Published online Feb 21, 2008. doi: 10.3748/wjg.14.1053
Revised: September 5, 2007
Published online: February 21, 2008
AIM: To investigate the polymorphic simple sequence repeat in intron 1 of the epidermal growth factor receptor gene (EGFR) (CA-SSRI), which is known to affect the efficiency of gene transcription as a putative target of the mismatch repair (MMR) machinery in colorectal tumors.
METHODS: The CA-SSR I genotype was analyzed in a total of 86 primary colorectal tumors, selected upon their microsatellite instability (MSI) status [42 with high frequency MSI (MSI-H) and 44 microsatellite stable (MSS)] and their respective normal tissue. The effect of the CA-SSR I genotype on the expression of the EGFR gene was evaluated in 18 specimens using quantitative real-time reverse transcription PCR and immunohistochemistry.
RESULTS: Mutations in CA-SSR I were detected in 86% (36 of 42) of MSI-H colorectal tumors and 0% (0 of 44) of MSS tumors, indicating the EGFR gene as a novel putative specific target of the defective MMR system (P < 0.001). Impaired expression of EGFR was detected in most of the colorectal tumors analyzed [6/12 (50%) at the mRNA level and 15/18 (83%) at the peptide level]. However, no association was apparent between EGFR expression and CA-SSR I status in tumors or normal tissues.
CONCLUSION: Our results suggest that CA-SSRI sequence does not contribute to the regulation of EGFR transcription in colon, and should thus not be considered as a promising predictive marker for response to EGFR inhibitors in patients with colorectal cancer.
-
Citation: Buisine MP, Wacrenier A, Mariette C, Leteurtre E, Escande F, Aissi S, Ketele A, Leclercq A, Porchet N, Lesuffleur T. Frequent mutations of the CA simple sequence repeat in intron 1 of
EGFR in mismatch repair-deficient colorectal cancers. World J Gastroenterol 2008; 14(7): 1053-1059 - URL: https://www.wjgnet.com/1007-9327/full/v14/i7/1053.htm
- DOI: https://dx.doi.org/10.3748/wjg.14.1053
The epidermal growth factor receptor EGFR (HER1/ERBB1) is a member of the erbB family. It plays a pivotal role in the control of processes occurring during oncogenesis and tumor progression, such as cell survival, proliferation, angiogenesis and metastasis (for review[1]). Overexpression of this receptor is observed in a large range of human cancers, correlating with an aggressive tumor phenotype, poor prognosis and decreased survival[23]. EGF, which is faintly expressed in normal colon, is seen overexpressed in approximatively 25%-77% of colorectal cancers depending on the technique used[2]. Overexpression of EGFR in colorectal tumors led to the development of targeted therapies against this receptor. Cetuximab (Erbitux®), a monoclonal antibody that specifically blocks the EGFR, has been recently approved for treatment of patients with chimiotherapy-refractory metastatic colorectal cancer. Clinical trials showed response rates of approximately 10% for cetuximab alone and 20% for cetuximab in combination with irinotecan[4]. A recent key finding was a lack of a close correlation between EGFR expression in colorectal tumors, at least as measured by immunohistochemistry, and clinical response[5]. There is therefore urgency to identify robust biological predictive markers for response to EGFR inhibitors.
Mechanisms underlying EGFR overexpression in colon are not fully understood. This overexpression can be attributed in part to gene amplification as previously shown in glioblastoma[6]. However, EGFR amplification is infrequent in colorectal cancers[7]. Furthermore, the EGFR gene contains a highly polymorphic simple sequence repeat in intron 1 (CA-SSRI), which consists of a variable number of CA dinucleotide repeats[8]. This sequence has been shown to modulate gene transcription in skin, mammary, head and neck, and gastric cell lines, with transcription activity declining with increasing number of CA repeats[910]. This finding was confirmed in breast tumors[11]. The mechanism is not fully understood, but it has been proposed that CA-SSR I could act like a joint, bringing the promoter in proximity to a putative repressor protein bound downstream of the repeat sequence[12]. Moreover, a recent study performed on head and neck cell lines reported that cell lines with a lower number of CA repeats are more sensitive to the inhibitory effect of erlotinib (Tarceva®), a small EGFR tyrosine kinase inhibitor[10]. Transferring these data to in vivo, this would lead to the hypothesis that patients would display different responses to therapy with EGFR inhibitors depending on the status of their tumor for CA-SSRI. Altogether, these data present preliminary evidence that the genotype of CA-SSR I determined in the tumor may be a promising marker to predict the clinical response to anti-EGFR therapy. However, very little is known regarding CA-SSR I in colorectal cancer.
Colorectal cancers can be subdivided into two major categories reflecting different mechanisms of carcinogenesis[13]. Almost 85% of colorectal tumors display chromosomal instability, which is characterized by aneuploidy, chromosomal rearrangements, allelic imbalance (AI) with amplifications and losses of heterozygosity (LOH). On the other hand, 12%-15% of colorectal tumors show high frequency microsatellite instability (MSI-H), which reflects a failure in the DNA mismatch repair (MMR) system. Most of them occur sporadically, but they can also occur (2%-5%) in the context of hereditary non-polyposis colorectal cancer (HNPCC). A great number of microsatellite repeat sequences distributed throughout the genome have been found (by targeted or systematic search) to be affected by MSI. However, only a small number are accepted as real targets of MSI, as they are assumed to play a role in the progression of MMR-deficient tumors[14–17]. Most of them have been described in coding sequences of genes implicated in the regulation of cell growth or cell survival. However, some non-coding repeat sequences affecting gene expression have also been proposed as targets of MMR[18].
In this context, we investigated the CA-SSR in intron 1 of EGFR in a total of 86 colorectal cancers selected upon their MSI status (42 MSI-H and 44 MSS) and in their respective normal tissue. Furthermore, we evaluated the influence of CA-SSRI mutations and length polymorphisms on EGFR expression and tested the hypothesis that CA-SSRI could be a target of MSI in MMR-deficient colorectal tumors.
A total of 86 colorectal cancers were included in the study. Cancers were selected among 250 colorectal tumors we previously analyzed for MSI using the Bethesda panel[14] or the pentaplex mononucleotide panel[19] in order to enrich our series into MSI-H tumors. They comprised 42 MSI-H tumors and 44 microsatellite stable (MSS) tumors. All were from patients who had undergone surgery for primary colorectal cancer in various hospitals from northern France. Patients followed the revised Bethesda criteria for MSI testing and identifying individuals with HNPCC (for review, see[20]).
CA-SSR I genotype analysis was conducted on tumoral tissue DNA and on respective constitutional DNA from normal colon taken at a distance from the tumor. DNA was extracted from formalin-fixed paraffin-embedded tissues using the EZ1 DNA Tissue kit with the BioRobot® EZ1 (Qiagen, Courtaboeuf, France). CA-SSR I was then amplified by PCR using previously described primers[10], the forward primer being labelled with 6-FAM at the 5’ end. Genotypes were resolved on Applied Biosystems 377XL analyzer using GeneScan software (Applied Biosystems, Courtaboeuf, France). The (CA)n dinucleotide repeat length was confirmed by direct sequencing of some samples. Alleles were compared between the tumor and the normal matching tissue. The tumor was considered as presenting MSI for CA-SSR I when additional alleles were observed compared to the matching normal tissue. The tumor was considered as presenting AI when the AI score was < 0.6 or > 1.67, as determined previously by Canzian et al[21]. AI score = T1 × N2 / T2 × N1, where T is tumor, N is normal and 1 is the area under the peak corresponding to the shorter allele, 2 is the area under the peak corresponding to the longer allele. This threshold allows for the possibility of contaminating normal tissue or tumor heterogeneity in some samples[21].
QRT-PCR analysis was conducted in 12 out of the 86 patients for whom quick-frozen tumor and normal colonic tissues were available in the tissue bank of Lille (CHRU de Lille). Total RNA was extracted from frozen tissues with the NucleoSpin® RNA II kit (Macherey-Nagel) and treated with RNase-free DNase to remove any contaminating genomic DNA. RNA (900 ng) was transcripted reversely using oligodT and the Advantage® RT-for-PCR kit (BD Biosciences Clontech) according to the manufacturer’s instructions. Expression of EGFR was investigated by QRT-PCR using SybrGreen methodology on a LightCycler (Roche) and previously described primers[22]. β-actin mRNA was used as the endogenous control gene. All real-time assays were performed in triplicate. Relative expression of EGFR in tumor samples (compared with their respective normal tissue) and in normal tissues (compared with normal tissues with a 16/16 genotype) was calculated using the 2-ΔΔCt method.
Immunohistochemical analysis was performed using the monoclonal anti-EGFR antibody Clone 31G7 from Zymed Laboratories (CliniSciences, Montrouge, France). The staining procedure was conducted using an automated immunostainer Benchmark XT (Ventana medical Systems, Strasbourg, France) and a three-step indirect process based on the biotin-streptavidin-peroxidase method. Briefly, after proteinase pretreatment (Ventana medical Systems), tissue sections were incubated with fresh 3% hydrogen peroxide in methanol to block endogenous peroxidase. The sections were then sequentially incubated with the mouse monoclonal anti-EGFR antibody (dilution 1:20, 20 min), a biotinylated rabbit anti-mouse antibody and streptavidine-peroxidase conjugate. The sections were then developed with diaminobenzidine and counterstained with hematoxylin. A section of esophagus known to express EGFR was used as a positive control.
Specimens were examined for the distribution of the immunostaining (membranous and/or cytoplasmic). Only cells showing membranous staining (alone or in association with cytoplasmic staining) were considered as positive and scored. EGFR expression was assessed according to the percentage of positive cells using the 0 to 3+ scale as follows: score 0, absence of positive cells; 1+, > 0%-1%; 2+, 2%-50%; 3+, > 50%.
Ordinal data were compared with the χ2 test or Fisher’s exact test, as appropriate. Continuous data, specifically the relationship between the number of CA repeats in normal tissues and EGFR mRNA expression levels was assessed by the Mann-Whitney U test. All statistical tests were two-sided with the threshold of significance set at P < 0.05.
Germline alleles determined in normal colonic tissues ranged in length from 14 to 21 CA repeats. Heterozygosity was 58% (50/86) for this series of patients. The most common alleles consisted of 16 CA repeats [85 of 172 chromosomes (49%)], 18 CA [31/172 (18%)], and 20 CA [38/172 (22%)]. The most common genotype was found to be homozygous for 16 CA repeats [24/86 (28%)] (Table 1).
| CA-SSR I status of the tumora | |||
| Genotypes | CA-SSR I MSI | CA-SSR I MSS | Total |
| in normal colon | (n = 36) | (n = 50) | (n = 86) |
| 14/20 | 1 (2.8) | 0 (0) | 1 (1.2) |
| 15/20 | 1 (2.8) | 0 (0) | 1 (1.2) |
| 16/16 | 10 (27.8) | 14 (28) | 24 (27.9) |
| 16/17 | 2 (5.6) | 2 (4.0) | 4 (4.7) |
| 16/18 | 4 (11.1) | 11 (22.0) | 15 (17.4) |
| 16/19 | 0 (0) | 1 (2.0) | 1 (1.2) |
| 16/20 | 6 (16.7) | 9 (18) | 15 (17.4) |
| 16/21 | 0 (0) | 2 (4.0) | 2 (2.3) |
| 17/20 | 0 (0) | 1 (2.0) | 1 (1.2) |
| 18/18 | 2 (5.6) | 2 (4.0) | 4 (4.7) |
| 18/19 | 1 (2.8) | 0 (0) | 1 (1.2) |
| 18/20 | 2 (5.6) | 3 (6.0) | 5 (5.8) |
| 18/21 | 1 (2.8) | 1 (2.0) | 2 (2.3) |
| 19/21 | 1 (2.8) | 0 (0) | 1 (1.2) |
| 20/20 | 4 (11.1) | 3 (6.0) | 7 (8.1) |
| 20/21 | 1 (2.8) | 0 (0) | 1 (1.2) |
| 21/21 | 0 (0) | 1 (2.0) | 1 (1.2) |
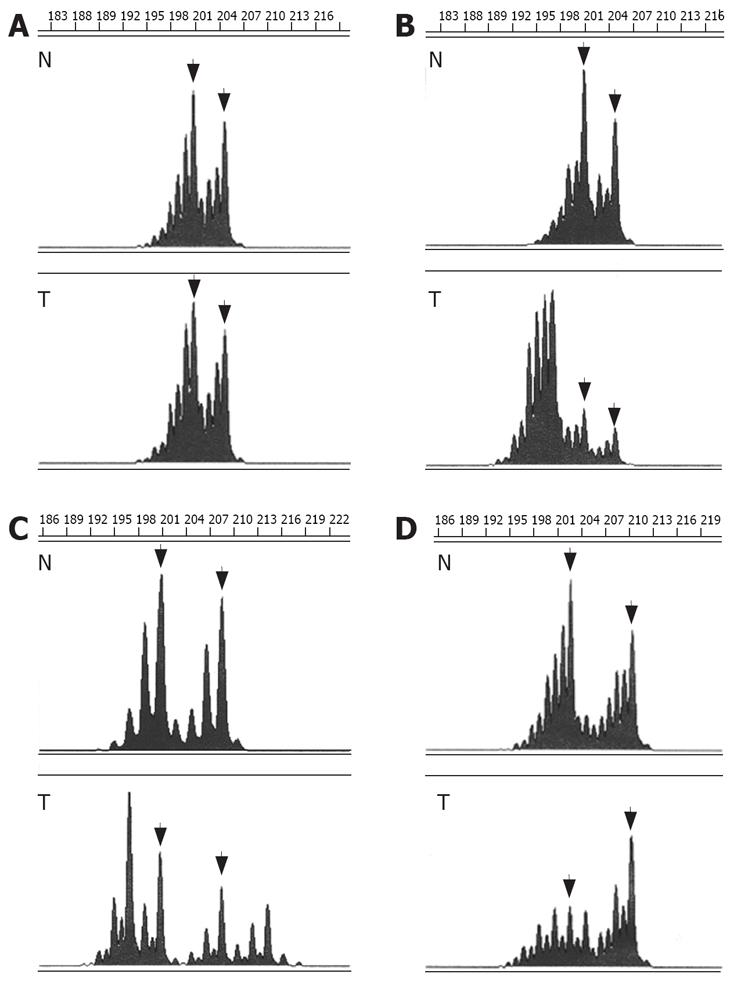
Comparison of alleles in MSS colorectal tumors and respective normal colonic tissues showed a perfect correlation of allele lengths. In contrast, instability of CA-SSR I was detected in 36 of the 42 (86%) MSI-H tumors (compared to 0 of the 44 (0%) MSS tumors, P < 0.001, χ2 test) (Table 2). Instability consisted in shortening of the CA-SSR I in 6 tumors (6/36, 17%), elongation of the CA-SSR I in 18 tumors (18/36, 50%), and combined shortening and elongation of the CA-SSR I in 12 tumors (12/36, 33%) (Figure 1). In most cases, tumors displayed multiple alleles in addition to normal germline alleles, hardly suggesting tumor cell heterogeneity. New alleles ranged in length from 12 CA repeats to 27 CA repeats for major alleles (from 12 CA repeats to 29 CA repeats for all alleles). Tumors from patients presenting at least one long allele with ≥ 20 CA in the germline were not more frequently unstable than tumors from patients with both alleles shorter (< 20 CA) [17/19 (89%] versus [19/23 (83%)], (P = 0.673, χ2 test) (Table 1), indicating that long CA-SSR I alleles are not particularly prone to MSI.
Moreover, comparison of alleles in tumors and respective normal tissues allowed detection of AI in 5 of the 30 (17%) MSS tumors from patients heterozygous for CA-SSR I and that were thus informative (AI scores = 0.47, 0.59, 2.27, 3.03, and 4.46, respectively) (Table 2).
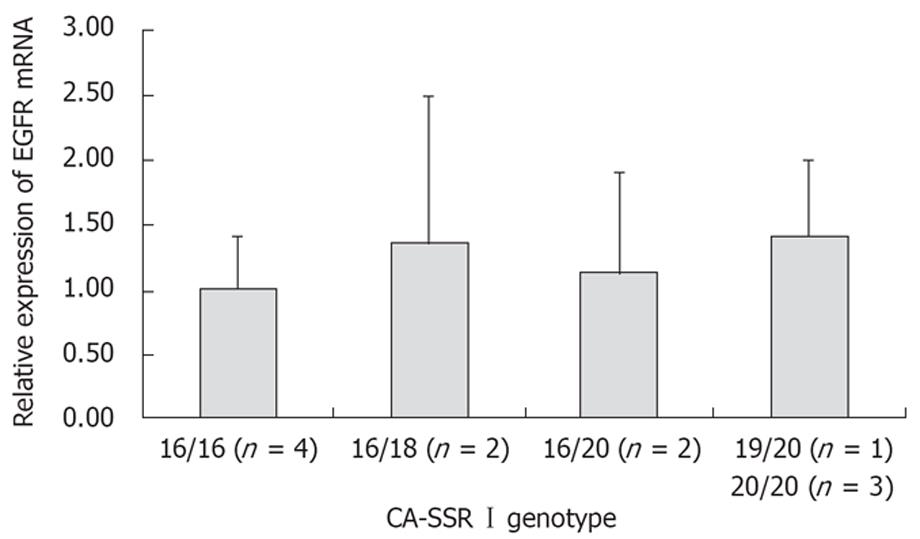
To investigate the influence of CA-SSR I mutations and allele lengths on EGFR gene transcription, we used QRT-PCR to evaluate EGFR mRNA expression in both tumor and normal tissues from 12 patients we previously analyzed for CA-SSR I genotype. Comparison between tumors and matching normal tissues showed impaired expression of EGFR mRNA in half of the cases (6/12 with fold change > 2 or < 0.5) (Table 3). However, no association was apparent between EGFR mRNA expression levels measured in tumors and the length of CA-SSR I alleles, the presence of CA-SSR I instability or AI. Furthermore, EGFR mRNA expression levels measured in normal tissues were similar among individuals, whatever their CA-SSR I genotype status, indicating no association between expression of EGFR mRNA and the number of CA repeats in normal colon (two by two comparisons, P > 0.468, Mann-Whitney U test) (Figure 2).
| CA-SSR genotypes1 | CA-SSR I status2 of the tumor | EGFR expression | |||
| Normal colon | Tumor | QRT-PCR3 | IHC4 | ||
| 1 | 16/16 | 16/20 | MSI | 0.87 | ++ m |
| 2 | 16/16 | 16/16 | MSS | 0.70 | ++ m |
| 3 | 16/18 | 16/18 | MSS | 2.68 | ++ m |
| 4 | 16/16 | 14/16/20/(21/22/23) | MSI | 5.10 | +++ m, c |
| 5 | 20/20 | 16/25/26/27 | MSI | 0.27 | ++ m, c |
| 6 | 20/20 | 20/20 | MSS | 0.76 | ++ m |
| 7 | 16/20 | 16/20 | MSS (AI +) | 1.11 | +++ m |
| 8 | 20/20 | 20/20 | MSS | 0.48 | ++ m |
| 9 | 16/20 | 16/20 | MSS | 3.68 | ++ m |
| 10 | 16/16 | 16/16 | MSS | 1.32 | ++ m |
| 11 | 16/18 | 16/18 | MSS (AI +) | 0.47 | + m |
| 12 | 19/21 | 19/21/(22/23) | MSI | 1,57 | +++ m, c |
| 13 | 16/18 | 14/16/18 | MSI | ND | ++ m, c |
| 14 | 14/20 | 12/14/20/(22/23) | MSI | ND | ++ m, c |
| 15 | 16/18 | 16/18/(23/24) | MSI | ND | +++ m, c |
| 16 | 16/20 | 16/20 | MSS | ND | - |
| 17 | 16/16 | 16/16 | MSS | ND | - |
| 18 | 18/18 | 18/18 | MSS | ND | ++ m |
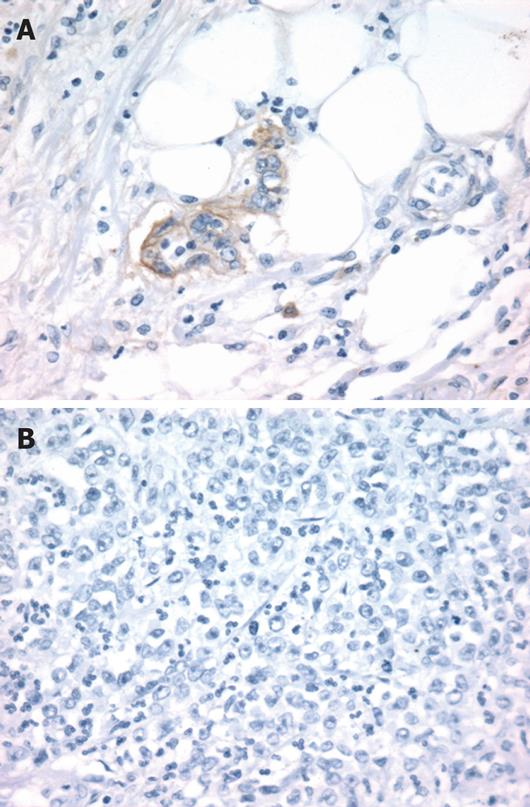
EGFR peptide expression was evaluated by immuno-histochemistry in 18 tumors previously analyzed for CA-SSR I genotype. Overexpression of EGFR with membranous staining or both membranous and cytoplasmic staining (score ≥ 2+) was observed in 15 out of the 18 (83%) tumors analyzed (Table 3, Figure 3). EGFR immunostaining level appeared to be not associated with the length of CA-SSRIalleles, the presence of CA-SSRIinstability or AI. Moreover, no association could be observed between EGFR immunostaining and EGFR mRNA expression levels. Noteworthy, cytoplasmic staining alone (without membranous staining) was observed in 6 of the 7 tumors (86%) with CA-SSRIinstability compared to 0 of the 11 tumors (0%) without CA-SSRIinstability (P < 0.001, χ2 test). This observation could not be related to a particular histopathological feature such as the degree of differentiation.
Here, we have analyzed primary colorectal tumors selected by their MSI status (42 MSI-H and 44 MSS) and their respective normal tissue for CA-SSR I mutations and length polymorphisms. Furthermore, we evaluated the effect of CA-SSR I genotype with respect to EGFR expression in colorectal tumors.
The distribution of CA-SSR I alleles and genotypes that were determined in normal tissues were similar to those reported previously in Caucasian reference pedigrees, with the most common alleles consisting of 16, 18, and 20 CA repeats and the most common genotype being homozygous for 16 CA repeats[8]. Interestingly, whereas allele lengths were perfectly conserved in MSS tumors, frequent instability of CA-SSR I was detected in MSI-H tumors. This instability consisted in shortening, elongation, or combined shortening and elongation of the CA-SSR I; it was independent of the length of alleles determined in normal tissues. The frequency of instability was particularly high, accounting for 86% of MSI-H colorectal tumors. EGFR CA-SSR I may thus be of interest as an alternative marker for the determination of the MSI status in colorectal tumors[14]. It is noteworthy that this study was performed on tumors without previous microdissection. Enriching for tumor cells using microdissection would still enhance the likelihood of detecting novel shifted alleles. One could argue that this study was performed on tumors of patients following the Bethesda criteria, and thus suspected to have a HNPCC syndrome, introducing some biases in the data. However, previous studies revealed similar frequencies in instability for a given marker in both sporadic and HNPCC tumors[16]. Moreover, 15 of 24 patients with MSI-H who were screened for germline mutations in MMR genes by direct sequencing and by multiplex ligation-dependant probe amplification displayed mutations in MLH1, MSH2 or MSH6, indicating that approximately 1/3 of MSI-H tumors tested here were likely to be sporadic (data not shown). Of note, CA-SSR I analysis of two MSI-H keratoacanthomas and one MSI-H endometrial tumor from three independent HNPCC patients showed CA-SSR I instability in one keratoacanthome indicating that CA-SSR I instability is probably not restricted to colorectal tumors (data not shown). To our knowledge, there has been no previous report demonstrating such alteration of EGFR CA-SSR I polymorphism in MSI-H tumors.
Most of the non-coding repeat mutations observed in MSI-H tumors are considered as background events, since they are unlikely to participate to oncogenesis processes. In this respect, mutation frequencies in these sequences, in the absence of selection pressure, are generally low[141517]. However, mutations in non-coding sequences may contribute to the manifestation of the malignant phenotype if they occur in gene regulatory regions that control levels of expression[1723]. For example, a T repeat in intron 4 of the MRE11 gene is frequently mutated in MSI-H tumors. These mutations have been shown to cause a splicing defect with reduced mRNA and protein levels[18]. Here, we have shown that 86% of MSI-H colorectal tumors harbor mutations of CA-SSR I, which is indicative of a possible positive selection pressure. Moreover, the CA-SSR I sequence has been shown in various tissues and cell lines to modulate EGFR gene transcription[9–11]. These findings led us to consider EGFR as a novel putative target gene of the defective MMR machinery and to hypothesize that EGFR CA-SSR I mutations, as well as CA-SSRIlength polymorphism, could alter the transcriptional activity of the EGFR gene and explain, at least in part, variations of EGFR expression between colorectal tumors.
In addition to the presence of frequent CA-SSRImutations in MSI-H tumors, CA-SSR I analysis allowed us to detect AI in 17% of MSS tumors. EGFR gene amplification has been recently shown to occur in 12% to 31% of colorectal tumors, as determined by chromogenic[7] and fluorescent in situ hybridization[24]. Therefore, the presence of AI in some MSS tumors probably reflects amplification of the EGFR gene in these tumors, as demonstrated previously for breast tumors[25]. Interestingly, Moroni et al[24] have shown that assessment of EGFR copy number might identify patients who are likely to respond to cetuximab.
In order to evaluate potential functional consequences of CA-SSR I mutations and polymorphisms, we investigated EGFR expression at both the mRNA and peptide levels. As expected, EGFR peptide was found to be overexpressed in most of the tumors analyzed (83% in the present series). EGFR mRNA expression was also altered in most colorectal tumors, as assessed by QRT PCR. However, EGFR mRNA and peptide expression appeared to be not associated with the CA-SSR I allele length, nor with the presence of MSI or AI. Our data regarding EGFR peptide expression are in accordance with those obtained by McKay et al[26] who showed no association between the distribution of CA-SSR I alleles in colorectal tumors and EGFR peptide expression. More intriguingly, the CA-SSR I genotype identified in normal colon samples was not predictive of EGFR mRNA expression levels. Our results differ from those obtained in other tissues. EGFR transcription activity has been shown in vitro and in vivo in mammary cell lines to decline with increasing number of CA repeats[9]. In accordance, EGFR peptide expression was subsequently found in breast tumors to correlate with the length of CA-SSR I alleles[11]. A direct correlation between EGFR expression levels and CA-SSR I has also been demonstrated in skin, gastric, and head and neck cell lines[910], but these findings remain to be confirmed in corresponding tissues. Altogether, our present data hardly suggest that in colorectal cancer and normal colon, unlike in breast cancer, CA-SSR I does not contribute to the regulation of EGFR transcription.
In conclusion, we have shown that the CA-SSR polymorphism in intron 1 of EGFR is frequently altered in MMR-deficient colorectal tumors, as well as in a subset of MSS tumors However, EGFR CA-SSR I polymorphisms and mutations seem to be not associated with EGFR expression. Our data indicate that the EGFR gene is more likely to be a ‘bystander’ rather than a ‘real functional’ target of the MMR machinery. Given the present data, CA-SSR I should not be considered as a promising marker for predicting the clinical response to EGFR inhibitors in patients with colorectal tumors.
Cetuximab (Erbitux®), a monoclonal antibody that specifically blocks the epidermal growth factor receptor (EGFR), has been recently approved for treatment of patients with chimiotherapy-refractory metastatic colorectal cancer. Clinical trials showed limited response rates for cetuximab alone or in combination with irinotecan. There is therefore urgency to identify robust biological predictive markers for response to EGFR inhibitors.
Our article focuses on genetic alterations of the EGFR gene in colorectal cancers.
The polymorphic simple sequence repeat in intron 1 of the EGFR gene (CA-SSR I) has been investigated in various cancers, including colorectal cancers. No abnormality has been found in this sequence, except in breast cancers in which allelic imbalance has been extensively described. The present study shows very frequent mutations of CA-SSR I in mismatch repair-deficient colorectal cancers, indicating the EGFR gene as a novel putative specific target of the defective mismatch repair system.
Impaired expression of EGFR was detected in most of colorectal tumors analyzed, but without apparent association with CA-SSRIstatus. Although these results are preliminary, they hardly suggest that CA-SSRIsequence does not contribute to the regulation of EGFR transcription in colon and should thus not be considered as a promising predictive marker for response to EGFR inhibitors.
The manuscript from Dr. Buisine et al presents an interesting review of mismatch repair deficient cancers looking at polymorphism and mutations of the CA simple sequence repeat in intron-1 of EGFR receptor.
| 1. | Camp ER, Summy J, Bauer TW, Liu W, Gallick GE, Ellis LM. Molecular mechanisms of resistance to therapies targeting the epidermal growth factor receptor. Clin Cancer Res. 2005;11:397-405. |
| 2. | Salomon DS, Brandt R, Ciardiello F, Normanno N. Epidermal growth factor-related peptides and their receptors in human malignancies. Crit Rev Oncol Hematol. 1995;19:183-232. |
| 3. | Nicholson RI, Gee JM, Harper ME. EGFR and cancer prognosis. Eur J Cancer. 2001;37 Suppl 4:S9-S15. |
| 4. | Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, Bets D, Mueser M, Harstrick A, Verslype C. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351:337-345. |
| 5. | Ciardiello F, Tortora G. Epidermal growth factor receptor (EGFR) as a target in cancer therapy: understanding the role of receptor expression and other molecular determinants that could influence the response to anti-EGFR drugs. Eur J Cancer. 2003;39:1348-1354. |
| 6. | Romeike BF, Jung V, Feiden W, Moringlane JR, Zang KD, Urbschat SM. Distribution of epidermal growth factor receptor protein correlates with gain in chromosome 7 revealed by comparative genomic hybridization after microdissection in glioblastoma multiforme. Pathol Res Pract. 2001;197:427-431. |
| 7. | Shia J, Klimstra DS, Li AR, Qin J, Saltz L, Teruya-Feldstein J, Akram M, Chung KY, Yao D, Paty PB. Epidermal growth factor receptor expression and gene amplification in colorectal carcinoma: an immunohistochemical and chromogenic in situ hybridization study. Mod Pathol. 2005;18:1350-1356. |
| 8. | Liu W, Innocenti F, Chen P, Das S, Cook EH Jr, Ratain MJ. Interethnic difference in the allelic distribution of human epidermal growth factor receptor intron 1 polymorphism. Clin Cancer Res. 2003;9:1009-1012. |
| 9. | Gebhardt F, Zanker KS, Brandt B. Modulation of epidermal growth factor receptor gene transcription by a polymorphic dinucleotide repeat in intron 1. J Biol Chem. 1999;274:13176-13180. |
| 10. | Amador ML, Oppenheimer D, Perea S, Maitra A, Cusati G, Iacobuzio-Donahue C, Baker SD, Ashfaq R, Takimoto C, Forastiere A. An epidermal growth factor receptor intron 1 polymorphism mediates response to epidermal growth factor receptor inhibitors. Cancer Res. 2004;64:9139-9143. |
| 11. | Buerger H, Gebhardt F, Schmidt H, Beckmann A, Hutmacher K, Simon R, Lelle R, Boecker W, Brandt B. Length and loss of heterozygosity of an intron 1 polymorphic sequence of egfr is related to cytogenetic alterations and epithelial growth factor receptor expression. Cancer Res. 2000;60:854-857. |
| 12. | Gebhardt F, Burger H, Brandt B. Modulation of EGFR gene transcription by secondary structures, a polymorphic repetitive sequence and mutations--a link between genetics and epigenetics. Histol Histopathol. 2000;15:929-936. |
| 13. | Ilyas M, Straub J, Tomlinson IP, Bodmer WF. Genetic pathways in colorectal and other cancers. Eur J Cancer. 1999;35:1986-2002. |
| 14. | Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248-5257. |
| 15. | Perucho M. Correspondence re: C.R. Boland et al., A National Cancer Institute workshop on microsatellite instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res., 58: 5248-5257, 1998. Cancer Res. 1999;59:249-256. |
| 16. | Duval A, Hamelin R. Mutations at coding repeat sequences in mismatch repair-deficient human cancers: toward a new concept of target genes for instability. Cancer Res. 2002;62:2447-2454. |
| 17. | Perucho M. Tumors with microsatellite instability: many mutations, targets and paradoxes. Oncogene. 2003;22:2223-2225. |
| 18. | Giannini G, Rinaldi C, Ristori E, Ambrosini MI, Cerignoli F, Viel A, Bidoli E, Berni S, D'Amati G, Scambia G. Mutations of an intronic repeat induce impaired MRE11 expression in primary human cancer with microsatellite instability. Oncogene. 2004;23:2640-2647. |
| 19. | Suraweera N, Duval A, Reperant M, Vaury C, Furlan D, Leroy K, Seruca R, Iacopetta B, Hamelin R. Evaluation of tumor microsatellite instability using five quasimonomorphic mononucleotide repeats and pentaplex PCR. Gastroenterology. 2002;123:1804-1811. |
| 20. | Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Ruschoff J, Fishel R, Lindor NM, Burgart LJ, Hamelin R. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261-268. |
| 21. | Canzian F, Salovaara R, Hemminki A, Kristo P, Chadwick RB, Aaltonen LA, de la Chapelle A. Semiautomated assessment of loss of heterozygosity and replication error in tumors. Cancer Res. 1996;56:3331-3337. |
| 22. | Pawlowski V, Revillion F, Hebbar M, Hornez L, Peyrat JP. Prognostic value of the type I growth factor receptors in a large series of human primary breast cancers quantified with a real-time reverse transcription-polymerase chain reaction assay. Clin Cancer Res. 2000;6:4217-4225. |
| 23. | Hienonen T, Sammalkorpi H, Enholm S, Alhopuro P, Barber TD, Lehtonen R, Nupponen NN, Lehtonen H, Salovaara R, Mecklin JP. Mutations in two short noncoding mononucleotide repeats in most microsatellite-unstable colorectal cancers. Cancer Res. 2005;65:4607-4613. |
| 24. | Moroni M, Veronese S, Benvenuti S, Marrapese G, Sartore-Bianchi A, Di Nicolantonio F, Gambacorta M, Siena S, Bardelli A. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. Lancet Oncol. 2005;6:279-286. |
| 25. | Tidow N, Boecker A, Schmidt H, Agelopoulos K, Boecker W, Buerger H, Brandt B. Distinct amplification of an untranslated regulatory sequence in the egfr gene contributes to early steps in breast cancer development. Cancer Res. 2003;63:1172-1178. |
| 26. | McKay JA, Murray LJ, Curran S, Ross VG, Clark C, Murray GI, Cassidy J, McLeod HL. Evaluation of the epidermal growth factor receptor (EGFR) in colorectal tumours and lymph node metastases. Eur J Cancer. 2002;38:2258-2264. |