Published online Mar 7, 2008. doi: 10.3748/wjg.14.1437
Revised: November 13, 2007
Published online: March 7, 2008
AIM: To investigate the role of Ras association domain family protein 1 isoform A (RASSF1A) in gastric tumorigenesis.
METHODS: Through over-expression of RASSF1A gene in the SGC7901 cell line which was induced by a lipofectamine-mediated gene transfer approach. Activator protein-1 (AP-1) DNA binding activity was measured by electrophoretic mobility shift assay (EMSA).
RESULTS: Compared with the control clones, cells over-expressing RASSF1A exhibited significant inhibition of cell growth with G1 cell cycle arrest in vitro and in vivo. The over-expression of RASSF1A significantly inhibited AP-1 activity in SGC7901 cells (0.981 ± 0.011 vs 0.354 ± 0.053, P < 0.001). In addition, both Western blot analysis and immunocytochemistry demonstrated that RASSF1A down-regulated the expression of c-Fos (0.975 ± 0.02 vs 0.095 ± 0.024, P < 0.001) but not c-Jun.
CONCLUSION: Over-expression of RASSF1A inhibits the growth of SGC7901 cells by negatively regulating the AP-1 activity, the latter in turn negatively signals cell proliferation.
- Citation: Deng ZH, Wen JF, Li JH, Xiao DS, Zhou JH. Activator protein-1 involved in growth inhibition by RASSF1A gene in the human gastric carcinoma cell line SGC7901. World J Gastroenterol 2008; 14(9): 1437-1443
- URL: https://www.wjgnet.com/1007-9327/full/v14/i9/1437.htm
- DOI: https://dx.doi.org/10.3748/wjg.14.1437
Gastric carcinoma is one of the most frequent tumors that seriously threaten people’s health in China[1]. Molecular genetics studies indicate that loss of 3p was observed in different types of solid tumors[2]. Frequent loss on 3p21-23 was detected in gastric cancer[34].
Ras association domain family protein 1 isoform A (RASSF1A), one transcript of RASSF1 gene, is a recently identified 3p21.3 tumor suppressor gene[5]. Loss of expression of RASSF1A was a frequent event in primary gastric carcinoma[67]. However, the exact role of RASSF1A in gastric tumorigenicity is largely unknown.
Activator protein-1 (AP-1) plays an important role in various human diseases and regulates the expression of multiple genes essential for cell proliferation, differentiation and apoptosis[8]. AP-1 is thought to serve as a nuclear target of Ras[9]. It is not known whether RASSF1A, as effectors of Ras signaling[10], could or could not inhibit the activity of AP-1.
In this study, we established gastric cancer cell lines stably over expressing RASSF1A. Characterization of these cells with regard to proliferation rate and tumorigenicity in vitro and in vivo was performed. AP-1 activity was measured by electrophoretic mobility shift assay (EMSA). Our results suggest that over-expression of RASSF1A exerts inhibitory effects on the transformed phenotype of gastric cancer cells and RASSF1A inhibits AP-1 activity.
The human gastric cancer cells, SGC7901 (Shanghai Cell Bank, Chinese Academy of Sciences), were maintained in RPMI 1640 medium (Life Technologies, Inc, Grand Island, NY) supplemented with 100 mL/L fetal bovine serum plus penicillin (50 IU/mL) and streptomycin (50 &mgr;g/mL) with passage every three days. Cultures were incubated in an incubator containing 5 mL/L CO2 at 37°C.
Plasmid pcDNA3.0-RASSF1A and pcDNA3.0 were gifts from Professor Michael White (Department of Cell Biology, UT Southwestern Medical Center, Dallas, TX 75390, USA). Cells were seeded in six-well plates to 70%-80% confluence. The cells were transfected with 4 &mgr;g/well plasmids using Lipofectamine 2000 (Invitrogen). After transfection for 6 h, the cells were transferred to normal medium and allowed to recover overnight. The cells were trypsinized and split 1:10, and then seeded into new six-well plates. 48 h after transfection, transfected cells were grown in RPMI containing G418 (Alexis Biochemicals) at 0.8 g/L until all of the nontransfected cells were dead (2 wk). Resistant clones were selected separately using cloning cylinders and maintained in RPMI containing 0.2 g/L G418 for further study. Meanwhile, SGC-7901 cells were transfected with the empty pcDNA3.0 vector as the control.
Nuclear and cytoplasm extracts were prepared as described by Dignam et al[11]. Confluent cells in 10 cm dishes were treated for various times with the indicated effectors. Cells were resuspended in 400 &mgr;L of buffer A [10 mmol/L HEPES (pH 7.9), 1.5 mmol/L MgCl2, 10 mmol/L KCl, 0.5 mmol/L DTT, 0.5 mmol/L phenylmethylsulfonyl fluoride, 1 &mgr;g/mL leupeptin, 1 &mgr;g/mL aprotinin, and 1 &mgr;g/mL pepstatin A], kept on ice for 15 min, lysed gently with 12.5 &mgr;L of 10% NP40, and centrifuged at 2000 r/min for 10 min at 4°C. The supernatant was collected and used as the cytoplasm extracts. The nuclei pellet was resuspended in 40 &mgr;L of buffer C [20 mmol/L HEPES (pH 7.9)], containing 1.5 mmol/L MgCl2, 450 mmol/L NaCl, 25% glycerol, 0.2 mmol/L EDTA, 0.5 mmol DTT, 0.5 mmol/L phenylmethylsulfonyl fluoride, 1 &mgr;g/mL leupeptin, 1 &mgr;g/mL aprotinin, and 1 &mgr;g/mL pepstatin A] and agitated for 30 min at 4°C, and the nuclear debris was spun down at 20 000 r/min for 15 min. The supernatant (nuclear extract) was collected and stored at -80°C until ready for analysis. Proteins were measured using the BCA kit (Pierce) according to the manufacturer’s protocol.
Eighty &mgr;g of cytoplasm proteins or 40 &mgr;g nuclear proteins were separated by 10% SDS-PAGE under reducing conditions, and transferred to a nitrocellulose membrane. The nitrocellulose membrane was then incubated with blocking buffer (TBST containing 5% non-fat milk) for 2 h at room temperature and with mouse monoclonal antibody against RASSF1A (Abcam, USA), c-Jun, c-Fos, CyclinD1 (Santa Cruz Biotechnology, USA) overnight at 4°C with gentle shaking. The membrane was washed with TBST twice for 5 min, and then incubated with rabbit anti-mouse IgG conjugated horseradish peroxidase diluted at 1:2000 (Santa Cruz Biotechnology, USA) for 2 h at room temperature. After washing, RASSF1A was detected using DAB reagents. The level of β-actin or tubulin was used as a control for equal loading of protein.
Total RNA from SGC7901 cells was obtained using a RNA Mini Kit (Qiagen, Inc). Two &mgr;g of total RNA extracted from each cell line were reverse-transcribed using a RevertAid First Strand cDNA Synthesis Kit (MBI). Five ng of reverse-transcribed cDNA per sample were used to perform PCR in triplicate samples for RASSF1A and β-actin as an internal control. Reactions were carried out under the following conditions: 95°C for 5 min, followed by 35 cycles at 94°C for 30 s, and 52°C for 40 s and 72°C for 60 s. The following primers were used: RASSF1A forward, 5’-TCTGGGGCGTCGTGAGTAAA-3’ reverse, 5’-CCACCACCAAGAACAGTCG-3’, β-actin forward, 5’-CCTTCCTGGGCATGGAGTCCT-3’, β-actin reverse 5’-GGAGCAATGATCTTGATCTT-3’, Three independent measurements were averaged and relative gene expression levels were calculated as a ratio to β-actin expression of each.
Cells were cultured in 96-well microtiter plates at a density of 1 × 104 cells per well. The surviving cells were measured by MTT assay at 1 d, 2 d, 3 d, 4 d, 5 d, 6 d, 7 d after seeding. 20 &mgr;L of 5 g/L MTT [3-(4,5-dimethyl-thiazolyl-2)-2.5-diphenyltetrazolium bromide, Fluka, Buchs, Switzerland] in PBS was added to each well and the cells were incubated for another 4 h at 37°C. The supernatant was removed, and 150 &mgr;L of DMSO was added to each well. The absorbency at a wavelength of 595 nm was measured with a micro ELISA reader (BioRad, CA, USA).
Cells were collected and fixed in 70% of ice-cold ethanol in phosphate buffer saline (PBS) and stored at -20°C. After resuspension, 100 &mgr;L RNAase I (1 g/L) and 100 &mgr;L propidium iodide (PI, 400 g/L, Sigma, USA) were added and incubated at 37°C for 30 min. Analysis of samples was performed by flow cytometry (Coulter Epics, XL, UK). The cell cycle phase distribution was calculated from the resultant DNA histogram using Multicycle AV software (Phoenix Flow System, San Diego, CA, USA).
Plating Efficiency was prepared as described by Hu et al[12]. Cells (1 × 102) were plated in 6-well plates. Colonies were scored at 14 d, fixed with 70% ethanol, stained with 5% Giemsa (Sigma), and counted under a microscope. Only those colonies containing at least 50 cells were considered to be viable survivors. The plating efficiency (PE) was calculated as follows: PE = (colonies formed/cells seeded) × 100%.
Single cell suspensions were trypsinized and collected. The cell viability was > 95% as determined by trypan blue staining. Cells (5 × 106) in a 0.1 mL volume of RPMI were inoculated s.c. into the right flank of 4-6 wk-old female BALB/c-nu/nu mice (Laboratory Animal Unit, Central South University). The mice were maintained under sterile conditions for 30 d. At the end of the experiment, the tumors were excised and the tumor weight was measured.
EMSA was prepared as described by Li et al[13]. Fifteen &mgr;g of nuclear proteins were incubated with 1 &mgr;g each of poly (dI-dC) in the presence of 30 fmol of digoxin (DIG)-labeled double-stranded AP-1 probe (5’-CGCTTGATG ACTCAGCCGGAA-3’, BoYa Biotechnology, China) for 15 min at room temperature in a total volume of 20 &mgr;L using DIG gel shift kit (Roche Diagnostics GmbH, Mannheim, Germany). Oligonucleotide competition experiments were performed in 50-fold excess of unlabeled oligonucleotides. DNA complexes were resolved from free probe with 4% nondenaturing polyacrylamide gels in 0.5 × Tris-borate-EDTA (pH 8.3) and visualized by fluorography.
Cells were grown to 70% confluency on 22 mm × 22 mm microscope coverslips, washed with PBS, then incubated for 1 h at room temperature with c-Jun, c-Fos monoclonal antibody (Santa Cruz Biotechnology, USA) at a final dilution of 1:200. Primary antibody was removed by repetitive washes with PBS and secondary antibody was added for 1 h at room temperature. Cells were washed in PBS and stained with DAB.
The data shown were mean values of at least three different experiments and expressed as mean ± SD. Student’s t test was used for comparison. P < 0.05 is considered statistically significant.
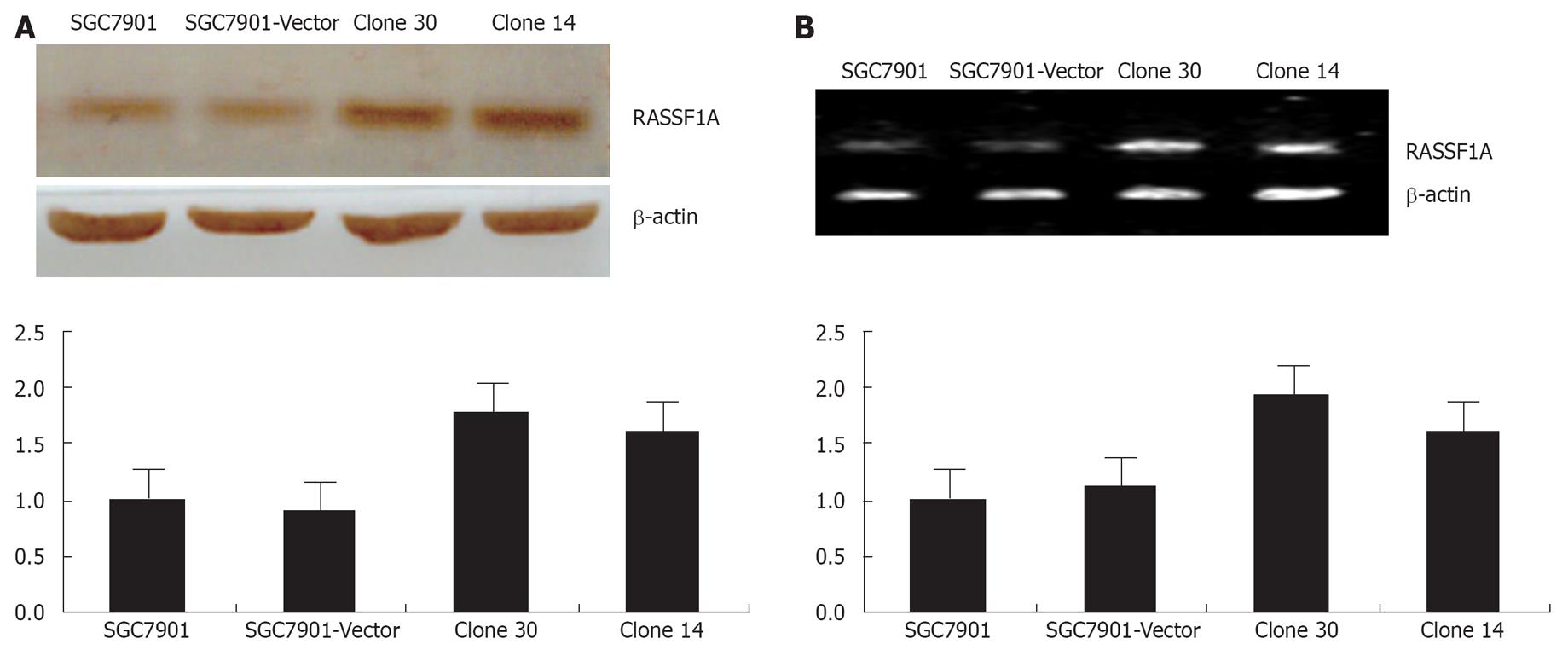
SGC-7901 human gastric cancer cells were transfected with the control pcDNA3.0, pcDNA3.0-RASSF1A plasmids, respectively. Empty vector pcDNA3.0-transfected cell clones were named vector control and, together with the parental SGC-7901 cells, served as controls in this study. After G418 selection, 4 clones of pcDNA3.0-RASSF1A cells were picked, spread, and collected, including clone14, clone 19, clone 30 and clone 33. The expression of RASSF1A in clone 30 and clone 14 were further detected by Western blot analysis and RT-PCR analysis. As shown in Figure 1A, cells transfected with the control vector did not alter RASSF1A expression when compared with the SGC-7901 cells, whereas introduction of pcDNA3.0-RASSF1A resulted in marked over-expression. The protein expression of RASSF1A was increased about 77.5%, 61.2%, when compared with the parental cells. As shown in Figure 1B, RASSF1A mRNA was increased by 80.2%, 74.7%, respectively, in the corresponding cells. Clone 30, which showed the highest degree of over-expressing RASSF1A, consequently was selected for further study. Thus, the over-expression of RASSF1A was obviously increased in our established transfected cells.
To characterize the gastric cancer cells stably over expressing RASSF1A protein, we first examined the possible effects on the rate of cell proliferation. Growth curves indicated that the vector control and parental cells displayed rapid growth rates, whereas the growth rate of the SGC7901-RASSF1A cells was significantly reduced (Figure 2A). The suppress rate was 28.13% at 48 h. The cell cycle distribution and apoptosis were determined by flow cytometry. As shown in Figure 2B, RASSF1A induced the cell cycle into G1 phase. Compared with parental and vector control cells, the percentage of G1 phase in RASSF1A transfected cells obviously increased (P < 0.05). And the apoptosis rate of cells expressing RASSF1A had a slight increase.
We analyzed the colony forming ability of the RASSF1A transfectants in planting because anchorage-independent growth often correlates with tumorigenicity. Plating efficiency in parent, vector control and RASSF1A transfected cells were 38.6% ± 1.5%, 39.75% ± 2.1% and 7.3% ± 0.6%, respectively (Figure 2C). SGC7901-RASSF1A cells displayed almost complete loss of colony-forming efficiency, and had a > 80% decrease when compared with the vector control and parental cells. In view of these results, we examined whether the RASSF1A over-expression in gastric cancer cells might affect their tumorigenicity in nude mice.
5 × 106 cells were injected s.c. into athymic nude mice and monitored for 30 d. The tumors appeared in three groups almost at the same time. At the end of the study, all of the tumors were removed and dissociated, and the weights of the tumors were measured. RASSF1A transfection revealed an obvious difference in tumor growth compared with vector control and parental cells during the observation period. The mean tumor weights in mice of parental, vector control cells and RASSF1A transfected cells were 1.4 g ± 0.26 g, 1.5 ± 0.32 g and 0.6 ± 0.1 g, respectively (Figure 2D).
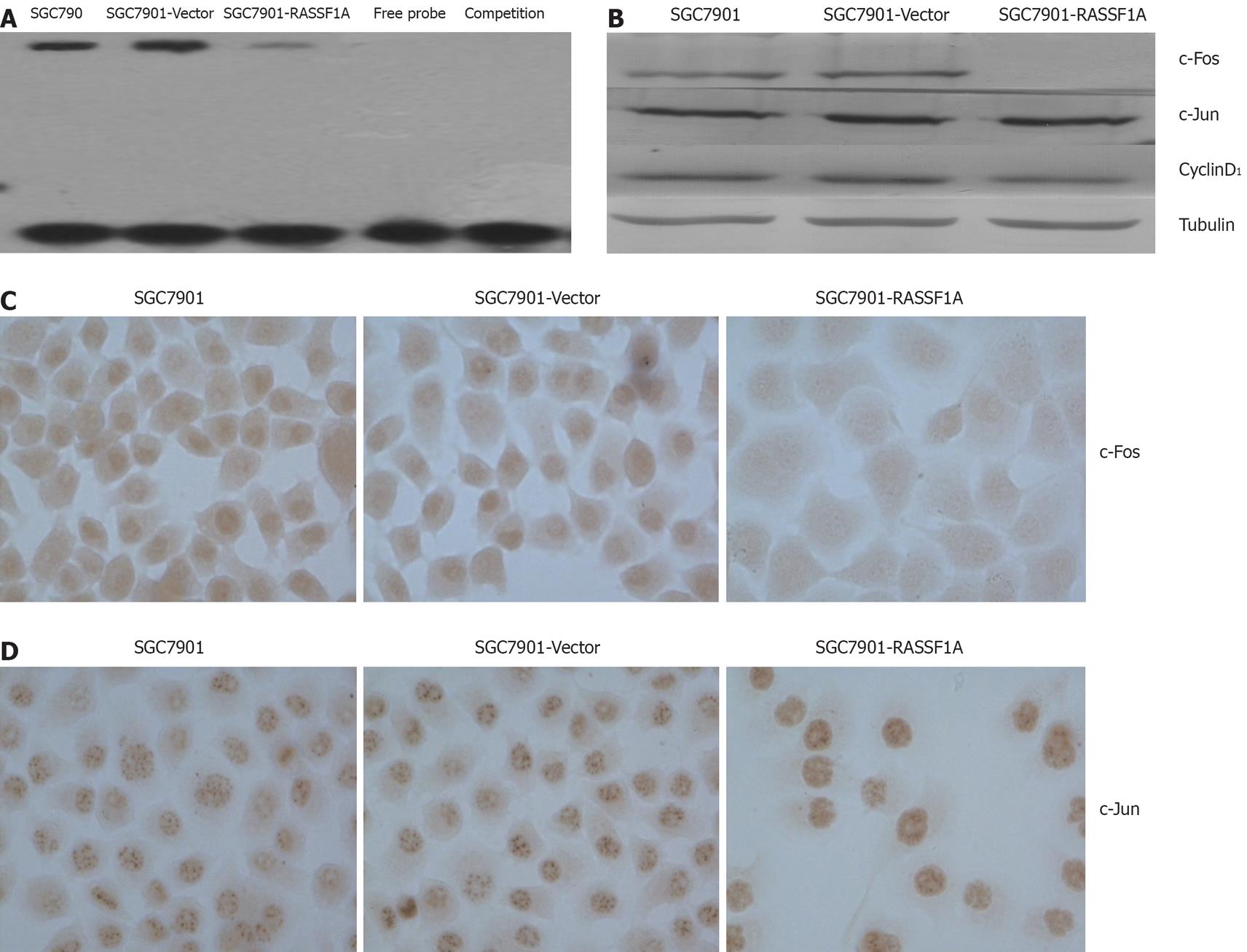
We determined the DNA binding activity of AP-1 in SGC7901 cells by electrophoresis mobility shift assay. The results showed that AP-1 DNA binding activity was significantly lower in SGC7901-RASSF1A cells than in vector control and parental cells (Figure 3A). Western blotting and immunocytochemistry data showed that c-Fos expression in the nuclear extract of SGC7901-RASSF1A cells was significantly inhibited, while, c-jun expression in SGC7901-RASSF1A cells had no significant change. Furthermore, cyclinD1 expression in nuclear extract was significantly lower in SGC7901-RASSF1A cells than in vector control and parental cells (Figure 3B -D).
In this report, we have demonstrated that RASSF1A inhibited proliferation of SGC7901 cells. The cell growth was reduced by 28.13% at 48 h as determined by the MTT assay. The alteration of cell malignant phenotype was obvious as a result of loss of anchorage-independent growth ability as measured by a plating efficiency test. The tumorigenicity in nude mice was reduced significantly (P < 0.01). RASSF1A over-expression induced cell arrest from 48.9% to 76.6% (P < 0.01) in the G1 population, and increased cell apoptosis rate from 0.78% to 2.33% (P < 0.01). These results are similar to the findings previously reported in other cancers, such as NSCLC[510], kidney[14], prostate[15], nasopharyngeal carcinoma[16], and glioma cell lines[17]. RASSF1A differentially regulated many genes identified as having relevance to tumorigenesis including involvement in transcription, cytoskeleton, signaling, cell cycle, cell adhesion, and apoptosis[18].
RASSF1A is predicted to encode a 39 kDa peptide that contains an N-terminal diacylglycerol (DAG)-binding domain, a Ras-association domain, a sequence PxxP and PEST sequences .The Ras-association domain is more than 50% identical and more than 70% similar to the carboxyl terminal 225 residues of mouse Nore1[10]. RASSF1 binds Ras in a GTP-dependent manner, both in vivo and directly in vitro[19]. It has also been shown to heterodimerize with Nore1[20]. The presence of a Ras-association domain in both RASSF1 isoforms suggests that these proteins may function as effectors of Ras signaling (or signaling of a Ras-like molecule) in normal cells. The fact that RASSF1A can function as a tumor suppressor gene implies that RASSF1 acts in opposition to Ras-effector pathways stimulating proliferation[10].
AP-1 is a transcription factor that consists of either a Jun-Jun homodimer or a Jun-Fos heterodimer. AP-1 regulates the expression of multiple genes essential for cell proliferation, differentiation and apoptosis[8]. Our results showed that RASSF1A dramatically decreased basal AP-1 activity. We further detected the expression of c-Jun/c-Fos in SGC7901. Surprisingly we failed to observe any significant change of expression of c-Jun in the present study. However, expression of c-Fos had significant change. This indicated that RASSF1A could inhibit the expression of c-Fos but not c-Jun. However, in lung cancer cells, RASSF1A reduced c-Jun phosphorylation, suppresses the c-Jun-NH2-kinase pathway and inhibits cell cycle progression[21]. We thought this difference may be due to the different histology and the role of c-Jun in RASSF1A-mediate growth inhibition in gastric carcinoma needs further investigation.
Shivakumar found that the exogenous expression of RASSF1A induced cell cycle arrest at the G1 phase by down-regulating CyclinD1[22]. In agreement with this report, we also observed that the ectopic expression of RASSF1A down-regulated CyclinD1. As a target of AP-1, CyclinD1 plays an important role in cell proliferation[23]. According to our results, we presumed that RASSF1A induced SGC7901 cell cycle arrest at the G1 phase by down-regulating CyclinD1 through inhibition of the activity of AP-1, but needs further investigation. Our current results indicate that inhibition of AP-1 activity contributes to RASSF1A mediated regulation of gastric carcinogenesis.
Thus, our data presented here clearly demonstrated that exogenous RASSF1A inhibits the growth of gastric carcinoma cells SGC7901 and RASSF1A gene may be a suppressor in gastric carcinogenesis. AP-1 may be involved in growth inhibition by the RASSF1A gene in the human gastric carcinoma cell line SGC7901.
Ras association domain family protein 1, isoform A (RASSF1A), one transcript of RASSF1 gene, is a recently identified 3p21.3 tumor suppressor gene and it is described as an effector of Ras signaling. Expression loss of RASSF1A was a frequent event in primary gastric carcinoma. However, the exact role of RASSF1A in gastric tumorigenicity is largely unknown.
Gastric carcinoma is one of the most frequent tumors that seriously threaten people’s health in China. Molecular genetics studies indicate that loss of 3p was observed in different types of solid tumors. Frequent loss on 3p21-23 was detected in gastric cancer. RASSF1A is predicted to encode a 39-kd peptide that contains an N-terminal diacylglycerol (DAG)-binding domain, a Ras-association domain, a sequence PxxP and PEST sequences. The presence of a Ras-association domain in both RASSF1 isoforms suggests that these proteins may function as effectors of Ras signaling (or signaling of a Ras-like molecule) in normal cells. The fact that RASSF1A can function as a tumor suppressor gene implies that RASSF1 acts in opposition to Ras-effector pathways stimulating proliferation.
Our study is believed to be the first to observe the role of RASSF1A in gastric cancer and the possible mechanisms involved.
Our research observed that RASSF1A suppressed the growth of the human gastric cancer cell line SGC7901 and RASSF1A inhibited AP-1 activity through down-regulated c-fos expression. This may have a significant clinical impact in the future.
Electrophoretic mobility shift assay (EMSA): The EMSA technique is based on the observation that protein: DNA complexes migrate more slowly than free DNA molecules when subjected to non-denaturing polyacrylamide or agarose gel electrophoresis. Because the rate of DNA migration is shifted or retarded upon protein binding, the assay is also referred to as a gel shift or gel retardation assay.
This is a well-written paper that suggests the role of RASSF1A in human gastric cancer. But the mechanism remains to be studied further.
| 1. | Yang L. Incidence and mortality of gastric cancer in China. World J Gastroenterol. 2006;12:17-20. |
| 2. | Kok K, Naylor SL, Buys CH. Deletions of the short arm of chromosome 3 in solid tumors and the search for suppressor genes. Adv Cancer Res. 1997;71:27-92. |
| 3. | Sakakura C, Mori T, Sakabe T, Ariyama Y, Shinomiya T, Date K, Hagiwara A, Yamaguchi T, Takahashi T, Nakamura Y. Gains, losses, and amplifications of genomic materials in primary gastric cancers analyzed by comparative genomic hybridization. Genes Chromosomes Cancer. 1999;24:299-305. |
| 4. | Yustein AS, Harper JC, Petroni GR, Cummings OW, Moskaluk CA, Powell SM. Allelotype of gastric adenocarcinoma. Cancer Res. 1999;59:1437-1441. |
| 5. | Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet. 2000;25:315-319. |
| 6. | Byun DS, Lee MG, Chae KS, Ryu BG, Chi SG. Frequent epigenetic inactivation of RASSF1A by aberrant promoter hypermethylation in human gastric adenocarcinoma. Cancer Res. 2001;61:7034-7038. |
| 7. | Kang GH, Lee S, Kim JS, Jung HY. Profile of aberrant CpG island methylation along multistep gastric carcinogenesis. Lab Invest. 2003;83:519-526. |
| 8. | Ashida R, Tominaga K, Sasaki E, Watanabe T, Fujiwara Y, Oshitani N, Higuchi K, Mitsuyama S, Iwao H, Arakawa T. AP-1 and colorectal cancer. Inflammopharmacology. 2005;13:113-125. |
| 9. | Rutberg SE, Adams TL, Glick A, Bonovich MT, Vinson C, Yuspa SH. Activator protein 1 transcription factors are fundamental to v-rasHa-induced changes in gene expression in neoplastic keratinocytes. Cancer Res. 2000;60:6332-6338. |
| 10. | Burbee DG, Forgacs E, Zochbauer-Muller S, Shivakumar L, Fong K, Gao B, Randle D, Kondo M, Virmani A, Bader S. Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J Natl Cancer Inst. 2001;93:691-699. |
| 11. | Jiang XH, Tu SP, Cui JT, Lin MC, Xia HH, Wong WM, Chan AO, Yuen MF, Jiang SH, Lam SK. Antisense targeting protein kinase C alpha and beta1 inhibits gastric carcinogenesis. Cancer Res. 2004;64:5787-5794. |
| 12. | Hu ZL, Wen JF, Xiao DS, Zhen H, Fu CY. Effects of transforming growth interacting factor on biological behaviors of gastric carcinoma cells. World J Gastroenterol. 2005;11:84-88. |
| 13. | Li B, Feng DY, Cheng RX, He QQ, Hu ZL, Zheng H, Wen JF. The effects of hepatitis C virus core protein on biological behaviors of human hepatocytes. Zhonghua Yixue Zazhi. 2005;85:1243-1248. |
| 14. | Dreijerink K, Braga E, Kuzmin I, Geil L, Duh FM, Angeloni D, Zbar B, Lerman MI, Stanbridge EJ, Minna JD. The candidate tumor suppressor gene, RASSF1A, from human chromosome 3p21.3 is involved in kidney tumorigenesis. Proc Natl Acad Sci USA. 2001;98:7504-7509. |
| 15. | Kuzmin I, Gillespie JW, Protopopov A, Geil L, Dreijerink K, Yang Y, Vocke CD, Duh FM, Zabarovsky E, Minna JD. The RASSF1A tumor suppressor gene is inactivated in prostate tumors and suppresses growth of prostate carcinoma cells. Cancer Res. 2002;62:3498-3502. |
| 16. | Chow LS, Lo KW, Kwong J, To KF, Tsang KS, Lam CW, Dammann R, Huang DP. RASSF1A is a target tumor suppressor from 3p21.3 in nasopharyngeal carcinoma. Int J Cancer. 2004;109:839-847. |
| 17. | Hesson L, Bieche I, Krex D, Criniere E, Hoang-Xuan K, Maher ER, Latif F. Frequent epigenetic inactivation of RASSF1A and BLU genes located within the critical 3p21.3 region in gliomas. Oncogene. 2004;23:2408-2419. |
| 18. | Agathanggelou A, Bieche I, Ahmed-Choudhury J, Nicke B, Dammann R, Baksh S, Gao B, Minna JD, Downward J, Maher ER. Identification of novel gene expression targets for the Ras association domain family 1 (RASSF1A) tumor suppressor gene in non-small cell lung cancer and neuroblastoma. Cancer Res. 2003;63:5344-5351. |
| 19. | Vos MD, Ellis CA, Bell A, Birrer MJ, Clark GJ. Ras uses the novel tumor suppressor RASSF1 as an effector to mediate apoptosis. J Biol Chem. 2000;275:35669-35672. |
| 20. | Ortiz-Vega S, Khokhlatchev A, Nedwidek M, Zhang XF, Dammann R, Pfeifer GP, Avruch J. The putative tumor suppressor RASSF1A homodimerizes and heterodimerizes with the Ras-GTP binding protein Nore1. Oncogene. 2002;21:1381-1390. |
| 21. | Whang YM, Kim YH, Kim JS, Yoo YD. RASSF1A suppresses the c-Jun-NH2-kinase pathway and inhibits cell cycle progression. Cancer Res. 2005;65:3682-3690. |
| 22. | Shivakumar L, Minna J, Sakamaki T, Pestell R, White MA. The RASSF1A tumor suppressor blocks cell cycle progression and inhibits cyclin D1 accumulation. Mol Cell Biol. 2002;22:4309-4318. |