INTRODUCTION
Proteasomes of mammalian cells exhibit a complex heterogeneity. Different proteasome variants exist and perform subtle activities. The 20S proteasome catalytic core binds to different activators (19S, 11S and PA200), and the resulting complexes perform specific functions, such as ATP-ubiquitin proteolysis, immunological response, cell cycle regulation, nuclear factor (NF) κB activation, response to hypoxia, and transcription[1]. Upon immune response, or interferon (IFN)γ treatment, the three catalytic subunits, β1, β2 and β5, are replaced by three different subunits, LMP2, LMP7 and MECL-1, respectively, which form the immunoproteasome[2]. Several chemical compounds, proteins and regulatory complexes, function as activators or inhibitors of the catalytic core 20S proteasome[3]. Some of them are termed gate-openers because they provoke the opening of the gate at the alpha-type subunits of the 20S proteasome[4], and facilitate the access of designated proteins to be degraded to the catalytic chamber formed by the beta-type subunits. The best known of these activators are the regulatory complex 19S, which is involved in the ubiquitin-proteasome pathway (UPP), the PA28α/β and PA28γ complexes, which, among other functions, are important for the generation of epitope peptides presented to the major histocompatibility complex[5], and PA200, which is implicated in DNA repair[6]. Recently, the hybrid form of the 26S proteasome, which contains both 19S and PA28 complexes associated at the two opposite sides of the 20S core particle, has also been characterized[7], and other activators and proteins that modulate the proteasome activity are yet to be discovered.
As complex as is the UPP, it does not work alone. These regulatory complexes and proteasome interacting proteins (PIPs) complement the UPP to perform specific functions. Proteasome failure occurs in the liver cells because chronic ethanol exposure interferes with the binding of the 20S catalytic core to its regulatory complex and to its PIPs. The 26S proteasome is responsible for the ATP-ubiquitin degradation of short-lived cellular proteins and the elimination of damaged and misfolded proteins, in addition to providing basic housekeeping functions[8–10]. Therefore, damaged and ubiquitinated proteins accumulate, causing protein aggregation, such as Mallory-Denk body (MDB) formation[11–13], when, because of chronic ethanol ingestion, the 20S is unable to bind to the 19S to form the 26S proteasome, which leads to cell death and tissue damage[14].
PROTEASOMES AND TRANSCRIPTION
The UPP plays an important role in a variety of cellular functions, primarily via its proteolytic activity. However, recent studies have implicated this pathway in the direct regulation of specific transcriptional factors[1516]. It is now believed that the proteolytic activity of the proteasome is critical in promoting the exchange of transcriptional factors on chromatin, and possibly facilitating multiple rounds of transcription initiation, hence controlling gene expression[17]. Proteasomes are also now widely accepted as being essential for promoting the exchange of transcriptional factors on chromatin[1819]. A growing body of evidence demonstrates that the components of the UPP are directly, and mechanistically, involved in transcription and in regulating the gene expression of the DNA repair system[2021], cell cycle[2223], and chromatin modifying enzymes[17]. Although the role of the UPP in regulating many transcription factors, such as p53, NFκB and hypoxia-inducible factor 1 α, is well established, the role of the UPP in regulating epigenetic mechanisms has only recently been given attention[24].
Ethanol-induced oxidative stress in nuclei is believed to damage DNA and proteins, thereby disrupting genomic integrity[25]. Oxidative stress has also been shown to cause adduct formation with lysine located in the N-terminal histone tails[26]. These lysine residues are the site of a number of post-translational modifications that regulate chromatin function. Therefore, chronic accumulation of oxidative stress adduct modifications may cause epigenetic changes, which may have important implications in terms of histone function and genomic integrity. Oxidatively damaged histones are degraded by a nuclear 20S proteasome in an ATP- and ubiquitin-independent manner[27], which indicates that damaged histones accumulate when the activity of the proteasome is inhibited and causes alteration of the epigenetic mechanisms.
However, the effect of ethanol-induced proteasome inhibition in the nucleus, and thus in regulating gene expression, is still not well known. Inhibition of proteasome function has been widely reported in models of alcoholic liver disease (ALD)[2829], but how proteasome dysfunction may enhance hepatotoxicity is not well defined. It has been reported that chronic ethanol exposure leads to post-translational modifications of the alpha type subunits of the 20S proteasome that constitute the opening of the gate to the catalytic chamber of the 20S proteasome[29], thus blocking the opening of the gate and causing proteasome activity to decrease. Recently, proteasome activity has been measured in the isolated nuclei from liver cells of rats fed ethanol chronically, and the proteasome was found to be significantly inhibited (personal observation, manuscript submitted to World Journal of Gastroenterology, 2008). The inhibition of proteasome activity in the nucleus is therefore etiologically involved in the accumulation of damaged proteins in the nucleus, and in the deregulation of transcription.
Thus, it seems reasonable to postulate that the nucleus should have a more important proteasomal activity than other cellular compartments. It is possible that proteasome inhibition, caused by chronic ethanol feeding in the nucleus, may be greater than the inhibition caused in the cytosolic compartment[29].
EPIGENETIC MECHANISMS AND PROTEASOME INHIBITION
Chronic ethanol exposure causes an alteration in liver cell memory by changing the epigenetic mechanisms, thus causing significant liver pathology that persists after ethanol withdrawal[30]. Dr. French’s group has shown that cells forming MDBs have a memory of the exposure to the drug even four months after drug withdrawal[31]. This memory is primarily associated with histone modifications that are recovered when s-adenosylmethionine (SAMe) is added to the drug-primed mouse diet[32].
Epigenetic mechanisms are involved in development, phenotype determination and maintenance, aging, and cancer. These epigenetic mechanisms are based on histone modifications controlled by histone acetyltransferases (HATs) and histone methyltransferases, and by DNA methylation, controlled by a regulated balance between DNA methyltransferase and demethylase. These epigenetic events can change rapidly in response to changes in cell signaling, which are initiated by environmental influences, such as inflammation, metabolism, or toxic injury. These changes cause the genome to respond globally or in a restricted manner, depending on the nature of the signals generated. During these responses, the histone modifications themselves are rapidly changed as well[33]. The dynamics of this process are pervasive and extremely complex, which makes it difficult to connect the epigenetic responses to the final manifestations. The identification of key epigenetic changes, which are linked to alcohol liver injury, is of high interest and is at its very beginning. Histone modifying enzymes are the key target to regulate the histone modifications. Histone deacetylase (HDAC) inhibitors and demethylating agents are currently used in clinical trials to treat cancer. HDAC inhibitors, like trichostatin A, repress the expression of genes that induce growth arrest, cellular differentiation and apoptosis[34]. Likewise, DNA methyltransferase inhibitors, like 5-azacytidine (Aza), are being used in clinical trials to treat cancer. Aza, a potent inhibitor of Dnmt 1 and Dnmt3a (not Dnmt3b), induces demethylation and reactivation of silenced tumor suppressor genes[35]. Moreover, proteasome and HDAC inhibitors may be a prominent combination in future cancer therapy[36], and specifically, in gastrointestinal cancer[37].
HISTONE MODIFICATIONS AND HISTONE MODIFYING ENZYMES
The nucleosome surface interacts with several histone modifying enzymes that are responsible for histone tail lysine or arginine modifications[38], operating as part of a predictive and initiated epigenetic code that defines patterns of gene expression[39]. Histone modifications control the accessibility of DNA elements for transcription factors, which influences gene expression. These modifications control the recruitment of proteins involved in DNA methylation and affect chromatin structure and function[40].
Histone modification alterations can change long-term effects on gene expression patterns by influencing chromatin structure throughout the cell cycle, and from one cell generation to the next[41]. For instance, trimethylation of H3K9me3 and depletion of acetylation leads to gene silencing, because H3K9me3 acts as a binding site for heterochromatin-forming protein HP1. In the nuclear extract from liver cells of rats fed ethanol chronically, H3K9me3 is decreased and H3K9 acetylation is increased[4243]. These histone modifications are linked to DNA modifications and affect gene expression because DNA methylation is dependent on prior histone methylation[44–47].
More epigenetic mechanisms still need to be elucidated with respect to alcoholism, particularly histone modifications, such as methylation, phosphorylation and ubiquitinylation of lysine and arginine residues. The task will be to identify the modifications and the specific modifying enzymes that regulate gene expression and account for the cellular memory observed when ethanol is fed chronically[30]. For example, it has been reported that a global increase in histone H3 lysine 9 acetylation, histone H3 lysine 4 dimethylation and H3 lysine 27 trimethylation is found in the liver of rats fed ethanol chronically[48], which correlates with an increase and a decrease, respectively, in transcriptionally active promoters. However, the genes linked to these changes are still to be identified. Microarray analysis has shown that hundreds of genes are up-regulated and hundreds are down-regulated when rats are fed ethanol for 1 mo, which indicates that ethanol causes several epigenetic changes that affect gene expression[25]. More specifically, gene expression of ALDH1a4[48] and ADH[49] are up-regulated, which suggests a change in the epigenetic events that regulate the expression of these genes in the liver of rats fed ethanol chronically. However, the linkage of ethanol-induced proteasome inhibition to the changes in the expression of these genes still needs to be determined. Data mining of microarray analysis of proteasome inhibition has shown that, similar to ethanol-treated rat liver, several genes are changed, which indicates an epigenetic phenomenon c proteasome inhibition[1750].
PROTEASOME INHIBITION AND REMETHYLATION PATHWAY
The changes in gene expression in the remethylation pathway, particularly for the enzymes that regenerate SAMe, cause deregulation of cellular methylation, thus resulting in abnormal cellular methylation.
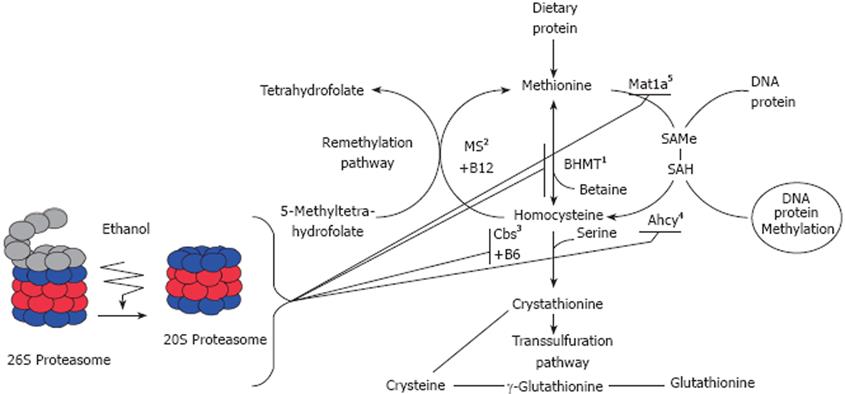
Hyperhomocysteinemia (HHcy) is a leading cause of liver injury in ALD. A lack of the activity of the enzymes responsible for homocysteine metabolism, particularly cystathionine b-synthase (Cbs) or 5,10-methylenetetrahydrofolate reductase, results in severe forms of HHcy. Additionally, nutritional deficiencies in B vitamin cofactors required for homocysteine metabolism, including folic acid, vitamin B6 (pyridoxal phosphate), and/or B12 (methylcobalamin), can induce HHcy (Figure 1).
Figure 1 Illustration of methionine metabolism enzymes system and the effects of proteasome inhibition in the remethylation pathway.
1-BHMT: Betaine homocysteine Methyltransferase; 2-MS: Methionine synthase; 3-Cbs: Cystathionine beta-Synthase; 4-Ahcy: S’-adenosylhomocysteine transferase; 5-Mat1a: Methionine adenosyltransferase.
Proteasome inhibition is believed to be involved etiologically in the changes in gene expression of these enzymes responsible for the hepatic transmethylation reactions. Indeed, analyses performed on the livers of rats given PS-341 (bortezomib, Velcade®), a dipeptide boronic acid used to inhibit proteasome activity, and used today as an anticancer agent in human myeloma[51], have shown down-regulation of gene expression for methionine metabolizing enzymes. Proteasome inhibition causes a decrease in methionine adenosyltransferase (MAT1a), in S-adenosylhomocysteine hydrolase gene expression, and a marked decrease in gene expression of betaine-homocysteine methyltransferase[5052] and Cbs[53], which affects cellular methylation. Rats fed ethanol chronically also exhibit a relatively low level of MAT1a, which is associated with low levels of SAMe and faster growth[54]. These observations indicate that SAMe is an attractive agent for both chemoprevention and treatment of alcohol associated liver cancer[54].
Proteasome inhibition, induced by ethanol feeding, is associated with histone modifications, and is involved in the regulation of histone modifying enzymes, such as the HAT p300[42]. It is believed that proteasome inhibition affects gene expression, particularly genes of the transmethylation pathway, via an epigenetic mechanism that involves modified histone and transcriptional factors.
CONCLUSION
Regulation of gene expression of the transmethylation pathway is one of the promising approaches to counteract proteasome inhibition effects. Investigating the changes in the remethylation pathway, particularly the enzymes that regenerate SAMe, the major methyl group donor, is essential to identify the key enzyme that is changed by chronic ethanol exposure and proteasome inhibition.
Supported by The NIH/NIAAA 8116 grant
Peer reviewer: Maria Concepción Gutiérrez-Ruiz, PhD, Departamento de Ciencias de la Salud, Universidad Autónoma Metropolitana-Iztapalapa, DCBS, Av San Rafael Atlixco 186, Colonia Vicentina, México, DF 09340, México
S- Editor Li LF L- Editor Kerr C E- Editor Yin DH