Published online Nov 21, 2009. doi: 10.3748/wjg.15.5442
Revised: October 15, 2009
Accepted: October 21, 2009
Published online: November 21, 2009
AIM: To develop short hairpin RNA (shRNA) against heparanase, and to determine its effects on heparanase expression and the malignant characteristics of gastric cancer cells.
METHODS: Heparanase-specific shRNA was constructed and transferred into cultured the gastric cancer cell line SGC-7901. Stable subclonal cells were screened by G418 selection. Heparanase expression was measured by reverse transcriptase-polymerase chain reaction (RT-PCR), real-time quantitative PCR and Western blotting. Cell proliferation was detected by 2-(4,5-dimethyltriazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) colorimetry and colony formation assay. The in vitro invasiveness and metastasis of cancer cells were measured by cell adhesion assay, wound healing assay and matrigel invasion assay. The angiogenesis capabilities of cancer cells were measured by tube formation of endothelial cells.
RESULTS: Stable transfection of heparanase-specific shRNA, but not of scrambled shRNA and mock vector, resulted in reduced mRNA and protein levels of heparanase. The shRNA-mediated knockdown of heparanase did not affect the cellular proliferation of SGC-7901 cells. However, the in vitro invasiveness and metastasis of cancer cells were decreased after knockdown of heparanase. Moreover, transfection of heparanase-specific shRNA decreased the in vitro angiogenesis capabilities of SGC-7901 cells.
CONCLUSION: Stable knockdown of heparanase can efficiently decrease the invasiveness, metastasis and angiogenesis of human gastric cancer cells. In contrast, stable knockdown of heparanase does not affect the cell proliferation.
-
Citation: Zheng LD, Jiang GS, Pu JR, Mei H, Dong JH, Hou XH, Tong QS. Stable knockdown of heparanase expression in gastric cancer cells
in vitro . World J Gastroenterol 2009; 15(43): 5442-5448 - URL: https://www.wjgnet.com/1007-9327/full/v15/i43/5442.htm
- DOI: https://dx.doi.org/10.3748/wjg.15.5442
Gastric cancer, a serious health problem, remains the second most common type of fatal cancer worldwide because of the invasiveness and metastasis of cancer cells[1,2]. Metastasis of tumor cells involves a multistep process, including detachment, invasion, migration, angiogenesis, adhesion to endothelial cells, extravasation, and regrowth in distant organs[3]. It has been established that the basement membrane (BM) and extracellular matrix (ECM) act as a barrier to prevent tumor cells from invasion and metastasis[4]. Heparanase (HPA), a mammalian endo-beta-D-glucuronidase, is capable of degrading the heparan sulfate proteoglycans (HSPGs) in the ECM and BM, and is proposed to have a promoting role in cancer invasion, angiogenesis and metastasis[5]. In advanced gastric cancer, expression of HPA was frequently observed and correlated with histopathological parameters reflecting the invasiveness, metastatic potential and prognosis of gastric cancers[6,7]. Thus HPA may represent an important target for treatment of gastric cancer.
Currently, a variety of HPA inhibitors have been developed, including competitive heparin sulfate-mimicking compounds, neutralizing anti-HPA antibodies, modified non-anticoagulant species of heparin, and inhibitory small molecules[8]. It has been reported that inhibiting the expression of HPA can lead to inhibition of tumor invasiveness, metastasis and angiogenesis[9,10]. However, because of the multiple biologic activities of these compounds, the mechanism of their antitumor activity and its relation to HPA inhibition are not straightforward[8-11]. Moreover, the pleiotropic interactions of these compounds with the ECM and the cell surface might produce nonspecific and undesirable effects[8-11]. Thus, novel approaches are needed to reduce the role of HPA in cancer progression[8].
In previous studies, genetic approaches targeting HPA have been regarded as a promising alternative. Antisense oligonucleotides, ribozymes, and short interference RNA (siRNA) have been developed to suppress HPA expression[12-15]. However, it still remains unknown whether stable transfection of short hairpin RNA (shRNA) can knockdown the HPA expression and decrease the invasiveness and metastasis of cancer cells. In the current study, the HPA-specific shRNA was constructed and transferred into cultured gastric cancer cells. We demonstrated, for the first time, that stable knockdown of HPA expression decreased the in vitro invasive, metastatic and angiogenetic capabilities of gastric cancer cells.
Human gastric cancer cell line SGC-7901 and human umbilical endothelial cell line (HUVEC) were purchased from the American Type Culture Collection and grown in RPMI1640 medium (Life Technologies, Inc., Gaithersburg, MD) supplemented with 10% fetal bovine serum (FBS, Life Technologies, Inc.), penicillin (100 U/mL) and streptomycin (100 g/mL). Cells were maintained at 37°C in a humidified atmosphere of 50 mL/L CO2.
Oligonucleotides encoding a shRNA specific for the heparanase encoding sequence were subcloned into pGenesil-1 (Genesil Biotechnology, Wuhan, China). Annealed oligonucleotides were cloned downstream of the U6 promoter (primer 1 sequence, 5'-GATCCGGAATCAACCTTTGAAGAGCTATGGACACCTTAGTTGGAAACTTCTCTTTTTTGAGCTCA-3'; primer 2 sequence, 5'-AGCTTGAGCTGAAAAAAGGAATCAACCTTTGAAGAGTGTCCATAGCCTTAGTTGGAAACTTCTCG-3'). The sequence 5'-GATCCAGCAUCGUACGUAGGCCA GCTATGGACATCGTAGCATGCATCCGGTCTTTTTTGAGCTCA-3' (sense), and 5'-AGCTTGAGCTGAAAAAAAGCAUCGUACGUAGGCCAGTGTCCATAGTCGTAGCATGCATCCGGTCG-3' (antisense) were used as a scrambled RNAi control. The constructs were verified by DNA sequencing. The plasmids pGenesil-1, pGenesil-scrambled, and pGenesil-HPA were transfected with Genesilencer Transfection Reagent (Genlantis, San Diego, CA), according to the manufacturer’s instructions. Stable cell lines were screened by administration of G418 (Invitrogen, Carlsbad, CA).
Total RNA was isolated with RNeasy Mini Kit (Qiagen Inc., Valencia, CA, USA). The reverse transcription reactions were conducted with Transcriptor First Strand cDNA Synthesis Kit (Roche, Indianapolis, IN, USA). The PCR primers were designed by Premier Primer 5.0 software as the following: for human HPA 5'-GAATGGACGGACTGCTAC-3'and 5'-CCAAAGAATACTTGCCTCA-3' amplifying a 261-bp fragment; for human GAPDH 5'-AGAAGGCTGGGGCTCATTT G-3' and 5'-AGGGGCCATCCACAGTCTTC-3' amplifying a 258-bp fragment. Real-time PCR with SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) was performed using ABI Prism 7700 Sequence Detector (Applied Biosystems). The fluorescent signals were collected during the extension phase, Ct values of the sample were calculated, and HPA transcript levels were analyzed by 2-ΔΔCt method.
Cellular protein was extracted with 1 × cell lysis buffer (Promega, Madison, WI, USA). Protein (50 μg) from each sample was subjected to 4%-20% pre-cast polyacrylamide gel (Bio-Rad, Hercules, CA, USA) electrophoresis and transferred to nitrocellulose membranes (Bio-Rad). For HPA (InSight Company, Rehovot, Israel) and GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA, USA) detection, the primary antibody dilutions were 1:500 and 1:1000, respectively, followed by 1:3000 dilution of goat anti-rabbit horseradish peroxidase-labeled antibody (Bio-Rad). ECL substrate kit (Amersham, Piscataway, NJ, USA) was used for the chemiluminescent detection of signals with autoradiographic film (Amersham).
Cell viability was monitored by the 2-(4,5-dimethyltriazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT, Sigma, St. Louis, MO, USA) colorimetric assay. Briefly, 20 μL of MTT (5 mg/mL) was added to each well. After 4 h incubation at 37°C, the cell supernatants were discarded, MTT crystals were dissolved with DMSO and the absorbance measured at 570 nm. Percent viability was defined as the relative absorbance of treated vs untreated control cells. All experiments were done with 6-8 wells per experiment and repeated at least 3 times.
The cells were seeded at a density of 300/mL on 35-mm dishes. Colonies were allowed to grow for 10-14 d. The medium was discarded and each well was carefully washed twice with phosphate buffered saline (PBS). The cells were fixed in methanol for 15 min, then stained with crystal violet for 20 min. Finally, positive colony formation (> 50 cells/colony) was counted. The survival fraction for cells was expressed as the ratio of plating efficiency of treated cells to that of untreated control cells.
2 × 104 cells were inoculated into each ECM-coated well of 96-well plates that were precoated with 100 μL of 20 μg/mL matrigel (BD Biosciences, Franklin Lakes, NJ, USA), and incubated at 37°C in serum-free complete medium (pH 7.2) for 2 h. After incubation, the wells were washed 3 times with PBS and the remaining cells were fixed in 4% paraformaldehyde for 20 min at room temperature. The cells were stained with 0.1% crystal violet and washed 3 times with PBS to remove free dye. After extraction with 10% acetic acid, absorbance of the samples was measured at 570 nm.
The cells were scraped with the fine end of 1-mL pipette tips (time 0). Plates were washed twice with PBS to remove detached cells, and incubated with the complete growth medium. Cell migration into the wound empty space was followed after 24 h and photographed.
The Boyden chamber technique (Transwell analysis) was performed. Briefly, the 8-μm pore size filters were coated with 100 μL of 1 mg/mL matrigel (dissolved in serum-free RPMI1640 medium). 600 μL of RPMI1640 medium containing 10% FBS was added to the lower chambers. Homogeneous single cell suspensions (1 × 105 cells/well) were added to the upper chambers and allowed to invade for 24 h at 37°C in a CO2 incubator. Migration was allowed to proceed for 24 h at 37°C. Cells remaining attached to the upper surface of the filters were carefully removed with cotton swabs. Migrated cells were stained with 0.1% crystal violet for 10 min at room temperature and examined by light microscopy. Quantification of migrated cells was performed according to published criteria[16].
Fifty microliters of growth factor-reduced matrigel was polymerized on 96-well plates. HUVEC cells were serum starved in RPMI1640 medium for 2 h. The endothelial cells were suspended in RPMI1640 medium preconditioned with subclonal SGC-7901 cells, added to the matrigel-coated wells at the density of 5 × 104 cells/well, and incubated at 37°C for 18 h. Tube formation was visualized using a Leitz inverted microscope equipped with a Sony color digital DXC-S500 camera. Quantification of antiangiogenic activity was calculated by measuring the length of tube walls formed between discrete endothelial cells in each well relative to the control.
Unless otherwise stated, all data are shown as mean ± standard error of the mean (SEM). Statistical significance (P < 0.05) was determined by the Student t-test or analysis of variance (ANOVA) followed by assessment of differences using SPSS 12.0 software (SPSS Inc., Chicago, IL, USA).
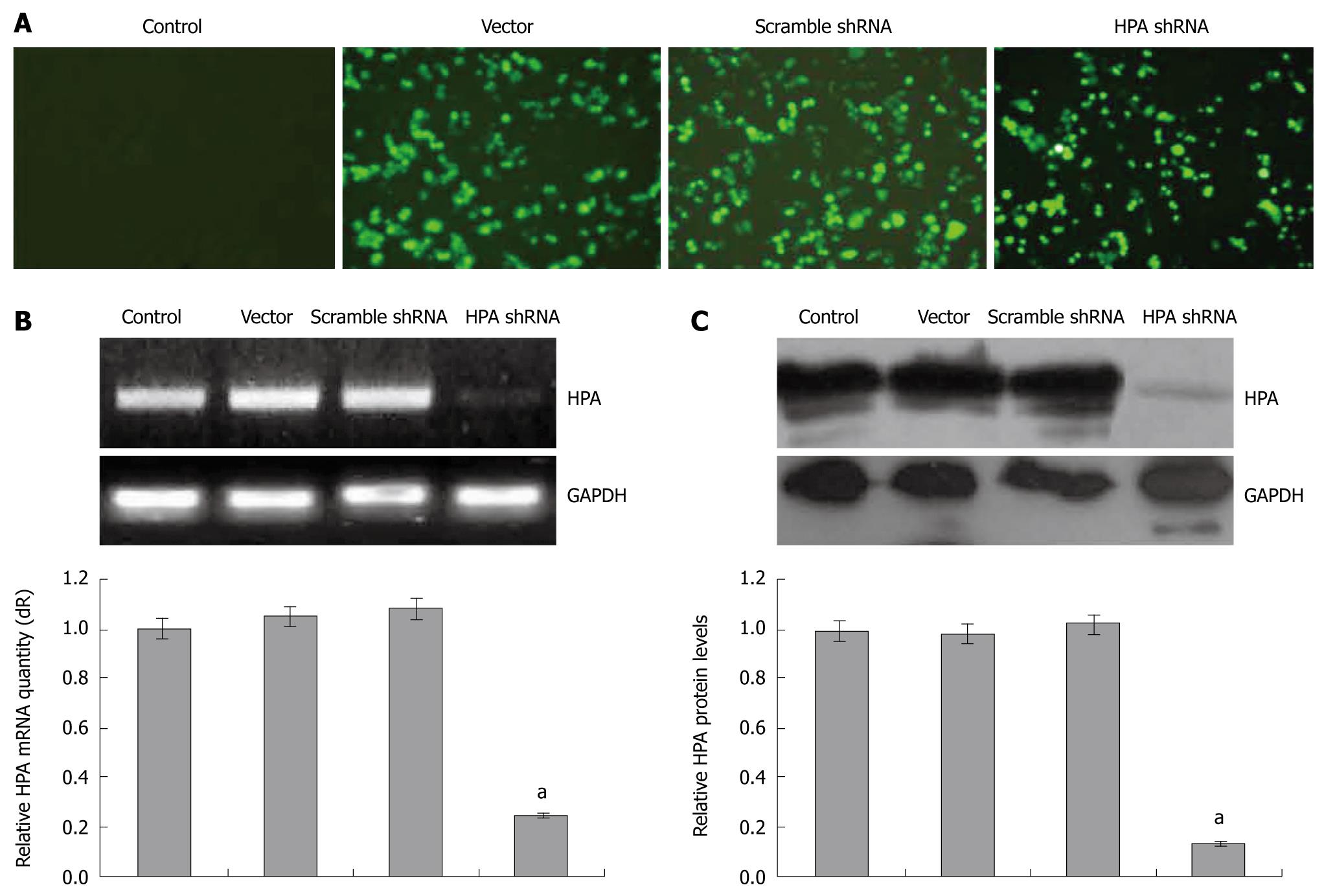
To examine the effects of shRNA on the expression of human HPA, gastric cancer SGC-7901 cells were transfected with pGenesil-1, pGenesil-scrambled, or pGenesil-HPA. The transfection efficiency was monitored by the enhanced green fluorescent protein (EGFP) reporter within these vectors. As shown in Figure 1A, 24 h after the Genesilencer-mediated transfection, EGFP was expressed within the cytoplasm of cancer cells. The subclonal cells were established by G418 selection. The mRNA and protein expression of HPA were examined by reverse transcriptase (RT)-PCR, real-time quantitative PCR and Western blot. As shown in Figure 1B and C, the HPA mRNA and protein could be detected in the parental cells, and stable transfection of the empty vector and scrambled shRNA did not affect the expression level of HPA. However, HPA was significantly decreased in the stable HPA shRNA-transfected cells. These results indicated that the HPA-specific shRNA used in this study was efficient in knockdown of the expression of HPA in gastric cancer cells.
The effects of stable transfection of HPA shRNA on cell proliferation of SGC-7901 cells were measured by MTT colorimetric assay. We found that stable transfection of HPA shRNA or scrambled shRNA, did not affect cell proliferation, when compared to the parental cells and mock group (Figure 2A). In addition, the colony formation assay further revealed that stable transfection of HPA shRNA did not influence the cell proliferation of cultured SGC-7901 cells (Figure 2B). These results indicated that knockdown of HPA did not affect the in vitro cell proliferation of gastric cancer cells.
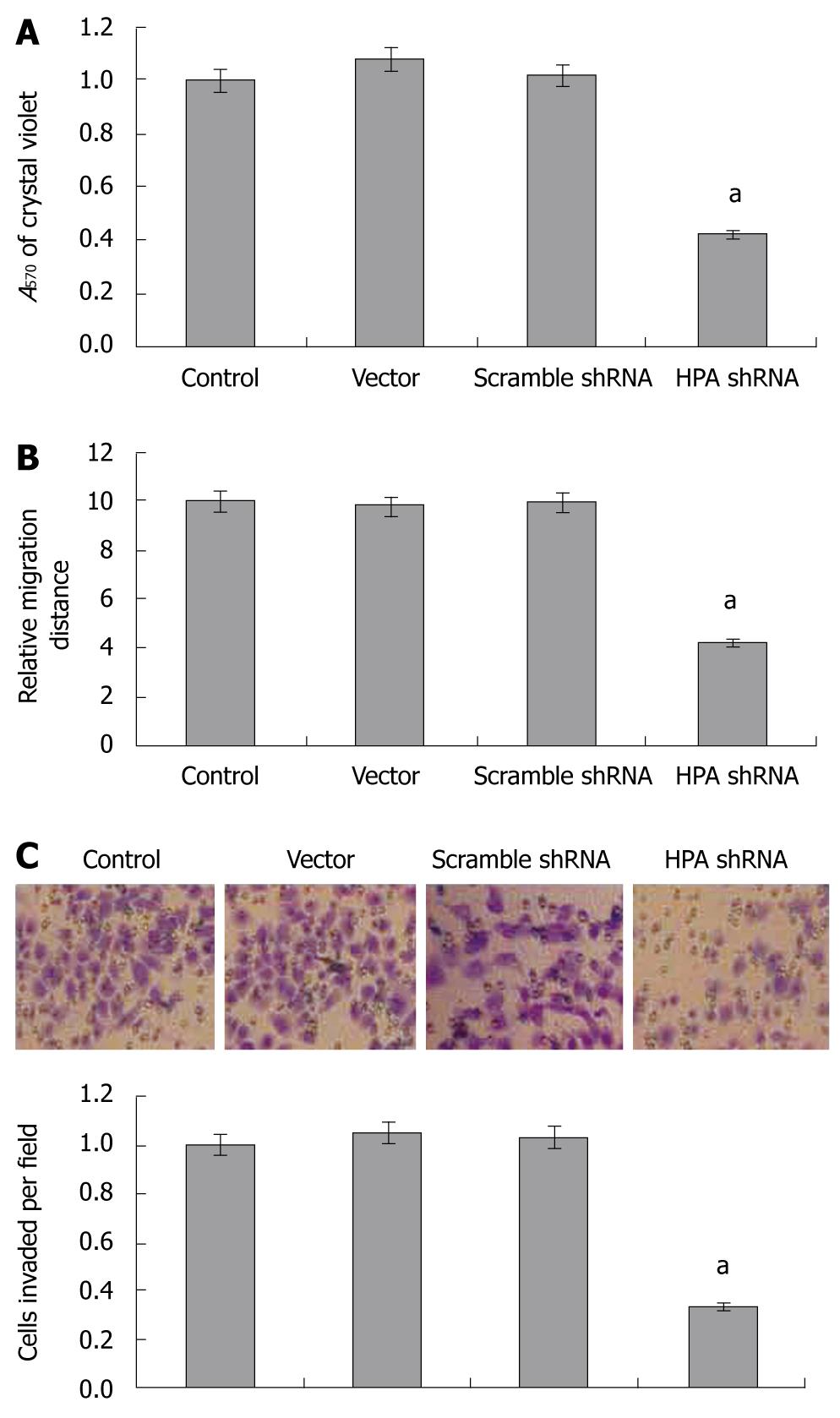
As cell adhesion, migration and invasiveness are 3 critical steps involved in metastasis, and combined with the evidence that HPA plays critical roles in invasiveness and metastasis of cancer cells, we examined the effects of HPA shRNA on these characteristics of gastric cancer cells. In the adhesion assay, SGC-7901 cells stably transfected with HPA shRNA exhibited markedly reduced ability to adhere to the precoated matrigel, when compared to parental cells and the mock group (Figure 3A). However, the cells stably transfected with scrambled shRNA and vector (mock) had similar ability to adhere as parental cells (Figure 3A). In addition, stable transfection of HPA shRNA into SGC-7901 cells resulted in an impaired migration capacity, when compared to the parental cells and mock group as evidenced by the scratch migration assay (Figure 3B). Moreover, stable transfection of HPA shRNA decreased the invasive capabilities of SGC-7901 cells, when compared to the parental cells and mock group as evidenced by transwell analysis (Figure 3C). These results suggested that HPA-specific shRNA decreased the adhesion, invasiveness and metastasis of gastric cancer cells in vitro.
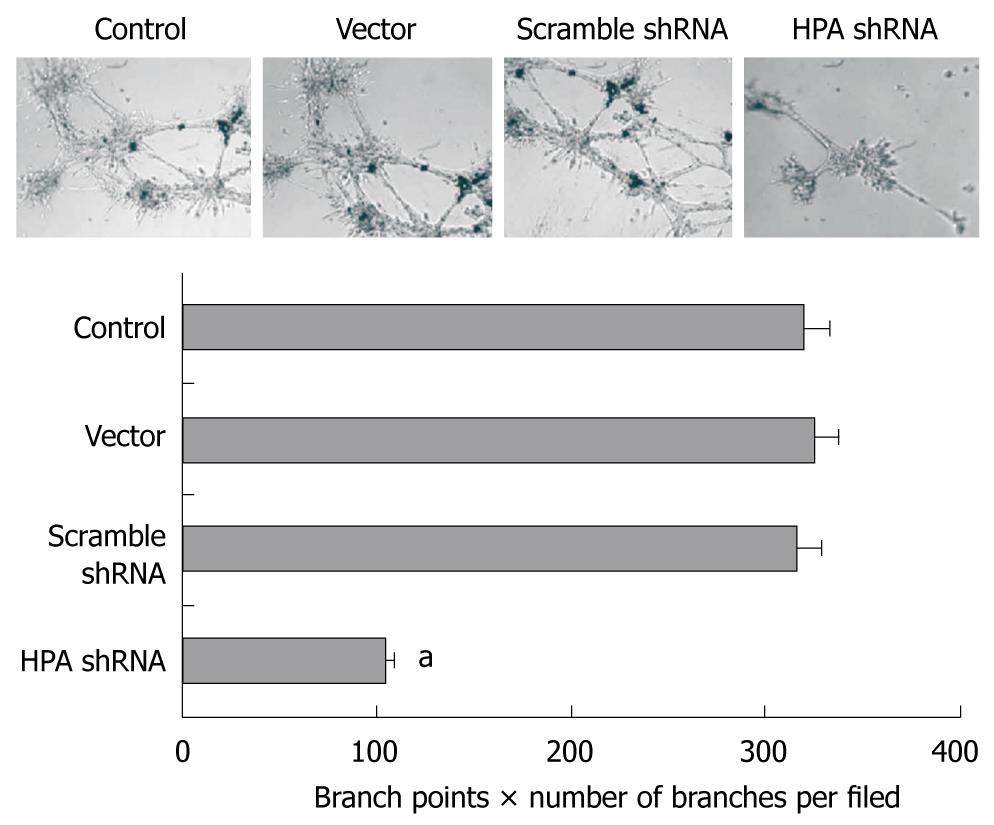
We further investigated the effects of HPA-specific shRNA on the in vitro angiogenesis capabilities of SGC-7901 cells. As shown in Figure 4, extensive tube formation of endothelial cells was observed in parental and mock cells. However, when the endothelial cells were treated with medium preconditioned with stable HPA shRNA-transfected SGC-7901 cells, tube formation was significantly decreased (Figure 4). These results indicate that transfection of HPA shRNA markedly decreased the angiogenesis of gastric cancer cells in vitro.
Invasiveness and metastasis is one of the characteristics of malignant cells[17]. Metastatic cancer cells invade parenchymal tissue and penetrate vascular channels to form satellite tumors in distant organs[17,18]. The ECM and BM provide a physical barrier to the migration of cancer cells[4]. HSPGs are the essential structural component of ECM and cell surfaces, and are composed of a protein core covalently linked to sulfated glycosaminoglycan[19]. The heparan sulfate side chains of HSPGs interact with other components of ECM, such as fibronectin, collagen, and laminin, to provide matrix assembly and stability[19]. HPA is the predominant enzyme responsible for degradation of heparan sulfate activity, which is involved in fundamental biologic processes associated with ECM remodeling and cell migration, such as development and morphogenesis, inflammation, angiogenesis, and cancer metastasis[20-22]. Many kinds of cells express HPA, such as metastatic tumor cells, proliferating endothelial cells, and activated leukocytes[23]. Numerous studies have indicated that HPA is highly expressed in tumors at both the mRNA and protein levels[24]. In addition, HPA plays an important role in sustaining the pathology of malignant tumors[24]. Expression of HPA is associated with the aggressiveness of tumor cell lines[24]. Increased levels of HPA were detected in the sera of animals bearing metastatic tumors, in the sera of patients with cancer, and in the urine of some patients with aggressive metastatic disease[25], reflecting an association between abnormal HPA secretion and metastatic disease. Thus, HPA is one of the targets for cancer therapy.
It has been established that many potent inhibitors, such as sulfated polysaccharides and heparin-mimicking polyanionic molecules, are capable of inhibiting HPA activity[8]. One representative of these inhibitors, PI-88, is being evaluated in a multicentre phase II clinical trial[26]. Heparin, other sulfated polysaccharides, and heparin-mimicking molecules that inhibit HPA enzymatic activity also reduce the incidence of metastasis in experimental animals[21]. However, because of the potential non-specific activity of these inhibitors and insufficient knowledge of the functional mechanisms of HPA, the application of HPA inhibitors in clinical trials may require further investigation, such as identification of the sugar residues in heparan sulfate adjacent to the HPA cleavage site and the crystallization and analysis of the 3-dimensional structure of the enzyme[8,27-30]. In hepatocellular carcinoma cell lines, antisense oligonucleotides decreased HPA expression and inhibited the invasiveness and angiogenesis of cancer cells[13]. Uno et al[31] constructed an adenoviral vector carrying an antisense full-length human HPA cDNA, and found that HPA expression, and the in vitro invasiveness and metastasis of human lung cancer A549 cells were specifically inhibited. Downregulation of HPA by overexpression of either anti-HPA ribozyme or RNA silencing vector has been shown to not only inhibit HPA activity but also impair tumor invasiveness and metastasis, resulting in improved survival of tumor-bearing mice[14,15]. In highly metastatic B16-BL6 mouse melanoma cells, transfection of mouse HPA-specific siRNA resulted in a decrease in the invasiveness and metastasis of tumor cells[14].
Previous studies indicated that the expression of HPA was frequently observed in advanced gastric cancers, and the frequency was significantly correlated with histopathological parameters reflecting invasive and metastatic potentials and prognosis of gastric cancers[6,7]. Our studies also demonstrated the overexpression of HPA protein in advanced gastric cancer (data not shown). However, it still remains largely unknown whether knockdown of HPA expression can decrease the invasiveness and metastasis of gastric cancer cells. In this study, we constructed HPA-specific shRNA and transfected into cultured SGC-7901 cells. The subclonal cells were generated for stable knockdown of HPA. We found that shRNA decreased HPA mRNA and protein in gastric cancer cells. Stable knockdown of HPA expression decreased the in vitro migration, invasiveness and metastasis of gastric cancer cells. However, stable knockdown of HPA expression did not affect the proliferation of SGC-7901 cells, which was consistent with previous findings in human SMMC7721 liver cancer cells and mouse B16 L6 melanoma cells[13,14].
Previous evidence showed that the cleavage of HSPGs by HPA may release heparan sulfate-bound cytokines and growth factors from cell surface or from the ECM, such as basic fibroblast growth factor and vascular endothelial growth factor[29,32]. Thus, HPA may facilitate tumor cell invasiveness and neovascularization, both critical steps in tumor progression[33]. A pronounced correlation between HPA expression and tumor microvessel density has been reported[34]. In this study, we found that the angiogenesis of gastric cancer cells in vitro was decreased by stable shRNA transfection. To our knowledge, the data described here represent the first successful application of shRNA-mediated gene silencing to stably reduce the levels of HPA in gastric cancer.
In summary, we have demonstrated that stable knockdown of HPA expression can efficiently inhibit the invasiveness, metastasis and angiogenesis of human gastric cancer cells. It is likely that the inhibition of HPA expression possibly depresses the degradation of ECM and BM, thus inhibiting the invasiveness and metastasis of gastric cancer. Therefore, our study suggests that HPA-specific shRNA may be of potential value as a novel therapeutic strategy in human gastric cancer. Combined with our identical results of stable transfection of HPA-specific shRNA into bladder cancer cell lines (data not shown), we believe that this HPA-specific shRNA may be also applicable for the therapies of other cancers overexpressing HPA.
Previous studies have indicated that the heparanase (HPA) is correlated with histopathological parameters and poor prognosis of gastric cancers. Although their efficiencies in inhibiting the expression of HPA, the traditional HPA inhibitors may produce nonspecific and undesirable effects. Thus, novel approaches are needed to inhibit the role of HPA in cancer progression.
In recent years, genetic approaches targeting HPA have been regarded as a promising alternative. Antisense oligonucleotides, ribozyme, and small RNA interference (siRNA) have been developed to decrease the HPA expression. However, it remains unknown whether stable transfection of short hairpin RNA (shRNA) can knockdown the HPA expression and decrease the invasiveness and metastasis of gastric cancer cells.
In the current study, the HPA-specific shRNA was constructed and transferred into cultured gastric cancer cells. The authors demonstrated, for the first time, that stable knockdown of HPA expression decreased the in vitro invasive, metastatic and angiogenetic capabilities of gastric cancer cells. However, stable knockdown of heparanase does not affect the proliferation of gastric cancer cells.
By understanding the effects of HPA-specific shRNA on the in vitro invasiveness, metastasis and angiogenesis capabilities of gastric cancer cells, this study may represent a future strategy for therapeutic intervention in the treatment of patients with gastric cancer. In addition, the HPA-specific shRNA may be also applicable for the therapies of other cancers overexpressing HPA.
HPA is a mammalian endo-b-D-glucuronidase that is capable of degrading the heparan sulfate proteoglycans in basement membrane and extracellular matrix, and plays important roles in cancer invasion, angiogenesis and metastasis.
The authors examined the effects of HPA-specific shRNA on the cultured gastric cancer cells. It revealed that stable transfection of HPA-specific shRNA decreased the mRNA and protein levels of HPA in SGC-7901 cells. The shRNA against HPA efficiently decreased the invasiveness, metastasis and angiogenesis of gastric cancer cells. The results are interesting and may represent a novel strategy for the treatment of gastric cancer.
| 1. | Terry MB, Gaudet MM, Gammon MD. The epidemiology of gastric cancer. Semin Radiat Oncol. 2002;12:111-127. |
| 2. | Li C, Oh SJ, Kim S, Hyung WJ, Yan M, Zhu ZG, Noh SH. Risk factors of survival and surgical treatment for advanced gastric cancer with large tumor size. J Gastrointest Surg. 2009;13:881-885. |
| 3. | Leber MF, Efferth T. Molecular principles of cancer invasion and metastasis (review). Int J Oncol. 2009;34:881-895. |
| 4. | Liotta LA, Stetler-Stevenson WG. Tumor invasion and metastasis: an imbalance of positive and negative regulation. Cancer Res. 1991;51:5054s-5059s. |
| 5. | Nakajima M, Irimura T, Di Ferrante N, Nicolson GL. Metastatic melanoma cell heparanase. Characterization of heparan sulfate degradation fragments produced by B16 melanoma endoglucuronidase. J Biol Chem. 1984;259:2283-2290. |
| 6. | Wang Z, Xu H, Jiang L, Zhou X, Lu C, Zhang X. Positive association of heparanase expression with tumor invasion and lymphatic metastasis in gastric carcinoma. Mod Pathol. 2005;18:205-211. |
| 7. | Takaoka M, Naomoto Y, Ohkawa T, Uetsuka H, Shirakawa Y, Uno F, Fujiwara T, Gunduz M, Nagatsuka H, Nakajima M. Heparanase expression correlates with invasion and poor prognosis in gastric cancers. Lab Invest. 2003;83:613-622. |
| 8. | Miao HQ, Liu H, Navarro E, Kussie P, Zhu Z. Development of heparanase inhibitors for anti-cancer therapy. Curr Med Chem. 2006;13:2101-2111. |
| 9. | Nakajima M, DeChavigny A, Johnson CE, Hamada J, Stein CA, Nicolson GL. Suramin. A potent inhibitor of melanoma heparanase and invasion. J Biol Chem. 1991;266:9661-9666. |
| 10. | Karoli T, Liu L, Fairweather JK, Hammond E, Li CP, Cochran S, Bergefall K, Trybala E, Addison RS, Ferro V. Synthesis, biological activity, and preliminary pharmacokinetic evaluation of analogues of a phosphosulfomannan angiogenesis inhibitor (PI-88). J Med Chem. 2005;48:8229-8236. |
| 11. | Miao HQ, Elkin M, Aingorn E, Ishai-Michaeli R, Stein CA, Vlodavsky I. Inhibition of heparanase activity and tumor metastasis by laminarin sulfate and synthetic phosphorothioate oligodeoxynucleotides. Int J Cancer. 1999;83:424-431. |
| 12. | Jingting C, Yangde Z, Yi Z, Huining L, Rong Y, Yu Z. Heparanase expression correlates with metastatic capability in human choriocarcinoma. Gynecol Oncol. 2007;107:22-29. |
| 13. | Zhang Y, Li L, Wang Y, Zhang J, Wei G, Sun Y, Shen F. Downregulating the expression of heparanase inhibits the invasion, angiogenesis and metastasis of human hepatocellular carcinoma. Biochem Biophys Res Commun. 2007;358:124-129. |
| 14. | Edovitsky E, Elkin M, Zcharia E, Peretz T, Vlodavsky I. Heparanase gene silencing, tumor invasiveness, angiogenesis, and metastasis. J Natl Cancer Inst. 2004;96:1219-1230. |
| 15. | Zhang ZH, Chen Y, Zhao HJ, Xie CY, Ding J, Hou YT. Silencing of heparanase by siRNA inhibits tumor metastasis and angiogenesis of human breast cancer in vitro and in vivo. Cancer Biol Ther. 2007;6:587-595. |
| 16. | Nicosia RF, Ottinetti A. Growth of microvessels in serum-free matrix culture of rat aorta. A quantitative assay of angiogenesis in vitro. Lab Invest. 1990;63:115-122. |
| 18. | Boyd D. Invasion and metastasis. Cancer Metastasis Rev. 1996;15:77-89. |
| 19. | Sanderson RD, Yang Y, Suva LJ, Kelly T. Heparan sulfate proteoglycans and heparanase--partners in osteolytic tumor growth and metastasis. Matrix Biol. 2004;23:341-352. |
| 20. | Zcharia E, Metzger S, Chajek-Shaul T, Friedmann Y, Pappo O, Aviv A, Elkin M, Pecker I, Peretz T, Vlodavsky I. Molecular properties and involvement of heparanase in cancer progression and mammary gland morphogenesis. J Mammary Gland Biol Neoplasia. 2001;6:311-322. |
| 21. | Vlodavsky I, Goldshmidt O, Zcharia E, Metzger S, Chajek-Shaul T, Atzmon R, Guatta-Rangini Z, Friedmann Y. Molecular properties and involvement of heparanase in cancer progression and normal development. Biochimie. 2001;83:831-839. |
| 22. | McKenzie EA. Heparanase: a target for drug discovery in cancer and inflammation. Br J Pharmacol. 2007;151:1-14. |
| 23. | Nasser NJ. Heparanase involvement in physiology and disease. Cell Mol Life Sci. 2008;65:1706-1715. |
| 24. | Vlodavsky I, Ilan N, Naggi A, Casu B. Heparanase: structure, biological functions, and inhibition by heparin-derived mimetics of heparan sulfate. Curr Pharm Des. 2007;13:2057-2073. |
| 25. | Nakajima M, Irimura T, Nicolson GL. Heparanases and tumor metastasis. J Cell Biochem. 1988;36:157-167. |
| 26. | Liu CJ, Lee PH, Lin DY, Wu CC, Jeng LB, Lin PW, Mok KT, Lee WC, Yeh HZ, Ho MC. Heparanase inhibitor PI-88 as adjuvant therapy for hepatocellular carcinoma after curative resection: a randomized phase II trial for safety and optimal dosage. J Hepatol. 2009;50:958-968. |
| 27. | Sasisekharan R, Shriver Z, Venkataraman G, Narayanasami U. Roles of heparan-sulphate glycosaminoglycans in cancer. Nat Rev Cancer. 2002;2:521-528. |
| 28. | Parish CR, Freeman C, Hulett MD. Heparanase: a key enzyme involved in cell invasion. Biochim Biophys Acta. 2001;1471:M99-M108. |
| 29. | Vlodavsky I, Friedmann Y. Molecular properties and involvement of heparanase in cancer metastasis and angiogenesis. J Clin Invest. 2001;108:341-347. |
| 30. | Vlodavsky I, Goldshmidt O, Zcharia E, Atzmon R, Rangini-Guatta Z, Elkin M, Peretz T, Friedmann Y. Mammalian heparanase: involvement in cancer metastasis, angiogenesis and normal development. Semin Cancer Biol. 2002;12:121-129. |
| 31. | Uno F, Fujiwara T, Takata Y, Ohtani S, Katsuda K, Takaoka M, Ohkawa T, Naomoto Y, Nakajima M, Tanaka N. Antisense-mediated suppression of human heparanase gene expression inhibits pleural dissemination of human cancer cells. Cancer Res. 2001;61:7855-7860. |
| 32. | Elkin M, Ilan N, Ishai-Michaeli R, Friedmann Y, Papo O, Pecker I, Vlodavsky I. Heparanase as mediator of angiogenesis: mode of action. FASEB J. 2001;15:1661-1663. |
| 33. | Vlodavsky I, Goldshmidt O, Zcharia E, Atzmon R, Rangini-Guatta Z, Elkin M, Peretz T, Friedmann Y. Mammalian heparanase: involvement in cancer metastasis, angiogenesis and normal development. Semin Cancer Biol. 2002;12:121-129. |
| 34. | Ilan N, Elkin M, Vlodavsky I. Regulation, function and clinical significance of heparanase in cancer metastasis and angiogenesis. Int J Biochem Cell Biol. 2006;38:2018-2039. |
Peer reviewer: Alain L Servin, PhD, Faculty of Pharmacy, French National Institute of Health and Medical Research, Unit 756, Rue J.-B. Clément, F-922296 Châtenay-Malabry, France
S- Editor Wang JL L- Editor Cant MR E- Editor Ma WH