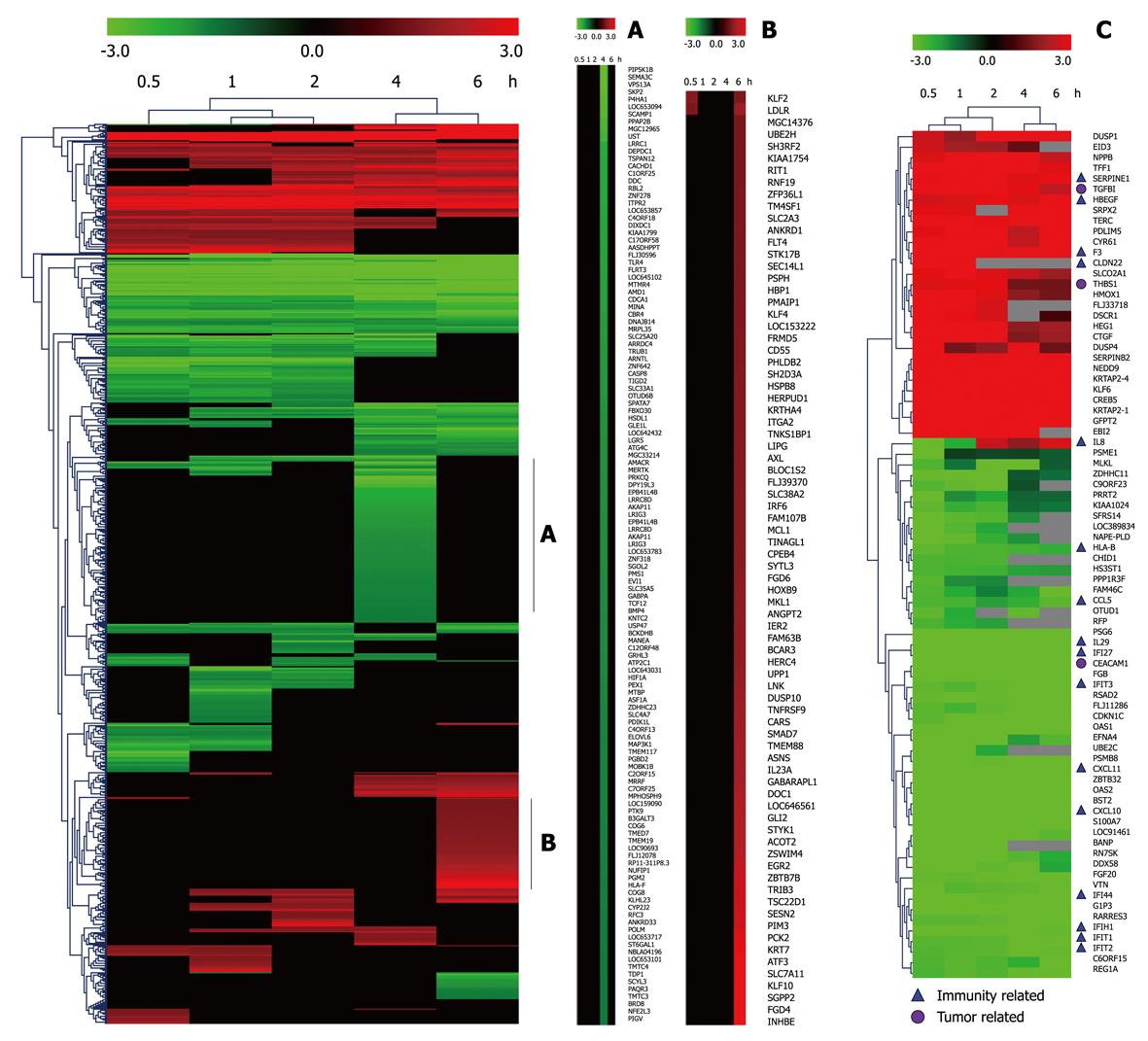
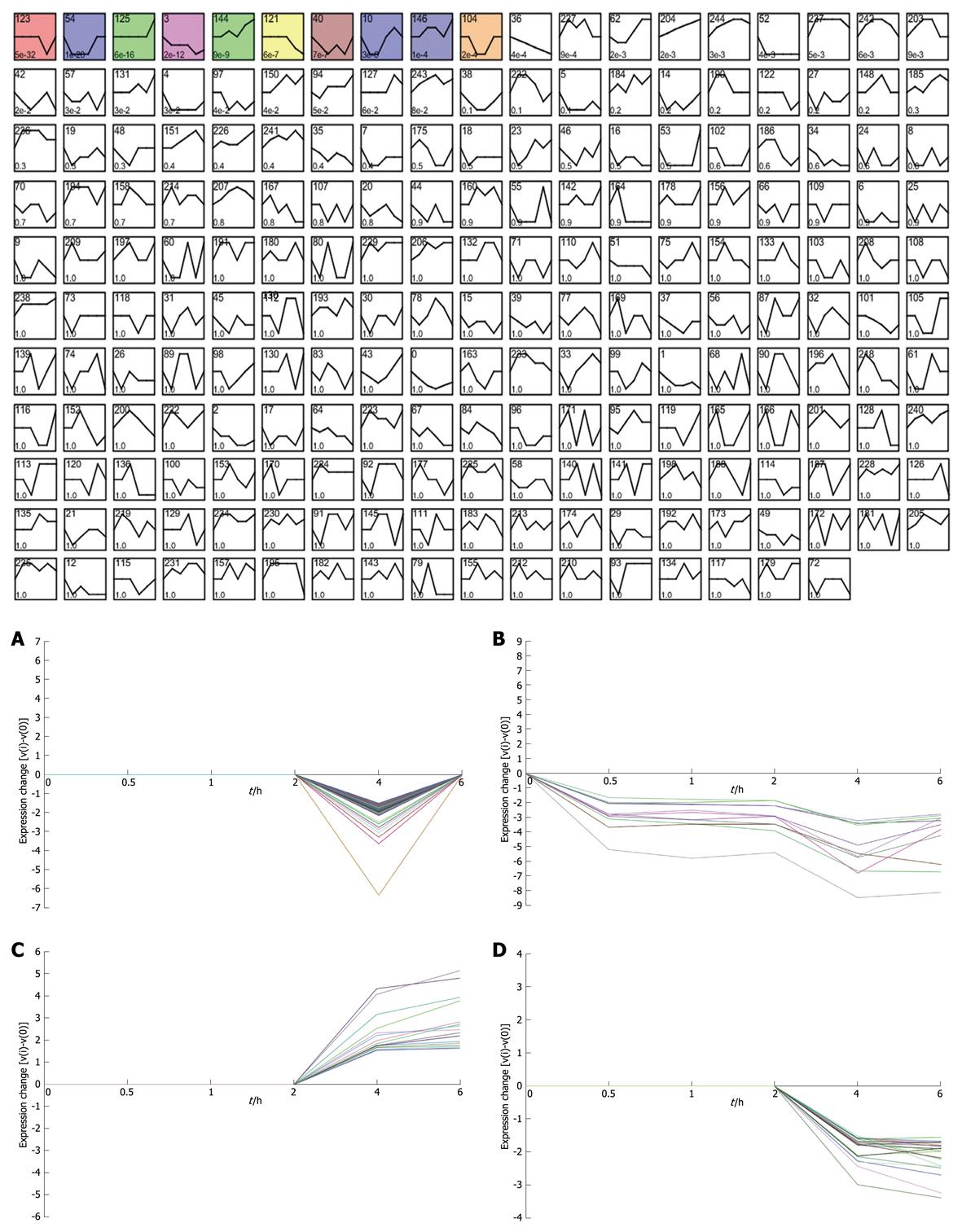
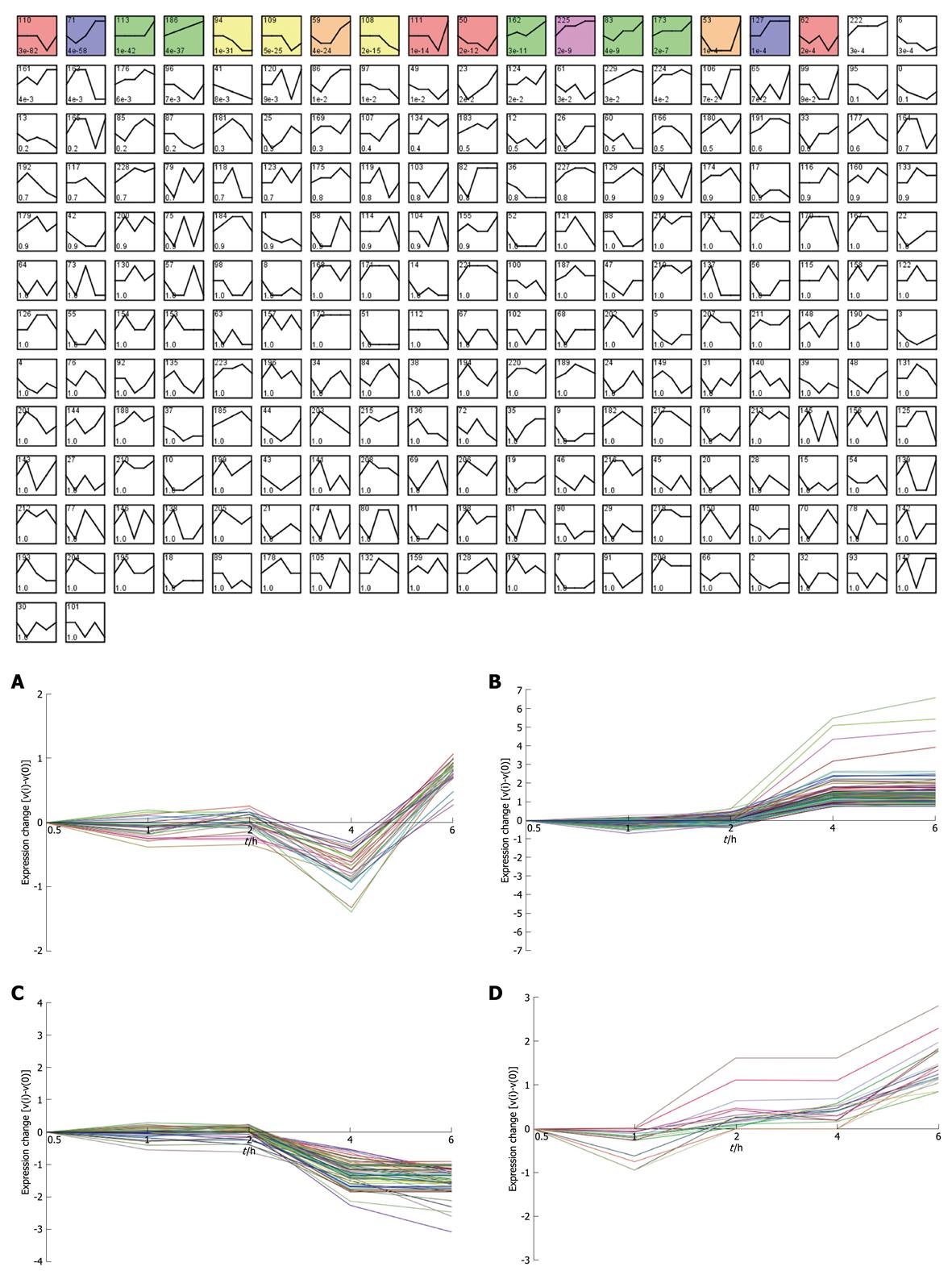
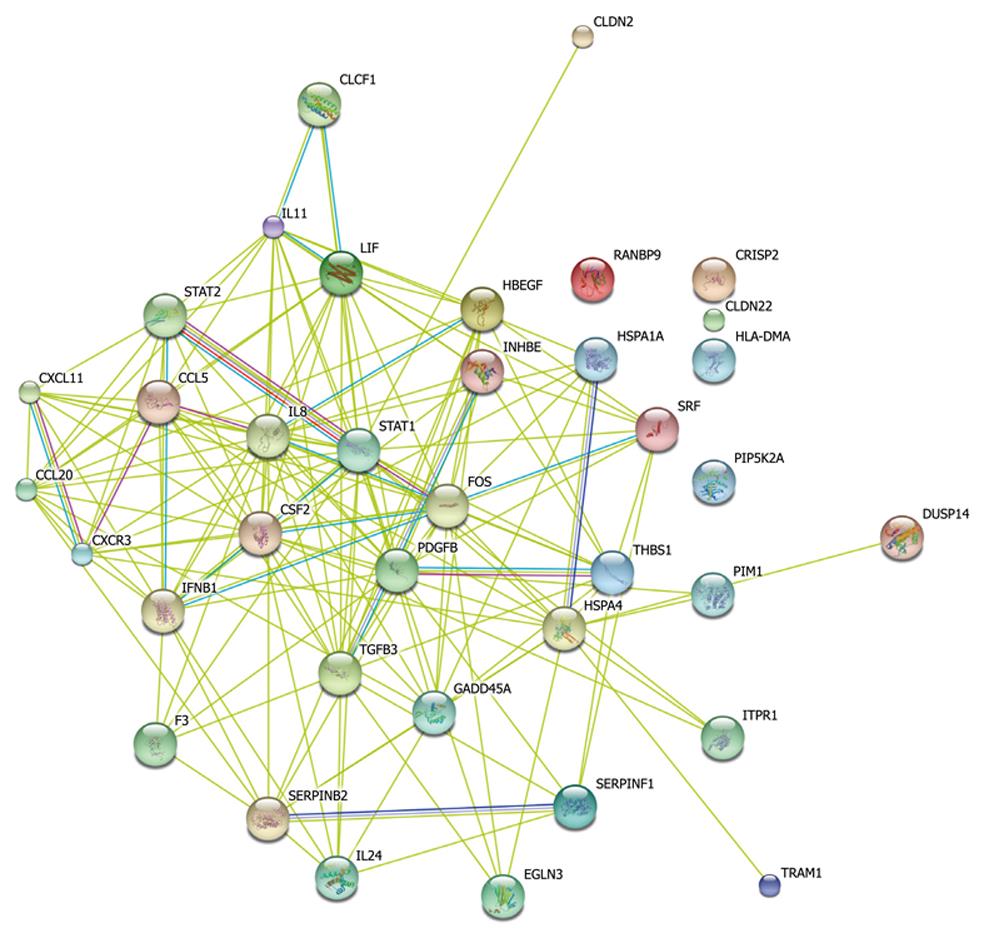
Taking dataset2 as input, hierarchical cluster analysis showed some differentially expressed genes down-regulated at 4 h and up-regulated at 6 h (Figure 1A and B). Eighty of the most differentially expressed genes were extracted by sorting their fold change and were hierarchically clustered as shown in Figure 1C. Immunity and tumor-related genes were labeled with triangles and circles, respectively. Ten significant profiles were obtained by STEM and four interesting profiles were shown with genes in detail (Figure 2 and Table 2). However, GO analysis did not provide significant terms. Taking dataset1 as input, the GO analysis results for the four profiles clustered are listed in Table 3 and Figure 3. Table 4 shows the GO distribution change of each time point by up-regulation and down-regulation, respectively. Analysis of KEGG pathways revealed many enrichment-related pathways including cell adhesion molecules, MAPK signaling, p53 signaling, and TGF-β signaling pathways, complement and coagulation cascades, and epithelial cell signaling in H. pylori infection. The top four significantly perturbed pathways are listed in Table 5. Related networks extracted from significant pathways are shown in Figure 4.
| Cluster ID | Symbol | |||||||||
| Profile 123 | C4ORF18 | USP47 | CYP2J2 | LGR5 | FLRT3 | LOC643031 | TMEM117 | CACHD1 | C12ORF48 | MTMR4 |
| RBL2 | ZDHHC23 | TTC13 | NUFIP1 | FLJ30596 | AASDHPPT | C2ORF15 | PGBD2 | LRRC8D | EVI1 | |
| SKP2 | ZNF318 | VPS13A | AMACR | ST6GAL1 | AMD1 | ELOVL6 | PGM2 | SLC35A5 | CBR4 | |
| EPB41L4B | C1ORF25 | C1GALT1 | ATG4C | MERTK | FANCL | LRIG3 | RHPN1 | PIP5K1B | SEMA3C | |
| P4HA1 | LOC653094 | SCAMP1 | PPAP2B | MGC12965 | UST | LRRC1 | DEPDC1 | DDC | ZNF278 | |
| ITPR2 | LOC653857 | DIXDC1 | KIAA1799 | C17ORF58 | TLR4 | LOC645102 | CDCA1 | MINA | DNAJB14 | |
| MRPL35 | SLC25A20 | ARRDC4 | TRUB1 | ARNTL | ZNF642 | CASP8 | TIGD2 | SLC33A1 | OTUD6B | |
| SPATA7 | FBXO30 | HSDL1 | GLE1L | LOC642432 | MGC33214 | PRKCQ | DPY19L3 | AKAP11 | LOC653783 | |
| SGOL2 | PMS1 | GABPA | TCF12 | BMP4 | KNTC2 | BCKDHB | MANEA | GRHL3 | ATP2C1 | |
| HIF1A | PEX1 | MTBP | ASF1A | SLC4A7 | PDIK1L | C4ORF13 | MAP3K1 | MOBK1B | MRRF | |
| C7ORF25 | MPHOSPH9 | LOC159090 | PTK9 | B3GALT3 | COG6 | TMED7 | TMEM19 | LOC90693 | FLJ12078 | |
| RP11-311P8.3 | ZNF181 | COG8 | KLHL23 | RFC3 | NBLA04196 | LOC653101 | TMTC4 | TDP1 | SCYL3 | |
| PAQR3 | TMTC3 | BRD8 | NFE2L3 | PIGV | TSPAN12 | |||||
| Profile 3 | PSG6 | FGB | CEACAM1 | CDKN1C | IFIT3 | RSAD2 | PSG7 | FLJ11286 | BTN3A2 | STAT1 |
| FLJ20035 | ||||||||||
| Profile 144 | EHD2 | RELB | COL16A1 | GDF15 | GNA15 | LETM2 | STX11 | FOSL1 | LOC647512 | SQSTM1 |
| C12ORF59 | ADM2 | DDIT3 | CHAC1 | CSF2 | DDIT4 | |||||
| Profile 12 | ZC3HAV1 | PSG9 | LYZ | FGG | PSG2 | PAGE4 | REG4 | GAD1 | PPM1H | TMEM70 |
| LRP8 | PAQR8 | SH3BGRL | MYLIP | ROR1 | C5ORF14 | SUSD4 | MGC3265 | CADPS2 | IDUA | |
| EPSTI1 | ||||||||||
| Profile | GO name | n | Corrected P value | Function code |
| 111 | Apical part of cell | 2 | 0.00842 | CC |
| 71 | Nucleic acid binding | 12 | 2.7E-4 | MF |
| Zinc ion binding | 23 | 0.00308 | MF | |
| Regulation of transcription | 22 | 0.01027 | BP | |
| Myeloid cell differentiation | 2 | 0.01577 | BP | |
| Nucleus | 39 | 2.9E-4 | CC | |
| Intracellular | 23 | 3.5E-4 | CC | |
| 108 | Small GTPase binding | 2 | 0.01173 | MF |
| Oxido-reductase activity | 6 | 0.02544 | MF | |
| GPI anchor biosynthetic process | 2 | 0.02591 | BP | |
| Female pregnancy | 3 | 0.02622 | BP | |
| Golgi membrane | 5 | 0.03987 | CC | |
| Cell surface | 3 | 0.03987 | CC | |
| 83 | DNA binding | 6 | 0.00577 | MF |
| Metal binding | 6 | 0.03346 | MF | |
| Nucleus | 10 | 0.01029 | CC |
| Time point (h) | Up-regulation | Down-regulation | ||||||||
| GO ID | GO name | Genes | P value | Code | GO ID | GO name | Genes | P value | Code | |
| 0.5 | GO:0008201 | Heparin binding | 5 | 7.1E-4 | MF | GO:0006955 | Immune response | 20 | 0.00000 | BP |
| GO:0008134 | Transcription factor binding | 4 | 0.01585 | MF | GO:0009615 | Response to virus | 10 | 0.00000 | BP | |
| GO:0003700 | Transcription activity | 10 | 0.02835 | MF | GO:0008150 | Biological process | 15 | 0.00896 | BP | |
| GO:0008083 | Growth factor activit | 4 | 0.03882 | MF | GO:0007267 | Cell-cell signaling | 10 | 0.00966 | BP | |
| GO:0005576 | Extracellular region | 15 | 0.02875 | CC | GO:0006935 | Chemotaxis | 6 | 0.01581 | BP | |
| GO:0005634 | Nucleus | 28 | 0.03452 | CC | GO:0006954 | Inflammatory response | 8 | 0.01581 | BP | |
| GO:0008285 | Negative regulation of cell proliferation | 7 | 0.03430 | BP | ||||||
| GO:0007275 | Multicellular organismal development | 16 | 0.03576 | BP | ||||||
| GO:0008009 | Chemokine activity | 7 | 0.00000 | MF | ||||||
| GO:0046870 | Cadmium ion binding | 3 | 0.00194 | MF | ||||||
| GO:0016779 | Nucleotidyl transferase activity | 5 | 0.02486 | MF | ||||||
| GO:0005576 | Extracellular region | 37 | 0.00000 | CC | ||||||
| GO:0005615 | Extracellular space | 14 | 2.0E-4 | CC | ||||||
| GO:0005634 | Nucleus | 56 | 7.0E-4 | CC | ||||||
| 1 | GO:0008201 | Heparin binding | 5 | 0.00265 | MF | GO:0008009 | Chemokine activity | 6 | 3.5E-4 | MF |
| GO:0003700 | Transcription factor activity | 13 | 0.00886 | MF | GO:0046870 | Cadmium ion binding | 3 | 0.00264 | MF | |
| GO:0005515 | Protein binding | 38 | 0.01716 | MF | GO:0003677 | DNA binding | 26 | 0.00264 | MF | |
| GO:0045766 | Positive regulation of angiogenesis | 3 | 0.01125 | BP | GO:0046872 | Metal ion binding | 36 | 0.01144 | MF | |
| GO:0001558 | Regulation of cell growth | 6 | 0.01502 | BP | GO:0008270 | Zinc ion binding | 34 | 0.02041 | MF | |
| GO:0006915 | Apoptosis | 8 | 0.02591 | BP | GO:0003674 | Molecular function | 15 | 0.02257 | MF | |
| GO:0008285 | Negative regulation of cell proliferation | 6 | 0.02591 | BP | GO:0003676 | Nucleic acid binding | 13 | 0.02257 | MF | |
| GO:0005634 | Nucleus | 36 | 0.00597 | CC | GO:0016779 | Nucleotidyl transferase activity | 4 | 0.02571 | MF | |
| GO:0005575 | Cellular component | 10 | 0.02160 | CC | GO:0005515 | Protein binding | 61 | 0.03204 | MF | |
| GO:0003704 | Specific RNA polymerase II transcription factor activity | 3 | 0.04080 | MF | ||||||
| GO:0009615 | Response to virus | 10 | 0.00000 | BP | ||||||
| GO:0006955 | Immune response | 18 | 0.00000 | BP | ||||||
| GO:0006355 | Regulation of transcription DNA-dependent | 39 | 4.0E-5 | BP | ||||||
| GO:0006350 | Transcription | 31 | 4.5E-4 | BP | ||||||
| GO:0008150 | Biological process | 18 | 0.00348 | BP | ||||||
| GO:0007267 | Cell-cell signaling | 11 | 0.00480 | BP | ||||||
| GO:0006954 | Inflammatory response | 8 | 0.03385 | BP | ||||||
| GO:0045087 | Innate immune response | 5 | 0.04274 | BP | ||||||
| GO:0005634 | Nucleus | 71 | 0.00000 | CC | ||||||
| GO:0005576 | Extracellular region | 37 | 1.1E-4 | CC | ||||||
| GO:0005615 | Extracellular space | 13 | 0.00474 | CC | ||||||
| GO:0005622 | Intracellular | 31 | 0.01344 | CC | ||||||
| GO:0005575 | Cellular component | 15 | 0.03381 | CC | ||||||
| 2 | GO:0003700 | Transcription factor activity | 18 | 1.4E-4 | MF | GO:0009615 | Response to virus | 10 | 0.00000 | BP |
| GO:0008201 | Heparin binding | 5 | 0.00193 | MF | GO:0006955 | Immune response | 16 | 0.00000 | BP | |
| GO:0043565 | Sequence-specific DNA binding | 10 | 0.01819 | MF | GO:0008150 | Biological process | 18 | 6.6E-4 | BP | |
| GO:0008083 | Growth factor activity | 5 | 0.01885 | MF | GO:0007267 | Cell-cell signaling | 10 | 0.01111 | BP | |
| GO:0005178 | Integrin binding | 3 | 0.02722 | MF | GO:0006954 | Inflammatory response | 8 | 0.01911 | BP | |
| GO:0008134 | Transcription factor binding | 4 | 0.02722 | MF | GO:0045087 | Innate immune response | 5 | 0.02866 | BP | |
| GO:0008009 | Chemokine activity | 3 | 0.02849 | MF | GO:0007565 | Female pregnancy | 5 | 0.03928 | BP | |
| GO:0046872 | Metal ion binding | 23 | 0.04806 | MF | GO:0005576 | Extracellular region | 36 | 1.0E-5 | CC | |
| GO:0045944 | Positive regulation of transcription from RNA polymerase II promoter | 7 | 0.00234 | BP | GO:0005615 | Extracellular space | 13 | 0.00113 | CC | |
| GO:0006955 | Immune response | 10 | 0.00470 | BP | GO:0005634 | Nucleus | 51 | 0.00899 | CC | |
| GO:0008285 | Negative regulation of cell proliferation | 7 | 0.00681 | BP | GO:0046870 | Cadmium ion binding | 3 | 0.01145 | MF | |
| GO:0000122 | Negative regulation of transcription from RNA polymerase II promoter | 6 | 0.00681 | BP | GO:0016831 | Carboxy-lyase activity | 3 | 0.02198 | MF | |
| GO:0006915 | Apoptosis | 9 | 0.00713 | BP | GO:0030674 | Protein binding bridging | 4 | 0.04373 | MF | |
| GO:0006954 | Inflammatory response | 7 | 0.00769 | BP | ||||||
| GO:0001558 | Regulation of cell growth | 5 | 0.00914 | BP | ||||||
| GO:0009611 | Response to wounding | 3 | 0.01457 | BP | ||||||
| GO:0005615 | Extracellular space | 12 | 8E-5 | CC | ||||||
| GO:0005634 | Nucleus | 42 | 2.4E-4 | CC | ||||||
| GO:0005576 | Extracellular region | 22 | 4E-4 | CC | ||||||
| GO:0030173 | Integral to Golgi membrane | 3 | 0.02101 | CC | ||||||
| 4 | GO:0008083 | Growth factor activity | 8 | 1.0E-5 | MF | GO:0009615 | Response to virus | 12 | 0.00000 | BP |
| GO:0005125 | Cytokine activity | 6 | 3.7E-4 | MF | GO:0007565 | Female pregnancy | 9 | 1.6E-4 | BP | |
| GO:0046983 | Protein dimerization activity | 6 | 0.00123 | MF | GO:0006955 | Immune response | 17 | 5.0E-4 | BP | |
| GO:0005100 | Rho GTPase activator activity | 3 | 0.00268 | MF | GO:0001525 | Angiogenesis | 7 | 0.02671 | BP | |
| GO:0008201 | Heparin binding | 4 | 0.00826 | MF | GO:0007267 | Cell-cell signaling | 10 | 0.02671 | BP | |
| GO:0003700 | Transcription factor activity | 13 | 0.00826 | MF | GO:0008150 | Biological process | 19 | 0.02928 | BP | |
| GO:0008047 | Enzyme activator activity | 3 | 0.01045 | MF | GO:0016477 | Cell migration | 5 | 0.03984 | BP | |
| GO:0005178 | Integrin binding | 3 | 0.01447 | MF | GO:0005576 | Extracellular region | 45 | 0.00000 | CC | |
| GO:0016563 | Transcription activator activity | 4 | 0.02237 | MF | GO:0005577 | Fibrinogen complex | 3 | 6.0E-4 | CC | |
| GO:0005515 | Protein binding | 33 | 0.03960 | MF | GO:0005615 | Extracellular space | 14 | 0.00843 | CC | |
| GO:0043565 | Sequence-specific DNA binding | 8 | 0.03960 | MF | GO:0031093 | Platelet α granule lumen | 4 | 0.01203 | CC | |
| GO:0006955 | Immune response | 11 | 2.4E-4 | BP | GO:0016020 | Membrane | 61 | 0.03962 | CC | |
| GO:0006915 | Apoptosis | 9 | 0.00440 | BP | GO:0005794 | Golgi apparatus | 15 | 0.03962 | CC | |
| GO:0030183 | B cell differentiation | 3 | 0.01798 | BP | ||||||
| GO:0045944 | Positive regulation of transcription from RNA polymerase II promoter | 5 | 0.03323 | BP | ||||||
| GO:0000079 | Regulation of cyclin-dependent protein kinase activity | 3 | 0.03704 | BP | ||||||
| GO:0007050 | Cell cycle arrest | 4 | 0.03704 | BP | ||||||
| GO:0008284 | Positive regulation of cell proliferation | 5 | 0.03704 | BP | ||||||
| GO:0007267 | Cell-cell signaling | 6 | 0.03704 | BP | ||||||
| GO:0001558 | Regulation of cell growth | 5 | 0.0407 | BP | ||||||
| 6 | GO:0005515 | Protein binding | 65 | 0.00000 | MF | GO:0046870 | Cadmium ion binding | 3 | 0.00135 | MF |
| GO:0003700 | Transcription factor activity | 24 | 1.0E-5 | MF | GO:0003674 | Molecular function | 13 | 0.00852 | MF | |
| GO:0008083 | Growth factor activity | 8 | 3.5E-4 | MF | GO:0008009 | Chemokine activity | 4 | 0.00852 | MF | |
| GO:0003714 | Transcription co-repressor activity | 7 | 3.5E-4 | MF | GO:0003950 | NAD+ADP-ribosyl transferase activity | 3 | 0.01169 | MF | |
| GO:0005125 | Cytokine activity | 8 | 3.5E-4 | MF | GO:0030674 | Protein binding bridging | 3 | 0.04041 | MF | |
| GO:0005100 | Rho GTPase activator activity | 4 | 3.5E-4 | MF | GO:0009615 | Response to virus | 12 | 0.00000 | BP | |
| GO:0003700 | Transcription factor activity | 7 | 6.2E-4 | MF | GO:0006955 | Immune response | 16 | 0.00000 | BP | |
| GO:0046983 | Protein dimerization activity | 7 | 6.9E-4 | MF | GO:0007565 | Female pregnancy | 9 | 0.00000 | BP | |
| GO:0008270 | Zinc ion binding | 32 | 0.00504 | MF | GO:0008150 | Biological process | 14 | 0.00612 | BP | |
| GO:0046872 | Metal ion binding | 32 | 0.00827 | MF | GO:0007267 | Cell-cell signaling | 8 | 0.00676 | BP | |
| GO:0005085 | Guanyl-nucleotide exchange factor activity | 5 | 0.02023 | MF | GO:0006952 | Defense response | 5 | 0.00728 | BP | |
| GO:0043565 | Sequence-specific DNA binding | 11 | 0.03272 | MF | GO:0030168 | Platelet activation | 3 | 0.01864 | BP | |
| GO:0008201 | Heparin binding | 4 | 0.03502 | MF | GO:0051258 | Protein polymerization | 3 | 0.03966 | BP | |
| GO:0005178 | Integrin binding | 3 | 0.04652 | MF | GO:0005576 | Extracellular region | 44 | 0.00000 | CC | |
| GO:0006915 | Apoptosis | 13 | 0.00173 | BP | GO:0005615 | Extracellular space | 13 | 6.0E-5 | CC | |
| GO:0006950 | Response to stress | 7 | 0.00173 | BP | GO:0005577 | Fibrinogen complex | 3 | 6.0E-5 | CC | |
| GO:0007050 | Cell cycle arrest | 6 | 0.00788 | BP | GO:0031093 | Platelet α granule lumen | 4 | 8.1E-4 | CC | |
| GO:0045944 | Positive regulation of transcription from RNA polymerase II promoter | 7 | 0.01021 | BP | ||||||
| GO:0045740 | Positive regulation of DNA replication | 3 | 0.01720 | BP | ||||||
| GO:0008360 | Regulation of cell shape | 4 | 0.02121 | BP | ||||||
| GO:0008285 | Negative regulation of cell proliferation | 8 | 0.02486 | BP | ||||||
| GO:0000122 | Negative regulation of transcription from RNA polymerase II promoter | 7 | 0.02486 | BP | ||||||
| GO:0009611 | Response to wounding | 3 | 0.02698 | BP | ||||||
| GO:0030183 | B cell differentiation | 3 | 0.02698 | BP | ||||||
| GO:0007229 | Integrin-mediated signaling pathway | 5 | 0.02698 | BP | ||||||
| GO:0006954 | Inflammatory response | 7 | 0.02698 | BP | ||||||
| GO:0007179 | Transforming growth factor β receptor signaling pathway | 4 | 0.02698 | BP | ||||||
| GO:0043066 | Negative regulation of apoptosis | 4 | 0.02698 | BP | ||||||
| GO:0006935 | Chemotaxis | 5 | 0.04499 | BP | ||||||
| GO:0007010 | Cytoskeleton organization and biogenesis | 5 | 0.04841 | BP | ||||||
| GO:0006955 | Immune response | 10 | 0.04843 | BP | ||||||
| GO:0005576 | Extra cellular region | 31 | 9.0E-5 | CC | ||||||
| GO:0005615 | Extra cellular space | 14 | 6.6E-4 | CC | ||||||
| GO:0005622 | Intracellular | 29 | 0.00843 | CC | ||||||
| GO:0005737 | Cytoplasm | 40 | 0.03660 | CC | ||||||
| Time point | 0.5 h | 1 h | 2 h | 4 h | 6 h |
| Gene mapping | |||||
| Up-regulation | CAM | P53 | MAPK | CAM | CAM |
| MAPK | TGF | ECHP | CY-CY | CY-CY | |
| P53 | MAPK | RCC | MAPK | JAK-STA | |
| TGF | CCC | P53 | JAK-STA | MAPK | |
| Down-regulation | APP | APP | APP | Phos | APP |
| Toll | CY-CY | CY-CY | APP | CY-CY | |
| CY-CY | Toll | Toll | Toll | Toll | |
| NKMC | Mela | Mela | Mela | Mela |
Relative expression levels of each time point were consistent with that of the microarray profile except at 0.5 h, for which a little higher fold-change was obtained in microarray data.
Some previous studies have reported that H. pylori type I strains that harbor the cag pathogenicity island (PAI) and cagA are associated with increased bacterial virulence and a more severe inflammatory response in gastric epithelial cells. These virulence factors have also been considered to be associated with induction of interleukin through an NF-κB-dependent pathway in host mucosa[16]. In addition, host protein phosphorylation, cytoskeletal rearrangement, and differential activation of MAP kinases have been described in host cells after infection of type I strains[1]. Although CagA and Cag PAI are considered to be factors highly involved in the development of gastritis and carcinoma, more complex as yet undiscovered mechanisms may exist between H. pylori and host cells. We aimed to take a global view of gene expression profiles of host response to infection in a time-series interaction model, which may help understand the pathogenesis of H. pylori related diseases.
Considering that only genes with fold changes > 1.5 were included in the analysis, the number of differential genes was only 808. This may lead to an ignorance for many important genes. Therefore, we initiated co-expression clustering analysis using STEM for both the 3577 differentially expressed genes (dataset1) and 808 genes (dataset2) with fold-change > 1.5. For the 808 genes, four significant clusters showed four different co-expression profiles (Figure 2). One hundred and twenty-six genes down-regulated at 4 h were clustered into profile 123, but no significant GO terms were enriched for these genes. In profile 3, some genes related to tumors were consistently down-regulated. For instance, cdkn1c had consistently decreased expression of theses genes, which may be involved in promotion of tumor formation. Profile 144 was mainly involved in factors regulating cell bioactivity and morphology such as rflb, gdf15, sqstm1 and adm2. DNA-damage-inducible transcript and csf2 also had increased gene expression at 4 and 6 h, suggesting that some potential mechanisms for cell differentiation and damage may be triggered beginning at 4 h. Hierarchically clustered results also showed two gene clusters with down-regulation at 4 h and up-regulation at 6 h. Analysis of all differentially expressed genes showed four interesting profiles whose GO distributions included nucleic acid binding, regulation of transcription, oxido-reductase activity etc. For the GO distribution of dataset1, profile 71 and profile 83 showed a similar co-expression profile as well as GO terms including nucleus, nucleic acid binding etc. (Table 3, Figure 3B and D). However, profile 83 showed an obvious and continuous up- regulated gene cluster. Profile 111 and 108 mainly focused on cell surface and showed a down-regulated gene cluster (Figure 3A and C). All profiles illustrated an obvious expressional change at 4 h. Statistically significant changes in gene ontology at each time point showed that apoptosis appeared from 1 h in up-regulated genes. At the same time, in down-regulated genes, chemokine activity became the most significant term (Table 4). This seemed consistent with results of the pathway analysis, which showed that the P53 signaling pathway became the most significantly perturbed pathway at 1 h in up-regulated genes. In down-regulated genes, the cytokine-cytokine receptor interaction pathway became more significant. Genes involving immune response and other responses to viruses were at the top of the GO list of down-regulated genes. This suggested an inhibition of immune response by H. pylori during early infection. Tumor-related pathways like P53 and MAPK may play an important role in determining the development of special phenotype and disease outcomes according to the results of pathway analysis. For the top 80 differentially expressed genes, 43 (54%) were related to immunity (29, 36%) and tumor development (14, 18%). Many immune factor-related down-regulated genes showed a consistently increasing expression levels. The cell adhesion molecules (CAM) pathway was the most significantly perturbed pathway at several time-points. The increased expression of CAM induced by H. pylori may contribute to cell adhesion, invasion and cell proliferation in gastric epithelial cells[17].
From the reconstructed simplified pathway, we can inspect some important nodes with several interaction edges like stat1, stat2, fos, csf2, pdgfb and ccl5 genes. These genes may be the trigger and linker of the pathway net during early infection, which however requires further studies. From Figure 4 and the expression value of each gene, we could learn that most immunity-related genes were down-regulated while many tumor-related genes were up-regulated. Il-24 is an important oncogene and could inhibit specifically the tumor growth. The protein encoded by this gene can induce apoptosis selectively in various cancer cells. Overexpression of this gene has been shown to lead to elevated expression of several GADD family genes, which correlates with the induction of apoptosis[18-20]. In this study, we examined il-24 levels which gradually increased more than two-fold from 2 to 6 h. At 6 h, there was a ten-fold change, indicating that after perturbation of P53 and MAPK, il-24 may participate in maintaining the immune defense against invading pathogens. We also examined an increased level of gadd45 which can stimulate DNA excision-repair in vitro and inhibits entry of cells into S phase. This gene is a member of a group of genes whose transcript levels are increased following stressful growth arrest and treatment with DNA-damaging agents. In the network, both c-Fos and c-Jun, two genes considered to mediate inflammation and carcinogenesis, have been found to be up-regulated, which is consistent with the results of this study[21].
We also analyzed expression profiles of some other important infection-related genes that were reported previously and may play an important role in H. pylori-induced diseases, although these genes were not clustered into a special profile in this study using the current analytical tools. MMP is a mucosal matrix metalloproteinase. Previous studies have demonstrated elevated MMP-9 levels in H. pylori-infected gastric mucosa, and eradication of H. pylori can significantly decrease MMP9 expression levels consistently[22,23]. MMP1 has been the subject of studies of inflammatory gene profiles in gastric mucosa[2,24]. MMP7 has been reported to be up-regulated in gastric cancer tissues[25,26]. However, few studies have reported on MMP24. In this study, the profile of MMP24 showed a consistent and increased level from 1 to 6 h, which suggested a similar function with MMP9 during H. pylori infection. Some other genes with similar expression profiles are il-27ra, il-32, il-23a, il-11, il-8 and ccl20. This gene cluster showed down-regulation or no change at the first two or three time points and up-regulation in the last two or three time points. Il-29, ccl5, cxcl10 and cxcl11 showed a consistent down-regulation at all time points with high fold-change. Expression of these genes suggested that the immune defense system may be suppressed during the first 1 or 2 h of H. pylori infection and some tumor-related genes and pathways were activated. After this short interaction and competition for about 2 h, the immune defense system may have regained the advantage with increasing expression levels of inflammatory and tumor suppressor factors. CagA translocation might occur 30 min after infection and may be at its maximum level in a time range of about 4-5 h[27,28]. In this study, the differentially expressed genes significantly increased at the time point of 4 h. This also suggested that it might be an important turning point between infection and host response. Although a model system of the AGS cell line infected with H. pylori was used to explore the host response[5,29], it should be noted that this is an isolated cell culture system, and cannot account for the varied effects of conditions in a human stomach. Therefore, the speculation generated from this study represents a valuable, but a simplified view of the situation. More researches are required to confirm these findings. In addition, we also compared our results with the genes with significant change after H. pylori infection in another report[30]. Several genes in that report are consistent with our results in dataset1 like socs2, stat6, ccl4, cxcl2, hla-dma, hsph1, plat, ifitm1, alox5, tlr4, faim3, cd47, ifngr1 and il8.
Only part of these genes showed a high fold change > 1.5 in differential expressions, including il8, faim3, tlr4, alox5, hla-dma, cxcl2 and ccl4.
In summary, the results from this sequential expression microarray have extended previous studies that were limited to the comparison of normal and diseased tissues. We took a global view on the genes and pathway net related to H. pylori infection, several co-expressional profiles and important new genes like mmp24 and il-24 involved in immune response and tumorigenesis during H. pylori infection were also identified. Our study also suggested that the outcome of H. pylori infection is probably involved in a complex mechanism, and is associated with a number of immune factors. Formation of tumors may be a result of an imbalance between bacterial attack and immune defense of host. We speculate that this competition may occur at 1-2 h after infection, and 4 h may be a first time point at which the balance is upset.
It has been indicated that Helicobacter pylori (H. pylori) infection may highly contribute to gastritis and carcinogenesis in the past two decades since it was recovered from human gastric mucosa in 1983, and many studies have focused on identification of both bacterial factors and host determinants that may contribute to the pathogenic mechanism.
Gene expression microarray has been widely used in identifying genes associated with H. pylori infection and gastric tumor. However, the time-series gene expression profile of H. pylori infection remains unexplored. In this study, the authors extended the knowledge of the dynamic interaction between H. pylori and host mucosa using a high density human gene microarray and flexible bioinformatics analysis.
Several important genes that have not been reported previously and a pathway net related to H. pylori infection were discovered by the sequential microarrays. Based on the co-expressional profile analysis during infection, a new speculation for the pathogenic mechanism has been set up.
This study has provided a systemic view of expression profile of time-series H. pylori infected AGS cells. The new identified genes and pathway net as well as the hypothesis could help researchers in this field further understand the potential mechanism associated with H. pylori infection and carcinogenesis, and provide important information for prevention and control of H. pylori related diseases.
The scientific and innovative contents as well as readability in this manuscript reflect the advanced levels of the clinical and basic researches in gastroenterology both at home and abroad.
| 1. | Segal ED, Lange C, Covacci A, Tompkins LS, Falkow S. Induction of host signal transduction pathways by Helicobacter pylori. Proc Natl Acad Sci USA. 1997;94:7595-7599. |
| 2. | Wen S, Felley CP, Bouzourene H, Reimers M, Michetti P, Pan-Hammarström Q. Inflammatory gene profiles in gastric mucosa during Helicobacter pylori infection in humans. J Immunol. 2004;172:2595-2606. |
| 3. | Kim JM, Kim JS, Jung HC, Oh YK, Chung HY, Lee CH, Song IS. Helicobacter pylori infection activates NF-kappaB signaling pathway to induce iNOS and protect human gastric epithelial cells from apoptosis. Am J Physiol Gastrointest Liver Physiol. 2003;285:G1171-G1180. |
| 4. | Conlin VS, Curtis SB, Zhao Y, Moore ED, Smith VC, Meloche RM, Finlay BB, Buchan AM. Helicobacter pylori infection targets adherens junction regulatory proteins and results in increased rates of migration in human gastric epithelial cells. Infect Immun. 2004;72:5181-5192. |
| 5. | Bach S, Makristathis A, Rotter M, Hirschl AM. Gene expression profiling in AGS cells stimulated with Helicobacter pylori isogenic strains (cagA positive or cagA negative). Infect Immun. 2002;70:988-992. |
| 6. | DeRisi J, Penland L, Brown PO, Bittner ML, Meltzer PS, Ray M, Chen Y, Su YA, Trent JM. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet. 1996;14:457-460. |
| 7. | Marchet A, Mocellin S, Belluco C, Ambrosi A, DeMarchi F, Mammano E, Digito M, Leon A, D'Arrigo A, Lise M. Gene expression profile of primary gastric cancer: towards the prediction of lymph node status. Ann Surg Oncol. 2007;14:1058-1064. |
| 8. | Wu CM, Lee YS, Wang TH, Lee LY, Kong WH, Chen ES, Wei ML, Liang Y, Hwang TL. Identification of differential gene expression between intestinal and diffuse gastric cancer using cDNA microarray. Oncol Rep. 2006;15:57-64. |
| 9. | Lim JW, Kim H, Kim KH. Cell adhesion-related gene expression by Helicobacter pylori in gastric epithelial AGS cells. Int J Biochem Cell Biol. 2003;35:1284-1296. |
| 10. | Chang YT, Wu MS, Chang YJ, Chen CC, Lin YS, Hsieh T, Yang PC, Lin JT. Distinct gene expression profiles in gastric epithelial cells induced by different clinical isolates of Helicobacter pylori--implication of bacteria and host interaction in gastric carcinogenesis. Hepatogastroenterology. 2006;53:484-490. |
| 11. | Chudin E, Kruglyak S, Baker SC, Oeser S, Barker D, McDaniel TK. A model of technical variation of microarray signals. J Comput Biol. 2006;13:996-1003. |
| 12. | Draghici S, Khatri P, Bhavsar P, Shah A, Krawetz SA, Tainsky MA. Onto-Tools, the toolkit of the modern biologist: Onto-Express, Onto-Compare, Onto-Design and Onto-Translate. Nucleic Acids Res. 2003;31:3775-3781. |
| 13. | Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, Georgescu C, Romero R. A systems biology approach for pathway level analysis. Genome Res. 2007;17:1537-1545. |
| 14. | Ernst J, Bar-Joseph Z. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics. 2006;7:191. |
| 15. | Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M. STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412-D416. |
| 16. | Censini S, Lange C, Xiang Z, Crabtree JE, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA. 1996;93:14648-14653. |
| 17. | Chen X, Leung SY, Yuen ST, Chu KM, Ji J, Li R, Chan AS, Law S, Troyanskaya OG, Wong J. Variation in gene expression patterns in human gastric cancers. Mol Biol Cell. 2003;14:3208-3215. |
| 18. | Papathanasiou MA, Kerr NC, Robbins JH, McBride OW, Alamo I Jr, Barrett SF, Hickson ID, Fornace AJ Jr. Induction by ionizing radiation of the gadd45 gene in cultured human cells: lack of mediation by protein kinase C. Mol Cell Biol. 1991;11:1009-1016. |
| 19. | Jackman J, Alamo I Jr, Fornace AJ Jr. Genotoxic stress confers preferential and coordinate messenger RNA stability on the five gadd genes. Cancer Res. 1994;54:5656-5662. |
| 20. | Carrier F, Smith ML, Bae I, Kilpatrick KE, Lansing TJ, Chen CY, Engelstein M, Friend SH, Henner WD, Gilmer TM. Characterization of human Gadd45, a p53-regulated protein. J Biol Chem. 1994;269:32672-32677. |
| 21. | Hofman VJ, Moreilhon C, Brest PD, Lassalle S, Le Brigand K, Sicard D, Raymond J, Lamarque D, Hébuterne XA, Mari B. Gene expression profiling in human gastric mucosa infected with Helicobacter pylori. Mod Pathol. 2007;20:974-989. |
| 22. | Lee LY, Wu CM, Wang CC, Yu JS, Liang Y, Huang KH, Lo CH, Hwang TL. Expression of matrix metalloproteinases MMP-2 and MMP-9 in gastric cancer and their relation to claudin-4 expression. Histol Histopathol. 2008;23:515-521. |
| 23. | Kubben FJ, Sier CF, Schram MT, Witte AM, Veenendaal RA, van Duijn W, Verheijen JH, Hanemaaijer R, Lamers CB, Verspaget HW. Eradication of Helicobacter pylori infection favourably affects altered gastric mucosal MMP-9 levels. Helicobacter. 2007;12:498-504. |
| 24. | Pillinger MH, Marjanovic N, Kim SY, Lee YC, Scher JU, Roper J, Abeles AM, Izmirly PI, Axelrod M, Pillinger MY. Helicobacter pylori stimulates gastric epithelial cell MMP-1 secretion via CagA-dependent and -independent ERK activation. J Biol Chem. 2007;282:18722-18731. |
| 25. | Ogden SR, Wroblewski LE, Weydig C, Romero-Gallo J, O'Brien DP, Israel DA, Krishna US, Fingleton B, Reynolds AB, Wessler S, Peek RM Jr. p120 and Kaiso regulate Helicobacter pylori-induced expression of matrix metalloproteinase-7. Mol Biol Cell. 2008;19:4110-4121. |
| 26. | Wei J, O'Brien D, Vilgelm A, Piazuelo MB, Correa P, Washington MK, El-Rifai W, Peek RM, Zaika A. Interaction of Helicobacter pylori with gastric epithelial cells is mediated by the p53 protein family. Gastroenterology. 2008;134:1412-1423. |
| 27. | Asahi M, Azuma T, Ito S, Ito Y, Suto H, Nagai Y, Tsubokawa M, Tohyama Y, Maeda S, Omata M. Helicobacter pylori CagA protein can be tyrosine phosphorylated in gastric epithelial cells. J Exp Med. 2000;191:593-602. |
| 28. | Odenbreit S, Gebert B, Püls J, Fischer W, Haas R. Interaction of Helicobacter pylori with professional phagocytes: role of the cag pathogenicity island and translocation, phosphorylation and processing of CagA. Cell Microbiol. 2001;3:21-31. |
| 29. | Cho SO, Lim JW, Kim KH, Kim H. Involvement of Ras and AP-1 in Helicobacter pylori-Induced Expression of COX-2 and iNOS in Gastric Epithelial AGS Cells. Dig Dis Sci. 2009;Epub ahead of print. |
| 30. | Kim KK, Kim HB. Protein interaction network related to Helicobacter pylori infection response. World J Gastroenterol. 2009;15:4518-4528. |
Peer reviewer: Dr. Yutao Yan, Medicine Department, Emory University, 615 Michael ST, Whitehead Building/265, Atlanta, GA 30322, United States
S- Editor Wang YR L- Editor Ma JY E- Editor Lin YP