Published online Oct 14, 2009. doi: 10.3748/wjg.15.4844
Revised: August 24, 2009
Accepted: August 31, 2009
Published online: October 14, 2009
AIM: To investigate the expression of Neurensin-2 (NRSN2) in hepatocellular carcinoma (HCC) and its prognostic values in predicting survival.
METHODS: The expression and prognostic significance of NRSN2 in HCC was examined by performing immunohistochemical analysis using a total of 110 HCC clinical tissue samples, and Western blotting analysis to further confirm the result.
RESULTS: Decreased NRSN2 expression was shown in 70.9% cases. Loss of NRSN2 expression in HCC was significantly related to tumor size (P = 0.006). Larger tumor size was related to negative expression of NRSN2. Patients showing negative NRSN2 expression had a significantly shorter overall survival than those with positive expression (P = 0.008). Multivariate Cox regression analysis indicated that NRSN2 expression level was an independent factor of survival (P = 0.013). Western blotting analysis further confirmed decreased expression of NRSN2 in tumor tissues compared with non-tumorous tissues.
CONCLUSION: Our study indicated that NRSN2 could be a tumor suppressor gene for HCC and a candidate biomarker for long-term survival in HCC.
-
Citation: Ma HQ, Liang XT, Zhao JJ, Wang H, Sun JC, Chen YB, Pan K, Xia JC. Decreased expression of
Neurensin-2 correlates with poor prognosis in hepatocellular carcinoma. World J Gastroenterol 2009; 15(38): 4844-4848 - URL: https://www.wjgnet.com/1007-9327/full/v15/i38/4844.htm
- DOI: https://dx.doi.org/10.3748/wjg.15.4844
| Clinicopathologic variables | n | NRSN2 expression | χ2 | P-value | |
| Negative | Positive | ||||
| All cases | 110 | 32 | 78 | ||
| Age | 0.037 | 0.848 | |||
| < 50 | 60 | 43 | 17 | ||
| ≥ 50 | 50 | 35 | 15 | ||
| Gender | 0.341 | 0.559 | |||
| Female | 14 | 9 | 5 | ||
| Male | 96 | 69 | 27 | ||
| Tumor size (cm) | 7.678 | 0.006a | |||
| < 5 | 37 | 20 | 17 | ||
| ≥ 5 | 73 | 58 | 15 | ||
| Histological differentiation | 0.879 | 0.258 | |||
| Well | 18 | 12 | 6 | ||
| Moderate | 69 | 49 | 20 | ||
| Poor | 23 | 17 | 5 | ||
| Liver cirrhosis | 0.176 | 0.675 | |||
| Yes | 55 | 38 | 17 | ||
| No | 55 | 40 | 15 | ||
| Metastasis | 0.152 | 0.697 | |||
| Yes | 16 | 12 | 4 | ||
| No | 94 | 66 | 28 | ||
| HBsAg status | 3.400 | 0.065 | |||
| Positive | 96 | 71 | 25 | ||
| Negative | 14 | 7 | 7 | ||
| Serum AFP (μg/L) | 0.360 | 0.549 | |||
| Positive (≥ 25) | 80 | 58 | 22 | ||
| Negative (< 25) | 30 | 20 | 10 | ||
| Variables | Relative risk (95% CI) | P-value |
| Univariate | ||
| Gender | 0.674 (0.308-1.474) | 0.320 |
| Age | 0.689 (0.423-1.123) | 0.133 |
| Tumor size | 1.752 (1.028-2.986) | 0.037a |
| Histological differentiation | 1.728 (0.855-3.495) | 0.083 |
| Liver cirrhosis | 1.689 (1.047-2.725) | 0.030a |
| HBsAg | 1.488 (0.680-3.252) | 0.316 |
| Serum AFP | 1.990 (1.105-3.585) | 0.019a |
| Metastasis | 1.368 (0.733-2.554) | 0.323 |
| NRSN2 | 0.470 (0.265-0.836) | 0.008a |
| Multivariate | ||
| Liver cirrhosis | 1.739 (1.076-2.808) | 0.024a |
| Serum AFP | 1.957 (1.086-3.528) | 0.025a |
| NRSN2 | 0.481 (0.270-0.854) | 0.013a |
Hepatocellular carcinoma (HCC) is the fifth most common cancer worldwide and the third most common cause of cancer-related death in the world[1,2]. In China, it is the second leading cause of cancer death among males[3]. Factors associated with increased risk of HCC include HBV infection, HCV infection, chronic alcohol consumption, cirrhosis, hemochromatosis and aflatoxin etc. Among them, chronic HBV infection is the most common cause of HCC especially in China. However, there are currently very limited therapeutic options for advanced or metastatic HCC. It is therefore critical to understand the genetic background and molecular pathogenic processes involved in the carcinogenesis of HCC, to aid the development of rational, targeted therapies[4,5].
In a recent publication, Zender et al[6] combined an integrated cancer genomic analysis, RNA interference (RNAi) technology and cancer-susceptible mouse models to discover and validate tumor suppressor genes contributing to HCC. The approach resulted in the functional validation of 13 tumor suppressor genes, including Exportin 4 (XPO4), DEAD box polypeptide 20 (DDX20), Gap junction protein, delta 4 (GJD4), Follistatin-like 5 (FSTL5) and Neurensin-2 (NRSN2) etc. Interestingly, the vast majority of these identified genes had not previously been linked to cancer.
To validate and further study the potential value of these tumor suppressors, we randomly selected NRSN2 from those genes showing a higher possibility of being tumor suppressors, and performed immunohistochemical analysis and Western blotting analysis to determine the expression of NRSN2 in HCC, further identified its relationship to clinicopathological features and evaluated its prognostic value to post-resectional survival in HCC.
A total of 110 HCC surgical resection specimens were collected at the Sun Yat-sen University Cancer Center between January 2001 and December 2002. The 110 patients included 96 males and 14 females with a median age of 45 years (range, 22-74 years). None of the patients had received radiotherapy or chemotherapy prior to surgery. Both tumor and corresponding non-tumorous tissues not less than 2 cm away from the HCC were sampled, respectively, and proved by pathological examination. All tissue samples were fixed in 10% formalin and embedded in paraffin, and consecutive 4 μm sections were cut. The histological types were assigned according to the criteria of the World Health Organization classification. The diagnosis of liver cirrhosis was based on the case records and pathological data of HCC patients from Sun Yat-sen University Cancer Center.
The sections were deparaffinized and rehydrated through graded ethanol, then endogenous peroxidase was inhibited with 0.3% hydrogen peroxide. For antigen retrieval, slides were boiled in EDTA (1 mmol/L, pH 8.0) for 15 min in a microwave oven. After rinsing with PBS, the sections were incubated with primary antibody (rabbit anti-NRSN2, Sigma-Aldrich, Inc. USA) in PBS (1:100) at 37°C for 3 h, and then incubated with horseradish peroxidase (ChemMate™ DAKO EnVision™ Detection Kit) at 37°C for 30 min. Finally, the visualization signal was developed with 3,3’-diaminobenzidine tetrahydrochloride (DAB) and then all of the slides were counterstained in hematoxylin. For negative controls, tissue sections were immunoreacted without anti-NRSN2 antibody under the same experimental conditions.
The total NRSN2 immunostaining score was calculated as the sum of the percent positivity of stained tumor cells and the staining intensity. The percent positivity was scored as “0” (< 5%, negative), “1” (5%-25%, sporadic), “2” (25%-50%, focal), “3” (> 50%, diffuse). The staining intensity was scored as “0” (no staining), “1” (weakly stained), “2” (moderately stained), and “3” (strongly stained). Both percent positivity of cells and staining intensity were decided under double-blind conditions. The final NRSN2 expression score was calculated using the value of percent positivity score × staining intensity score, which ranged from 0 to 9. We defined NRSN2 expression level as follow: “-” (score 0-1), “+” (score 2-3), “++” (score 4-6) and “+++” (score > 6).
The frozen HCC samples including tumor or non-tumorous tissue were homogenated in a RIPA lysis buffer, and lysates were cleared by centrifugation (14 000 rpm) at 4°C for 30 min. About 40 μg protein samples were run on a 15% SDS-PAGE gel and transferred to PVDF membrane. After blocking non-specific binding sites for 60 min with 5% non-fat milk, the membranes were incubated overnight at 4°C with primary polyclonal antibody against NRSN2 (at 1:200 dilution). The membrane was then washed three times with TBST for 10 min and probed with HRP-conjugated secondary antibody (at 1:2 000 dilution) for 30 min at room temperature. After being washed three times, the membrane was developed by an enhanced chemiluminescence system (ECL, Pierce).
Quantitative values were expressed as mean ± SD or median (range). Categorical variables were enumeration data from counting the number of samples. The χ2 test for proportion and Pearson’s correlation were used to analyze the relationship between NRSN2 expression and various clinicopathologic characteristics. For survival analysis, the main outcome was overall survival rates which were calculated from the date of surgery to the date of death. Follow-up time was censored if the patient was lost to follow-up. Survival curves were calculated using the Kaplan-Meier method and compared by the log-rank test. Cox proportional-hazard analysis was used for univariate and multivariate analysis to explore the effect of clinicopathological variables and NRSN2 expression on survival. The SPSS 16.0 software (SPSS Inc., Chicago, IL, USA) was used for all statistical analyses and a P-value < 0.05 was considered significant.
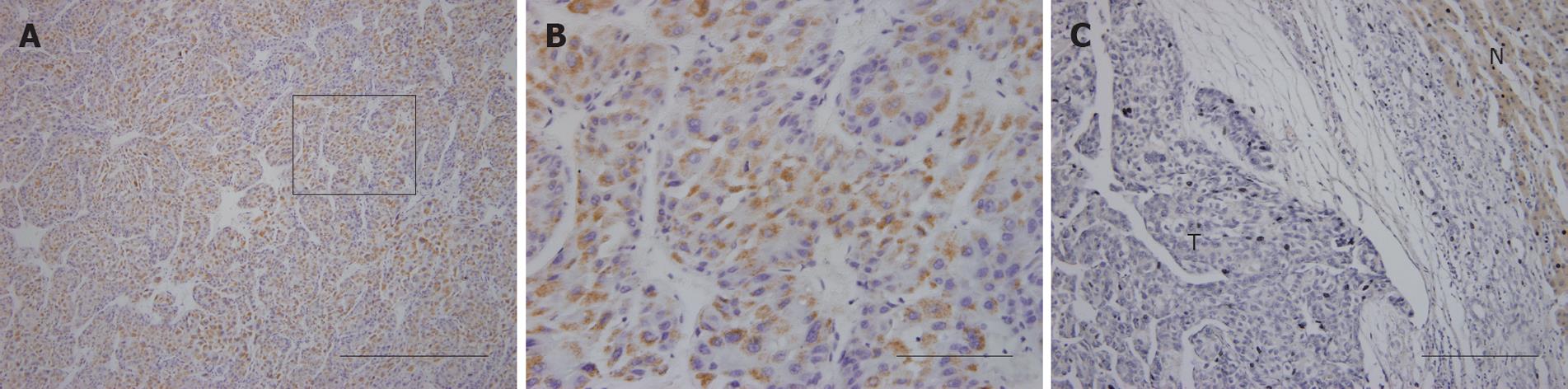
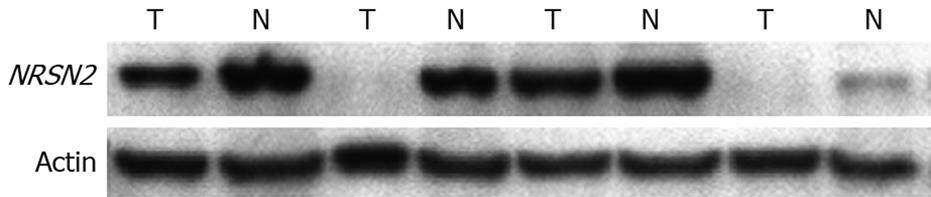
NRSN2 immunostaining was mostly in the cytoplasm. Overall, 32 of 110 (29.1%) cases had positive expression (NRSN2 + or ++) in tumor cells (Figure 1A and B), 78 of 110 (70.9%) cases had negative expression (NRSN2-). In cases with adjacent hyperplastic tissue, we often observed a sharp contrast between infiltrative tumor areas of negative staining and the adjacent non-tumorous tissue of positive staining (Figure 1C). In addition, we further performed Western blotting analysis to detect the expression of NRSN2 and the result was consistent with that of immunohistochemical analysis (Figure 2). Correlations between the expression of NRSN2 and various clinicopathologic parameters are listed in Table 1. The NRSN2 expression was significantly related to tumor size (P = 0.006). Negative expression of NRSN2 was associated with larger tumor size. However, there was no statistically significant difference between NRSN2 expression and age, gender, histological differentiation, liver cirrhosis, metastasis, HBsAg status, or serum AFP.
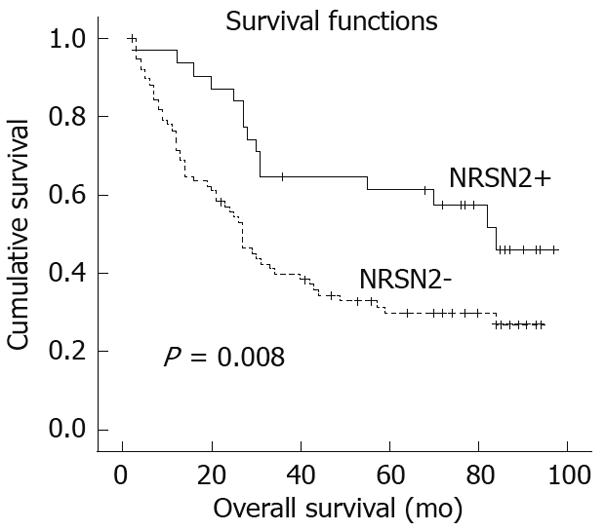
The 5-year overall survival rates were 61.2% and 29.8%, respectively, in patients with positive and negative NRSN2 expression. Patients showing negative NRSN2 expression had a significantly shorter overall survival than those with positive expression (P = 0.008, log-rank test; Figure 3). Univariate Cox regression analysis also identified that clinical variables including tumor size, liver cirrhosis, serum AFP and NRSN2 expression were significantly associated with overall survival (Table 2). Furthermore, to evaluate the potential of NRSN2 expression as an independent predictor for overall survival of HCC, multivariate Cox regression analyses (Forward: LR) were performed. While the others failed to demonstrate independence, liver cirrhosis, serum AFP level and NRSN2 expression may play a role in predicting the overall survival in HCC (P = 0.024, 0.025, and 0.013, respectively, Table 2).
HCC is one of the most deadly human carcinomas. Even with improved diagnosis and compositive therapy, the prognosis of HCC remains dismal[7]. Therefore, prognostic molecular biomarkers are invaluable for the clinician to evaluate patients and to aid in tumor control.
Recently, Zender et al[6] introduced a new, effective and high-yield approach for identifying liver tumor suppressors. They first used representational oligonucleotide microarray analysis (ROMA), a high-resolution array-based comparative genomic hybridization (CGH) platform, to narrow down the field of potential candidate genes. To further accelerate the study of cancer genes in vivo, they adapted stable RNAi technology, utilizing microRNA-based short hairpin RNAs (shRNAs), that are potent triggers of the RNAi machinery and can efficiently suppress gene expression when expressed from a single genomic copy[8,9], to downregulate tumor suppressor genes in mice[10]. In addition, to facilitate a more rapid and cost-effective analysis of cancer gene action in vivo, they developed a ‘‘mosaic’’ mouse model of HCC[11]. In this mouse model, HCCs with different oncogenic lesions can be rapidly produced by genetic manipulation of cultured embryonic liver progenitor cells (hepatoblasts) followed by their retransplantation into the livers of recipient mice[11,12].
According to Zender[6], they selected 16 shRNAs targeting 14 different genes for validation. The result showed that many of the candidate shRNAs triggered tumor growth above background, with those targeting Xpo4, Ddx20, Gjd4, Fstl5, and Nrsn2 showing the most prominent acceleration of tumor growth. Interestingly, most of these genes previously have not been linked to cancer.
To validate and further study the potential value of tumor suppressors in the results of Zender, we randomly selected NRSN2 as a sample, and performed immunohistochemical analysis and Western blotting analysis to determine the expression of NRSN2 in HCC and evaluate its potential clinical relevance.
To our knowledge, there are no studies of NRSN2 in cancer, not to mention HCC. NRSN2 encodes a 21 983 Da protein composed of 204 amino acids, belongs to the vesicular membrane protein (VMP) family, and shows a high sequence homology to Neurensin-1. So far, there is no definite function for NRSN2. After retrieval in UniProt Knowledgebase, it may play a role in maintenance and/or transport of vesicles, according to its sequence similarities with Neurensin-1, and it is uncertain whether Met-1 or Met-2 is the initiator (http://www.uniprot.org/uniprot/Q9GZP1#section_comments).
In the present paper, using immunohistochemistry technology, we demonstrated that negative NRSN2 protein expression was found in 70.9% of HCC samples (78/110). This was significantly associated with larger tumor size (P = 0.006) and was significantly correlated with poor patient outcome (P = 0.008). Regardless of whether NRSN2 has prognostic significance, decreased expression of NRSN2 was observed in larger tumors, which does support the hypothesis that NRSN2 may play a role in inhibiting tumor progression. Metastasis is one of the characteristics of progression of HCC. However, in our studies, no significant correlation was observed between NRSN2 expression and metastasis, indicating that some other mechanisms may be more important in moderating the metastasis of HCC, since hepatocarcinogenesis is a multi-step and complex process associated with accumulation of genetic and epigenetic changes[13]. Further multivariate Cox regression analysis indicated that NRSN2 expression level was an independent factor of survival and may constitute a prognostic factor for patients with HCC after surgery. Western blotting analysis further confirmed decreased expression of NRSN2 in tumor tissues compared with paired non-tumorous tissues.
At the same time, we also performed some experiments with the gene XPO4 to investigate its expression and prognostic values in HCC, and the result was similar to that of NRSN2 (data not shown). All these data supported the result of Zender and further validated the utility of this new approach. Therefore, we have reason to believe other genes identified by Zender are tumor suppressors, although more studies are still needed to further confirm this.
What’s more, in the study of Zender, NRSN2 was also found to be frequently deleted in human breast cancer, indicating that NRSN2 may be relevant to other epithelial malignancies as a tumor suppressor. However, to elucidate the molecular mechanism role of NRSN2 in liver carcinogenesis and its relation with other tumor types, more studies still should be done.
Hepatocellular carcinoma (HCC) is one of the most deadly human carcinomas. Even with improved diagnosis and compositive therapy, the prognosis of HCC remains dismal. Therefore, prognostic molecular biomarkers are invaluable for the clinician to evaluate patients and to aid in tumor control.
A research article published on November 26, 2008 in “Cell” introduced a new, effective and high-yield approach for identifying liver tumor suppressors by combing an integrated cancer genomic analysis, RNA interference (RNAi) technology and cancer-susceptible mouse models. The approach resulted in identification of 13 tumor suppressor genes, including Exportin 4 (XPO4), DEAD box polypeptide 20 (DDX20), Gap junction protein, delta 4 (GJD4), Follistatin-like 5 (FSTL5) and Neurensin-2 (NRSN2) etc. Interestingly, the vast majority of these identified genes had not previously been linked to cancer. Therefore, more work is needed to validate and further study the potential value of these tumor suppressors.
In the present study, for the first time, by using immunohistochemistry technology, NRSN2 expression was found to be decreased in 70.9% of cases (n = 110). Loss of NRSN2 expression in HCC was significantly related to tumor size (P = 0.006). Larger size tumors were related to negative expression of NRSN2. Patients showing negative NRSN2 expression had a significantly shorter overall survival than those with positive expression (P = 0.008). Multivariate Cox regression analysis indicated that NRSN2 expression level was an independent factor of survival. Western blotting analysis further confirmed decreased expression of NRSN2 in tumor tissues compared with paired non-tumorous tissues.
Because NRSN2 down-regulation frequently occurs in HCC, the authors propose that NRSN2 may be a candidate tumor suppressor gene for HCC, and may be used as a candidate biomarker for long-term survival in HCC.
NRSN2 encodes a 21 983 Da protein composed of 204 amino acids, belongs to the vesicular membrane protein (VMP) family, and shows a high sequence homology to Neurensin-1. So far, there is no definitive function for NRSN2.
Ma et al investigated the expression of NRSN2 in 110 cases of HCC immunohistochemically and by Western blotting, and found a correlation between its negative expression and tumor progression. Although the function of NRSN2 has not been clarified, their result was interesting and this molecule might be a new prognostic marker of HCC.
| 2. | Parkin DM, Bray F, Ferlay J, Pisani P. Estimating the world cancer burden: Globocan 2000. Int J Cancer. 2001;94:153-156. |
| 3. | Tung-Ping Poon R, Fan ST, Wong J. Risk factors, prevention, and management of postoperative recurrence after resection of hepatocellular carcinoma. Ann Surg. 2000;232:10-24. |
| 4. | Roberts LR, Gores GJ. Hepatocellular carcinoma: molecular pathways and new therapeutic targets. Semin Liver Dis. 2005;25:212-225. |
| 5. | Teufel A, Staib F, Kanzler S, Weinmann A, Schulze-Bergkamen H, Galle PR. Genetics of hepatocellular carcinoma. World J Gastroenterol. 2007;13:2271-2282. |
| 6. | Zender L, Xue W, Zuber J, Semighini CP, Krasnitz A, Ma B, Zender P, Kubicka S, Luk JM, Schirmacher P. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135:852-864. |
| 7. | Qin LX, Tang ZY. The prognostic molecular markers in hepatocellular carcinoma. World J Gastroenterol. 2002;8:385-392. |
| 8. | Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, Lowe SW. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet. 2005;37:1289-1295. |
| 9. | Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37:1281-1288. |
| 10. | Hemann MT, Fridman JS, Zilfou JT, Hernando E, Paddison PJ, Cordon-Cardo C, Hannon GJ, Lowe SW. An epi-allelic series of p53 hypomorphs created by stable RNAi produces distinct tumor phenotypes in vivo. Nat Genet. 2003;33:396-400. |
| 11. | Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, Fan ST, Luk JM, Wigler M, Hannon GJ. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253-1267. |
| 12. | Zender L, Xue W, Cordón-Cardo C, Hannon GJ, Lucito R, Powers S, Flemming P, Spector MS, Lowe SW. Generation and analysis of genetically defined liver carcinomas derived from bipotential liver progenitors. Cold Spring Harb Symp Quant Biol. 2005;70:251-261. |
| 13. | Aravalli RN, Steer CJ, Cressman EN. Molecular mechanisms of hepatocellular carcinoma. Hepatology. 2008;48:2047-2063. |
Peer reviewers: Hidetsugu Saito, MD, PhD, Assistant Professor, Department of Internal Medicine, School of Medicine, Keio University, 35 Shinanomachi, Shinjuku-ku, Tokyo 1608582, Japan; Akihito Tsubota, Assistant Professor, Institute of Clinical Medicine and Research, Jikei University School of Medicine, 163-1 Kashiwa-shita, Kashiwa, Chiba 277-8567, Japan
S- Editor Cheng JX L- Editor O'Neill M E- Editor Lin YP