Published online May 21, 2008. doi: 10.3748/wjg.14.3074
Revised: April 27, 2008
Published online: May 21, 2008
AIM: To evaluate the diagnostic role of serum RASSF1A promoter hypermethylation in gastric and colorectal adenocarcinoma.
METHODS: Methylation-specific polymerase chain reaction (MSPCR) was used to examine the promoter methylation status of the serum RASSF1A gene in 47 gastric adenocarcinoma patients, 45 colorectal adenocarcinoma patients, 60 patients with benign gastrointestinal disease (30 with benign gastric disease and 30 with benign colorectal disease), and 30 healthy donor controls. A paired study of RASSF1A promoter methylation status in primary tumor, adjacent normal tissue, and postoperative serum were conducted in 25 gastric and colorectal adenocarcinoma patients who later were underwent surgical therapy.
RESULTS: The frequencies of detection of serum RASSF1A promoter hypermethylation in gastric (34.0%) and colorectal (28.9%) adenocarcinoma patients were significantly higher than those in patients with benign gastric (3.3%) or colorectal (6.7%) disease or in healthy donors (0%) (P < 0.01). The methylation status of RASSF1A promoter in serum samples was consistent with that in paired primary tumors, and the MSPCR results for RASSF1A promoter methylation status in paired preoperative samples were consistent with those in postoperative serum samples. The serum RASSF1A promoter hypermethylation did not correlate with patient sex, age, tumor differentiation grade, surgical therapy, or serum carcinoembryonic antigen level. Although the serum RASSF1A promoter hypermethylation frequency tended to be higher in patients with distant metastases, there was no correlation between methylation status and metastasis.
CONCLUSION: Aberrant CpG island methylation within the promoter region of RASSF1A is a promising biomarker for gastric and colorectal cancer.
-
Citation: Wang YC, Yu ZH, Liu C, Xu LZ, Yu W, Lu J, Zhu RM, Li GL, Xia XY, Wei XW, Ji HZ, Lu H, Gao Y, Gao WM, Chen LB. Detection of
RASSF1A promoter hypermethylation in serum from gastric and colorectal adenocarcinoma patients. World J Gastroenterol 2008; 14(19): 3074-3080 - URL: https://www.wjgnet.com/1007-9327/full/v14/i19/3074.htm
- DOI: https://dx.doi.org/10.3748/wjg.14.3074
Gastric and colorectal cancers are two of the most common causes of cancer-related death worldwide. Development of efficient diagnostic methods to enable their early detection plays an essential role in increasing the survival rate of patients with these diseases. Although endoscopy is considered the most sensitive screening tool for gastric and colorectal cancers, its use is limited due to its considerable cost and risk, and patients’ lack of acceptance of the invasive procedure. Therefore, reliable noninvasive test, preferably blood test, for screening and diagnostic purposes are obviously needed.
Conventional tumor markers in serum, such as carcinoembryonic antigen (CEA), are generally insensitive for screening purposes[1]. Consequently, novel serum biomarkers are clearly needed for the early detection of gastric and colorectal cancers.
Aberrant DNA methylation, a feature of many human cancers, frequently occurs as an early event in tumorigenesis and is characterized by general hypomethylation and regional hypermethylation[2]. The hypermethylation of CpG islands within the promoter and/or upstream exon regions is an important epigenetic mechanism underlying the inactivation of tumor-suppressor genes (TSGs)[3]. It was reported that quite a few TSGs, including the Ras association domain family 1A (RASSF1A) gene, are epigenetically silenced by aberrant promoter hypermethylation in gastric and colorectal cancer[4–10]. RASSF1A is a newly identified candidate TSG located in the 3p21.3 region[11], and promoter hypermethylation of RASSF1A, which is its most common inactivation mechanism, has been observed in many human solid tumors, including gastric and colorectal cancers[11–17].
It has been long known that the serum level of free DNA is increased in cancer patients, which is believed to be released from cancer cells[1819]. It was reported that genetic and epigenetic alterations in serum DNA (such as point mutation, gene amplification, loss of heterozygosity, microsatellite instability, and aberrant methylation) are identical to those found in primary human cancers[20–24]. Because the promoter methylation status of TSGs in primary tumors and matched serum samples was consistent with each other[42526], promoter hypermethylation of TSGs in serum DNA may become a promising biomarker for gastric and colorectal cancers.
In the present study, we attempted to identify the RASSF1A promoter methylation status both in serum DNA and in available paired tumor genomic DNA from patients with gastric and colorectal adenocarcinomas by using methylation-specific polymerase chain reaction (MSPCR). We also analyzed the correlation between serum RASSF1A gene promoter hypermethylation and patients’ clinicopathologic parameters to further evaluate the clinical significance of this molecular change.
This study included 47 gastric adenocarcinoma patients and 45 colorectal adenocarcinoma patients diagnosed at Departments of General Surgery, Gastroenterology, and Medical Oncology of Jinling Hospital (Nanjing, China) between August 1, 2006 and November 30, 2007. All diagnoses were based on pathologic evidence, and only patients with adenocarcinoma, the most common histologic type of gastric and colorectal cancer, were included. The clinicopathologic characteristics of these patients are summarized in Table 1.
| Characteristics | Patients (n) | ||
| Gastric cancer | Colorectal cancer | ||
| Sex | Male | 29 | 24 |
| Female | 18 | 21 | |
| Age (yr) | ≤ 60 | 21 | 31 |
| > 60 | 26 | 14 | |
| Differentiation grade | G1/Broders’ I | 2 | 4 |
| G2/Broders’ II | 23 | 34 | |
| G3/Broders’ III & IV | 22 | 7 | |
| Stage | TNM I/Duke’s A | 4 | 5 |
| TNM II/Duke’s B | 15 | 16 | |
| TNM III/Duke’s C | 16 | 14 | |
| TNM IV/Duke’s D | 12 | 10 | |
The control population consisted of 60 patients with benign gastrointestinal diseases (30 with benign gastric disease and 30 with benign colorectal disease, such as chronic gastritis, gastric ulcer, benign polyp, nonmalignant adenoma, and ulcerative colitis; data not shown) and 30 healthy donors.
Gastric adenocarcinoma was staged according to the sixth edition of the TNM staging system[27], and colorectal adenocarcinoma was staged according to the Duke’s staging system. Gastric and colorectal adenocarcinoma differentiation was graded according to the World Health Organization grading system and the Broders’ grading system, respectively.
Our study was approved by the ethical committee of the hospital and informed consent was obtained from all patients.
Five mL of peripheral venous blood was collected from each patient 1 day after the patients were admitted to our hospital. At this time, the patients did not start their treatment. Any previous treatment (surgery and/or chemotherapy), if given, was discontinued at least 4 wk earlier. Fresh tumor tissue and paired adjacent normal tissue were obtained from 16 gastric and 9 colorectal adenocarcinoma patients who later were underwent to surgical therapy in Jinling Hospital. An additional 5 mL peripheral venous blood was collected from these 25 patients 4 wk after surgery for a comparative study. All blood samples were kept in tubes containing clot activator at 4°C for 2 h, and samples were centrifuged at 3000 r/min for 10 min to isolate sera. Thirty serum samples from healthy donors were obtained from the Blood Center of Jinling Hospital as normal controls. All serum and tissue samples were stored at -80°C until use.
Serum DNA, extracted with the QIAamp blood mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions, was stored at -80°C until use. Genomic DNA isolated from tissue samples was prepared using standard phenol/chloroform extraction protocols.
The extracted DNA was modified according to Herman et al[28] with minor modifications, to convert all unmethylated cytosines to uracils. Briefly, 1 &mgr;g of genomic DNA, or serum DNA extracted from 5 mL blood plus 1 &mgr;g of salmon sperm carrier DNA (Sigma, St. Louis, MO, USA), in a total volume of 50 &mgr;L, were denatured by NaOH (0.3 mol/L final concentration) at 40°C for 15 min. After 30 &mgr;L of freshly prepared 10 mmol/L hydroquinone (Sigma) and 520 &mgr;L of freshly prepared 3 mol/L sodium bisulfite (Sigma) at pH 5.0 were added, the samples were incubated under mineral oil at 55°C in darkness for 14 h. The modified DNA was purified using the Wizard DNA clean-up system (Promega, Madison, WI, USA), following its manufacturer’s protocol. Modification was completed by NaOH (0.3 mol/L final concentration) treatment for 10 min at room temperature, followed by ethanol precipitation. The modified DNA was resuspended in sterile deionized water (100 &mgr;L for genomic DNA and 25 &mgr;L for serum DNA) and used immediately or stored at -80°C.
Two sets of primers, described elsewhere[29], were used to discriminate between methylated and unmethylated alleles (Table 2). The PCR system has been described previously[30]. Briefly, the PCR mixture containing 2.5 &mgr;L of 10 × reaction buffer (100 mmol/L Tris-HCl (pH 8.3), 500 mmol/L KCl, 15 mmol/L MgCl2), 10 &mgr;L of modified DNA, 15 pmol of each primer (Shenery Biocolor, Shanghai, China), 2 &mgr;L of deoxynucleotide triphosphates (200 &mgr;mol/L each, final concentration), and 1 U TaKaRa Taq™ polymerase (Hot Start Version, TaKaRa, Shiga, Japan) was adjusted by H2O to a final volume of 25 &mgr;L. The cycling conditions consisted of an incubation period at 95°C for 15 min, 40 cycles of denaturation at 94°C for 30 s, annealing at 64°C or at 59°C for 50 s (Table 2), extension at 72°C for 30 s, and a final extension at 72°C for 10 min. PCR products were separated in 2% agarose gel and visualized under UV illumination.
| Primer | Sequence (5’-3’) | Amplicon location1 | Annealing temperature | Product size (bp) |
| MF | GGGTTTTGCGAGAGCGCG | 17 882-18 050 | 64°C | 169 |
| MR | GCTAACAAACGCGAACCG | |||
| UF | GGTTTTGTGAGAGTGTGTTTAG | 17 883-18 051 | 59°C | 169 |
| UR | CACTAACAAACACAAACCAAAC |
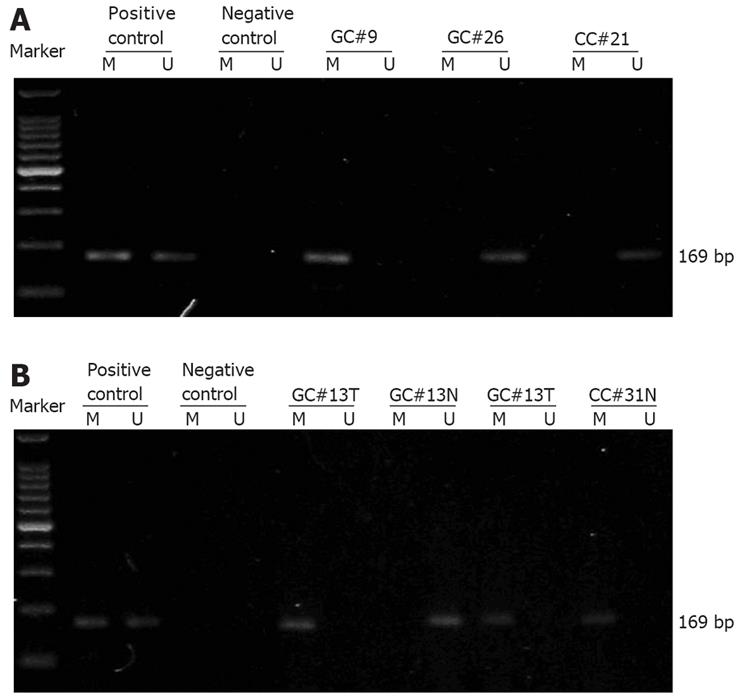
Lymphocyte DNA, original or methylated in vitro by excessive CpG (Sss I) methylase (New England Biolabs, Beverly, MA, USA), was used as an unmethylated and methylated DNA positive control, respectively (Figure 1A). Water blank was used as a negative control.
We analyzed the correlation between methylation status of serum RASSF1A promoter and clinicopathologic parameters. Chi-square test or Fisher’s exact test was conducted to examine the association of two categorical variables using SAS software (SAS Institute, Cary, NC, USA). All statistical tests were two-sided, and P < 0.05 was considered statistically significant.
First we analyzed the methylation status of CpG islands within the RASSF1A promoter in serum DNA from 47 gastric adenocarcinoma patients, 45 colorectal adenocarcinoma patients, 60 benign gastrointestinal disease patients (30 with benign gastric disease and 30 with benign colorectal disease), and 30 healthy donors. Hypermethylation of the RASSF1A promoter was detected in 16 gastric adenocarcinoma patients, 13 colorectal adenocarcinoma patients, 1 benign gastric disease patient (chronic fundal gastritis), and 2 benign colorectal disease patients (both colon adenomas). The representative agarose gel electrophoresis results are shown in Figure 1A. The frequencies of detection of serum RASSF1A promoter hypermethylation in gastric (34.0%) and colorectal (28.9%) adenocarcinoma patients were significantly higher than those in benign gastric disease patients (3.3%), benign colorectal disease patients (6.7%) and healthy donors (0%), respectively (P < 0.01).
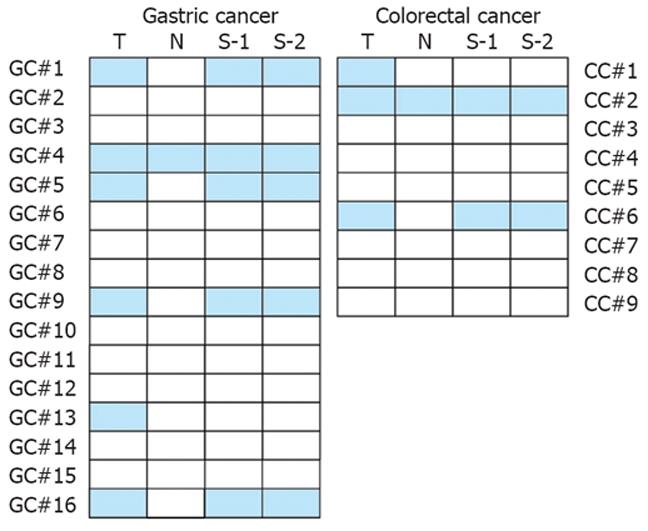
Next we compared the RASSF1A promoter methylation status in paired tissue and serum samples from 16 gastric and 9 colorectal adenocarcinoma patients who later were underwent to surgical resection in Jinling Hospital. For each patient, the RASSF1A promoter methylation status was analyzed in tumor tissue, adjacent normal tissue, preoperative serum, and postoperative serum collected 4 wk after surgery. The representative agarose gel electrophoresis results and the paired MSPCR results are shown in Figure 1B and Figure 2, respectively. In seven patients, the RASSF1A promoter hypermethylation was detected both in cancer tissue samples and in serum samples. In two patients, the hypermethylated RASSF1A promoter was present in tumor tissue samples but not in paired serum samples. The RASSF1A promoter hypermethylation was never detected in serum samples if it was not present in tumor tissue. In addition, the preoperative and postoperative serum RASSF1A promoter methylation status remained unchanged in all patients.
We further analyzed the relationship between serum RASSF1A promoter methylation status and clinicopathologic features in gastric and colorectal adenocarcinoma patients. The results are listed in Table 3. As indicated in the table, there was no correlation between RASSF1A promoter methylation status and patients’ sex, age, tumor differentiation grade, or serum CEA levels. No difference in serum RASSF1A promoter hypermethylation frequencies was detected between postoperative patients and those whose tumor was not resected. Although the serum RASSF1A promoter hypermethylation frequency tended to be higher in patients with distant metastases, no correlation between methylation status and metastasis was found.
| Clinicopathologic parameters | Gastric cancer | Colorectal cancer | |||||
| RASSF1A promoter status | P value | RASSF1A promoter status | P value | ||||
| M | U | M | U | ||||
| Sex | Male | 9 | 20 | 0.58071 | 7 | 17 | 0.96491 |
| Female | 7 | 11 | 6 | 15 | |||
| Age (yr) | ≤ 60 | 8 | 13 | 0.59821 | 8 | 23 | 0.50242 |
| > 60 | 8 | 18 | 5 | 9 | |||
| Differentiation grade | G1/Broders’ I | 0 | 2 | 0.22801 | 1 | 3 | 0.98301 |
| G2/Broders’ II | 6 | 17 | 10 | 24 | |||
| G3/Broders’ III & IV | 10 | 12 | 2 | 5 | |||
| Surgical resection | Yes | 7 | 20 | 0.22031 | 5 | 10 | 0.73252 |
| No | 9 | 11 | 8 | 22 | |||
| Distant metastasis | Yes | 7 | 5 | 0.07462 | 5 | 5 | 0.12372 |
| No | 9 | 26 | 8 | 27 | |||
| Serum CEA | Elevated | 7 | 7 | 0.23652 | 6 | 7 | 0.12322 |
| Normal | 3 | 10 | 3 | 14 | |||
RASSF1A protein is actively involved in microtube regulation, genomic stability maintenance, cell-cycle regulation, apoptosis modulation, cell motility and invasion control[31–39]. The frequent inactivation of TSG RASSF1A due to aberrant promoter methylation has been reported in various tumor types[13], suggesting that it plays a pivotal role in human cancer development. It was reported that RASSF1A is inactivated by promoter hypermethylation in gastric and colorectal cancer, but the frequencies of aberrant RASSF1A methylation vary widely[8914–164041]. In addition, serum promoter methylation of RASSF1A in gastric and colorectal cancer has not been extensively studied, and few comparative studies using both primary tumor and serum samples are available. To our knowledge, there is only one related study with a limited sample size[10]. In the present study, we identified the RASSF1A promoter methylation status both in serum DNA and in available paired tumor genomic DNA from patients with gastric and colorectal adenocarcinoma, showing that serum RASSF1A promoter hypermethylation is a potential biomarker for gastric and colorectal cancer diagnosis.
In the present study, serum RASSF1A promoter hypermethylation was detected in 34.0% of patients with gastric adenocarcinoma and in 28.9% of those with colorectal adenocarcinoma. The frequencies were slightly higher than those reported by Tan et al[10] (25% in gastric cancer and 24% in colorectal cancer, respectively). The serum RASSF1A promoter hypermethylation frequencies in gastric and colorectal adenocarcinoma patients were significantly higher than those in patients with benign gastric or colorectal disease or in healthy donors (P < 0.01). The sensitivity of serum RASSF1A promoter hypermethylation in detecting gastric and colorectal cancer is relatively low. Perhaps a simultaneous analysis of the methylation status of a panel of TSGs would be more sensitive in detecting gastric and colorectal cancer. On the other hand, the specificity of serum RASSF1A promoter hypermethylation was very high (approximate 98.3%). Since clinical tests with a high specificity are usually useful in confirming the diagnosis, serum RASSF1A promoter methylation status is a potential marker for the diagnosis of gastric and colorectal cancer.
We also compared the RASSF1A promoter methylation status in paired tissue and serum samples from 25 gastric and colorectal adenocarcinoma patients. For the seven patients with hypermethylated RASSF1A promoter detected in their serum samples, RASSF1A promoter hypermethylation was also present in the primary tumor, which supports the presumption that circulating DNA in peripheral blood of cancer patients reflects the epigenetic change in the primary tumor. In two patients, however, hypermethylated RASSF1A promoter could be detected in the primary tumor samples but not in the paired serum samples, suggesting that not all cancer patients have detectable tumor-originating DNA in their peripheral blood.
RASSF1A promoter hypermethylation was detected in adjacent normal tissue from 2 patients, which can be explained by the invisible invasion of the primary tumor to the adjacent tissue. Another possible reason is the presence of aberrant promoter methylation of TSGs in precancerous lesions adjacent to the primary tumor. Lee et al[9] reported that RASSF1A promoter hypermethylation occurs in 2.1% of colorectal adenomas, and Derks et al[42] found that aberrant RASSF1A promoter methylation is present in 19.1% of nonprogressed adenomas and in 24.4% of progressive adenomas. In our study, we also detected methylated RASSF1A promoter in the serum from one patient with chronic fundal gastritis and two patients with colon adenoma, believed to be precancerous lesions in gastric and colon cancer, respectively. These findings suggest that aberrant promoter hypermethylation of RASSF1A might be an early event in the development of gastric and colorectal cancer. Therefore, identification of serum RASSF1A promoter methylation status may contribute to the early diagnosis of gastric and colorectal cancer.
In the present study, no association was observed between RASSF1A promoter methylation status and patients’ sex, age, tumor differentiation grade, distal metastasis, or surgical therapy. We also compared the methylation status of RASSF1A promoter in preoperative and postoperative serum samples from patients who were underwent to surgical therapy in our hospital, and the status remained unchanged in all patients. Theoretically, when the primary tumor is resected, tumor-specific methylated DNA would decrease considerably in peripheral blood. However, this does not seem to be the case. Fiegl et al[43] monitored the serum RASSF1A promoter methylation status in 148 breast cancer patients for up to 1 year after surgery, and only 21 patients showed positive to negative transition in MSPCR analysis of serum RASSF1A promoter. A possible source of persistently present methylated copy after surgery is the micrometastases that may present before surgery.
We investigated whether serum RASSF1A promoter hypermethylation is correlated with elevated serum CEA levels and found that there is no correlation between them. Koike et al[44] reported that the detection rate of TSG (p16, E-cadherin, and RARβ) hypermethylation is higher than that of conventional tumor marker (CEA and CA19-9) abnormalities in the serum from gastric cancer patients, and that there is no correlation between them. Since serum CEA and TSG hypermethylation are not correlated, a combinational analysis of serum RASSF1A promoter methylation status and serum CEA level may be useful in the diagnosis of gastric and colorectal cancer.
In conclusion, serum RASSF1A promoter hypermethylation is common in gastric and colorectal adenocarcinoma and aberrant CpG island methylation within the promoter region of RASSF1A is a promising biomarker for such cancers.
RASSF1A inactivation by promoter hypermethylation in gastric and colorectal cancer has been reported. However, serum promoter methylation of RASSF1A in gastric and colorectal cancer has not been extensively studied. Particularly, comparative studies using both primary tumor and serum samples are indicated can evaluate the diagnostic role of serum RASSF1A promoter hypermethylation in gastric and colorectal cancer.
Circulating nucleotide acid is a hotspot in the early diagnosis of cancer. Characterization of molecular changes in serum DNA reflecting the genetic and epigenetic alterations in primary tumor would provide an alternative approach to the early detection of cancer.
This is the first comprehensive study on RASSF1A promoter hypermethylation status both in tumor and normal tissue samples and in pre- and post serum samples from gastric and colorectal cancer patients. Our results indicate that aberrant hypermethylation of RASSF1A promoter is a promising serum biomarker for gastric and colorectal cancer diagnosis.
A combined study on promoter hypermethylation of a panel of relevant tumor suppressor genes in serum samples may have a bright future in the early diagnosis of gastric and colorectal cancer.
In DNA, methylation is the addition of a methyl group to a cytosine residue to convert it to 5-methylcytosine. DNA methylation is the main epigenetic modification in humans, and changes in methylation patterns play an important role in tumorigenesis. In particular, hypermethylation of normally unmethylated CpG islands in the promoter region of tumor suppressor genes correlates with their loss of expression and may confer growth advantages to those cells that favor cancer development.
This paper is very interesting. The study is well designed. The authors evaluated the role of serum RASSF1A promoter hypermethylation in diagnosing gastric and colorectal adenocarcinoma, showing that aberrant CpG island methylation within the promoter region of RASSF1A is a promising biomarker for gastric and colorectal cancer.
| 1. | Macdonald JS. Carcinoembryonic antigen screening: pros and cons. Semin Oncol. 1999;26:556-560. |
| 2. | Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415-428. |
| 3. | Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141-196. |
| 4. | Lee TL, Leung WK, Chan MW, Ng EK, Tong JH, Lo KW, Chung SC, Sung JJ, To KF. Detection of gene promoter hypermethylation in the tumor and serum of patients with gastric carcinoma. Clin Cancer Res. 2002;8:1761-1766. |
| 5. | Kim H, Kim YH, Kim SE, Kim NG, Noh SH, Kim H. Concerted promoter hypermethylation of hMLH1, p16INK4A, and E-cadherin in gastric carcinomas with microsatellite instability. J Pathol. 2003;200:23-31. |
| 6. | Tamura G. Alterations of tumor suppressor and tumor-related genes in the development and progression of gastric cancer. World J Gastroenterol. 2006;12:192-198. |
| 7. | Zhao YF, Zhang YG, Tian XX, Juan Du, Jie Zheng. Aberrant methylation of multiple genes in gastric carcinomas. Int J Surg Pathol. 2007;15:242-251. |
| 8. | Xu XL, Yu J, Zhang HY, Sun MH, Gu J, Du X, Shi DR, Wang P, Yang ZH, Zhu JD. Methylation profile of the promoter CpG islands of 31 genes that may contribute to colorectal carcinogenesis. World J Gastroenterol. 2004;10:3441-3454. |
| 9. | Lee S, Hwang KS, Lee HJ, Kim JS, Kang GH. Aberrant CpG island hypermethylation of multiple genes in colorectal neoplasia. Lab Invest. 2004;84:884-893. |
| 10. | Tan SH, Ida H, Lau QC, Goh BC, Chieng WS, Loh M, Ito Y. Detection of promoter hypermethylation in serum samples of cancer patients by methylation-specific polymerase chain reaction for tumour suppressor genes including RUNX3. Oncol Rep. 2007;18:1225-1230. |
| 11. | Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet. 2000;25:315-319. |
| 12. | Pfeifer GP, Dammann R. Methylation of the tumor suppressor gene RASSF1A in human tumors. Biochemistry (Mosc). 2005;70:576-583. |
| 13. | Dammann R, Schagdarsurengin U, Seidel C, Strunnikova M, Rastetter M, Baier K, Pfeifer GP. The tumor suppressor RASSF1A in human carcinogenesis: an update. Histol Histopathol. 2005;20:645-663. |
| 14. | Byun DS, Lee MG, Chae KS, Ryu BG, Chi SG. Frequent epigenetic inactivation of RASSF1A by aberrant promoter hypermethylation in human gastric adenocarcinoma. Cancer Res. 2001;61:7034-7038. |
| 15. | Wagner KJ, Cooper WN, Grundy RG, Caldwell G, Jones C, Wadey RB, Morton D, Schofield PN, Reik W, Latif F. Frequent RASSF1A tumour suppressor gene promoter methylation in Wilms' tumour and colorectal cancer. Oncogene. 2002;21:7277-7282. |
| 16. | van Engeland M, Roemen GM, Brink M, Pachen MM, Weijenberg MP, de Bruine AP, Arends JW, van den Brandt PA, de Goeij AF, Herman JG. K-ras mutations and RASSF1A promoter methylation in colorectal cancer. Oncogene. 2002;21:3792-3795. |
| 17. | Oliveira C, Velho S, Domingo E, Preto A, Hofstra RM, Hamelin R, Yamamoto H, Seruca R, Schwartz S Jr. Concomitant RASSF1A hypermethylation and KRAS/BRAF mutations occur preferentially in MSI sporadic colorectal cancer. Oncogene. 2005;24:7630-7634. |
| 18. | Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646-650. |
| 19. | Stroun M, Anker P, Maurice P, Lyautey J, Lederrey C, Beljanski M. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology. 1989;46:318-322. |
| 20. | Camps C, Sirera R, Bremnes R, Blasco A, Sancho E, Bayo P, Safont MJ, Sanchez JJ, Taron M, Rosell R. Is there a prognostic role of K-ras point mutations in the serum of patients with advanced non-small cell lung cancer? Lung Cancer. 2005;50:339-346. |
| 21. | Gotoh T, Hosoi H, Iehara T, Kuwahara Y, Osone S, Tsuchiya K, Ohira M, Nakagawara A, Kuroda H, Sugimoto T. Prediction of MYCN amplification in neuroblastoma using serum DNA and real-time quantitative polymerase chain reaction. J Clin Oncol. 2005;23:5205-5210. |
| 22. | Cuda G, Gallelli A, Nistico A, Tassone P, Barbieri V, Tagliaferri PS, Costanzo FS, Tranfa CM, Venuta S. Detection of microsatellite instability and loss of heterozygosity in serum DNA of small and non-small cell lung cancer patients: a tool for early diagnosis? Lung Cancer. 2000;30:211-214. |
| 23. | Nawroz-Danish H, Eisenberger CF, Yoo GH, Wu L, Koch W, Black C, Ensley JF, Wei WZ, Sidransky D. Microsatellite analysis of serum DNA in patients with head and neck cancer. Int J Cancer. 2004;111:96-100. |
| 24. | Fujiwara K, Fujimoto N, Tabata M, Nishii K, Matsuo K, Hotta K, Kozuki T, Aoe M, Kiura K, Ueoka H. Identification of epigenetic aberrant promoter methylation in serum DNA is useful for early detection of lung cancer. Clin Cancer Res. 2005;11:1219-1225. |
| 25. | Ramirez JL, Sarries C, de Castro PL, Roig B, Queralt C, Escuin D, de Aguirre I, Sanchez JM, Manzano JL, Margeli M. Methylation patterns and K-ras mutations in tumor and paired serum of resected non-small-cell lung cancer patients. Cancer Lett. 2003;193:207-216. |
| 26. | Yamaguchi S, Asao T, Nakamura J, Ide M, Kuwano H. High frequency of DAP-kinase gene promoter methylation in colorectal cancer specimens and its identification in serum. Cancer Lett. 2003;194:99-105. |
| 27. | Sobin LH, Wittekind C. TNM Classification of Malignant Tumours, 6th Edition. New York: Wiley-Liss 2002; . |
| 28. | Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821-9826. |
| 29. | Burbee DG, Forgacs E, Zochbauer-Muller S, Shivakumar L, Fong K, Gao B, Randle D, Kondo M, Virmani A, Bader S. Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J Natl Cancer Inst. 2001;93:691-699. |
| 30. | Wang Y, Yu Z, Wang T, Zhang J, Hong L, Chen L. Identification of epigenetic aberrant promoter methylation of RASSF1A in serum DNA and its clinicopathological significance in lung cancer. Lung Cancer. 2007;56:289-294. |
| 31. | Donninger H, Vos MD, Clark GJ. The RASSF1A tumor suppressor. J Cell Sci. 2007;120:3163-3172. |
| 32. | Liu L, Tommasi S, Lee DH, Dammann R, Pfeifer GP. Control of microtubule stability by the RASSF1A tumor suppressor. Oncogene. 2003;22:8125-8136. |
| 33. | Vos MD, Martinez A, Elam C, Dallol A, Taylor BJ, Latif F, Clark GJ. A role for the RASSF1A tumor suppressor in the regulation of tubulin polymerization and genomic stability. Cancer Res. 2004;64:4244-4250. |
| 34. | Shivakumar L, Minna J, Sakamaki T, Pestell R, White MA. The RASSF1A tumor suppressor blocks cell cycle progression and inhibits cyclin D1 accumulation. Mol Cell Biol. 2002;22:4309-4318. |
| 35. | Whang YM, Kim YH, Kim JS, Yoo YD. RASSF1A suppresses the c-Jun-NH2-kinase pathway and inhibits cell cycle progression. Cancer Res. 2005;65:3682-3690. |
| 36. | Vos MD, Ellis CA, Bell A, Birrer MJ, Clark GJ. Ras uses the novel tumor suppressor RASSF1 as an effector to mediate apoptosis. J Biol Chem. 2000;275:35669-35672. |
| 37. | Vos MD, Dallol A, Eckfeld K, Allen NP, Donninger H, Hesson LB, Calvisi D, Latif F, Clark GJ. The RASSF1A tumor suppressor activates Bax via MOAP-1. J Biol Chem. 2006;281:4557-4563. |
| 38. | Matallanas D, Romano D, Yee K, Meissl K, Kucerova L, Piazzolla D, Baccarini M, Vass JK, Kolch W, O'neill E. RASSF1A elicits apoptosis through an MST2 pathway directing proapoptotic transcription by the p73 tumor suppressor protein. Mol Cell. 2007;27:962-975. |
| 39. | Dallol A, Agathanggelou A, Tommasi S, Pfeifer GP, Maher ER, Latif F. Involvement of the RASSF1A tumor suppressor gene in controlling cell migration. Cancer Res. 2005;65:7653-7659. |
| 40. | To KF, Leung WK, Lee TL, Yu J, Tong JH, Chan MW, Ng EK, Chung SC, Sung JJ. Promoter hypermethylation of tumor-related genes in gastric intestinal metaplasia of patients with and without gastric cancer. Int J Cancer. 2002;102:623-628. |
| 41. | Ye M, Xia B, Guo Q, Zhou F, Zhang X. Association of diminished expression of RASSF1A with promoter methylation in primary gastric cancer from patients of central China. BMC Cancer. 2007;7:120. |
| 42. | Derks S, Postma C, Moerkerk PT, van den Bosch SM, Carvalho B, Hermsen MA, Giaretti W, Herman JG, Weijenberg MP, de Bruine AP. Promoter methylation precedes chromosomal alterations in colorectal cancer development. Cell Oncol. 2006;28:247-257. |
| 43. | Fiegl H, Millinger S, Mueller-Holzner E, Marth C, Ensinger C, Berger A, Klocker H, Goebel G, Widschwendter M. Circulating tumor-specific DNA: a marker for monitoring efficacy of adjuvant therapy in cancer patients. Cancer Res. 2005;65:1141-1145. |
| 44. | Koike H, Ichikawa D, Ikoma H, Tani N, Ikoma D, Otsuji E, Okamoto K, Ueda Y, Kitamura K, Yamagishi H. Comparison of serum aberrant methylation and conventional tumor markers in gastric cancer patients. Hepatogastroenterology. 2005;52:1293-1296. |