Published online Feb 14, 2007. doi: 10.3748/wjg.v13.i6.950
Revised: December 28, 2006
Accepted: January 6, 2007
Published online: February 14, 2007
AIM: To investigate the feasibility of detecting methylated fecal DNA as a screening tool for colorectal carcinoma (CRC) and precancerous lesions.
METHODS: Methylated secreted frizzled-related protein gene 2 (SFRP2), hyperplastic polyposis protein gene (HPP1) and O6-methylguanine-DNA methyltransferase gene (MGMT) in stools from 52 patients with CRC, 35 patients with benign colorectal diseases and 24 normal individuals were analyzed using methylation-specific PCR.
RESULTS: Methylated SFRP2, HPP1 and MGMT were detected in 94.2%, 71.2%, 48.1% of CRC patients and 52.4%, 57.1%, 28.6% of adenoma patients, respectively. The overall prevalence of fecal DNA with at least one methylated gene was 96.2% and 81.8% in patients with CRC and precancerous lesions, respectively. In contrast, only one of the 24 normal individuals revealed methylated DNA. These results indicated a 93.7% sensitivity and a 77.1% specificity of the assay for detecting CRC and precancerous lesions.
CONCLUSION: Methylation testing of fecal DNA using a panel of epigenetic markers may be a simple and promising non-invasive screening method for CRC and precancerous lesions.
- Citation: Huang ZH, Li LH, Yang F, Wang JF. Detection of aberrant methylation in fecal DNA as a molecular screening tool for colorectal cancer and precancerous lesions. World J Gastroenterol 2007; 13(6): 950-954
- URL: https://www.wjgnet.com/1007-9327/full/v13/i6/950.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i6.950
Colorectal carcinoma (CRC) is one of the leading causes of cancer-related death in the world. Detection of early-stage cancer and precancerous lesions seems to be a key measure to reduce its death rate[1]. Most deaths due to CRC are preventable through screening, which involves the search for neoplasms (cancer and precancerous lesions) in asymptomatic individuals. Several CRC screening tests, including fecal occult-blood testing (FOBT), barium enema, colonoscopy and sigmoidoscopy, have been available for years. However, none of these methods has been established as a well-accepted screening tool, because of their low compliance rates or low sensitivity and/or specificity. More optimized screening methods should be established with a high sensitivity and specificity for early-stage cancers and precancerous lesions.
Epigenetic gene silencing is increasingly recognized to play a crucial role in human tumors[2]. One of the principal epigenetic mechanisms known to be involved in carcinogenesis is the methylation of cytosine residues in CpG-rich sequences (CpG islands) located within the promoter regions of genes regulating cell proliferation, apoptosis, and DNA repair. DNA methylation within gene promoters results in aberrant gene silencing, which can make an important contribution to the emergence of neoplasias. A number of genes can now be hypermethylated in colorectal tumors[3]. Aberrant promoter methylation often occurs very early during CRC carcinogenesis[4]. Moreover, it has been shown recently that DNA hypermethylation can be detected in DNA from stools of patients with colorectal tumor, suggesting that fecal DNA methylation analysis might provide a valuable approach to non-invasive screening for early colorectal lesions[5-8]. Lenhard et al[9] reported that a single methylation marker hypermethylated in cancer 1 (HIC1) can be used to detect CRC. Methylated HIC1 has been found in 42% and 31% of fecal samples from patients with CRC and adenoma, respectively. Aberrant methylation of vimentin gene was also reported in fecal DNA from 43 out of 94 CRC patients, with a sensitivity of 46% and a specificity of 90%, respectively[10]. These studies suggested that a panel of methylated genes might be required to improve the detection sensitivity for CRC. However, to date, methylation markers have not been assessed thoroughly in stools. Few studies have been performed on methylated fecal DNA in CRC patients in parallel with healthy individuals and patients with benign colorectal diseases, and most of these investigations only analyzed a small number of samples.
In the present study, we analyzed methylation of three genes: secreted frizzled-related protein gene 2 (SFRP2), hyperplastic polyposis protein gene (HPP1) and O6-methylguanine-DNA methyltrans- ferase gene (MGMT) in stools from patients with CRC and benign colorectal diseases as well as from normal controls, and evaluated the feasibility of detecting hypermethylation in stools as a non-invasive screening tool for CRC and precancerous lesions.
Fifty-two CRC patients, 35 patients with benign colorectal diseases (including 21 with adenomas, 8 with hyperplastic polyps and 6 with ulcerative colitis) and 24 patients with endoscopically normal colons, undergoing colonoscopy for routine clinical indications in the 4th Affiliated Hospital of Soochow University, were enrolled in this study (Table 1). In addition, fifteen CRC tissue samples and 8 normal colon samples that were resected at the time of surgery or colonoscopy were obtained from Wuxi Tumor Resource Bank. The study was approved by the ethical committee of our institution.
| Characteristics | Colorectal cancern = 52 | Benign controln = 35 | Normal controln = 24 |
| Sex | |||
| Male | 31 | 20 | 13 |
| Female | 21 | 15 | 11 |
| Age | |||
| Mean ± SD | 63.5 ± 11.8 | 60.0 ± 10.3 | 59.6 ± 8.2 |
| Range | 25-81 | 31-79 | 35-68 |
| Dukes stage | |||
| A + B | 27 | Na | Na |
| C + D | 25 | Na | Na |
| Nodal status | |||
| Positive | 23 | Na | Na |
| Negative | 29 | Na | Na |
| Tumor size | |||
| > 5 cm | 19 | Na | Na |
| ≤ 5 cm | 33 | Na | Na |
| Tumor locus | |||
| Rectum | 24 | Na | Na |
| Distal colon | 16 | Na | Na |
| Proximal colon | 12 | Na | Na |
Stool specimens were collected by patients and sent to laboratory within 2 h after defecation, aliquoted and stored at -80°C until analysis.
DNA isolation: DNA was isolated from stool samples (200 mg) using the QIAamp DNA Stool Mini Kit (Qiagen). Genomic DNA was purified from colon tissue samples using the QIAamp DNA Mini Kit. The extracted DNA was quantitated by UV spectrum and stored at -20°C.
Methylation-specific PCR (MSP): Stool DNA was modified with sodium bisulfite as previously described[11]. Blood DNA treated in vitro with SssI methyltransferase was used as a positive control for methylated alleles, and placental DNA was used as a negative control for MSP assay.
The bisulfite-modified DNA was subjected to PCR in a blinded manner using primer pairs designed to amplify specifically the methylated or unmethylated alleles of respective genes (Table 2). PCR products were analyzed by 2.5% agarose gel electrophoresis. To verify the reliability of MSP, ten PCR products from methylated amplicon of SFRP2 were randomly selected and bidirectionally sequenced.
| Primer | Sequences (5’-3’) | Annealingtemperature | Productsize (bp) |
| SRFP2-MF | GGGTCGGAGTTTTTCGGAGTTGCGC | 62°C | 138 |
| SRFP2-MR | CCGCTCTCTTCGCTAAATACGACTCG | ||
| SRFP2-UF | TTTTGGGTTGGAGTTTTTTGGAGTTGTGT | 58°C | 145 |
| SRFP2-UR | AACCCACTCTCTTCACTAAATACAACTCA | ||
| HPP1-MF | TTTAGCGGACGATTTTTTCGTTTCG | 57°C | 122 |
| HPP1-MR | AACGACGACGATAACAATAA | ||
| HPP1-UF | TTTAGTGGATGATTTTTTTGTTTTG | 57°C | 122 |
| HPP1-UR | AACAACAACAATAACAATAA | ||
| MGMT-MF | TTTCGACGTTCGTAGGTTTTCGC | 56°C | 81 |
| MGMT-MR | GCACTCTTCCGAAAACGAAACG | ||
| MGMT-UF | TTTGTGTTTTGATGTTTGTAGGTTTTTGT | 59°C | 93 |
| MGMT-UR | AACTCCACACTCTTCCAAAAACAAAACA |
Statistical probabilities were analyzed by 2 × 2 contingency tables using binomial distribution of differences, Fisher's exact test or Pearson’s chi-square test. Calculations were performed using SPSS10.0 software. P < 0.05 was considered statistically significant.
Methylated SFRP2, HPP1 and MGMT were found in 14, 11 and 8 of 15 CRC tissue samples, respectively. In contrast, none of the 8 samples from normal colonic epithelia revealed any detectable methylated DNA.
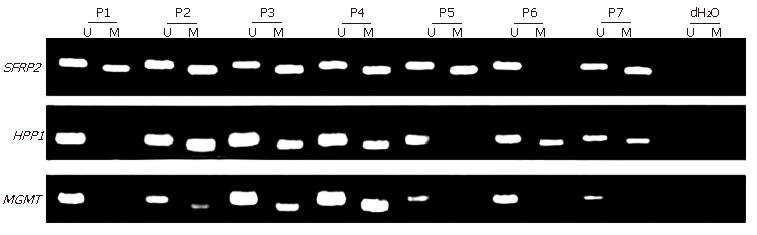
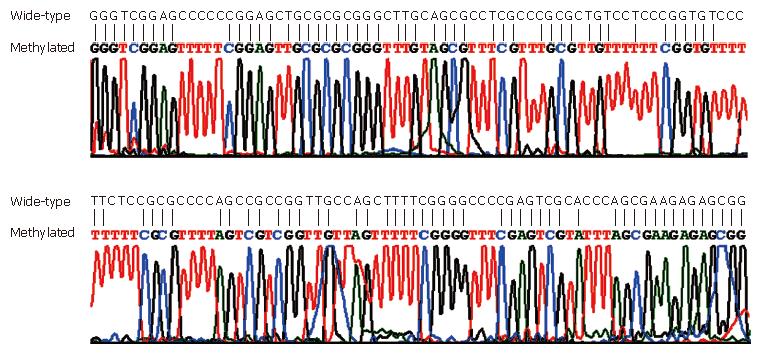
MSP was performed on all the 111 stool samples, including 15 fecal samples from CRC patients with matched CRC tissue samples, to analyze SFRP2, HPP1 and MGMT (Figure 1). The conversion rate of unmethylated cytosines by bisulfite was measured by sequencing random PCR products from methylated MSP for SFRP2 (Figure 2). A total of 10 PCR products were sequenced bidirectionally, revealing only 1 unconverted non-CpG cytosine. A total of 39 non-CpG cytosines were present in the genomic sequence matching with the amplicon of SFRP2. By, assuming error-free PCR, > 99.7% of unmethylated cytosines were converted, which assured the reliability of the positive results of MSP. In addition, the sequencing results revealed that CpGs in the amplicon were uniformly methylated (Figure 2), indicating that complete methylation or hypermethylation occurred at SFRP2.
The MSP results of fecal DNA are listed in Table 3. Of the 10 patients with advanced adenomas (characterized by size > 1 cm, villous histology, and high-grade dysplasia), 8 had detectable promoter hypermethylation in at least one gene. The sensitivity and specificity of MSP for detecting advanced adenoma were 80% and 83.7%, respectively. Of the 6 patients with ulcerative colitis (UC), one was positive for SFRP2 and HPP1. Colonoscopy with biopsy for this patient revealed high-grade dysplasia. One colonoscopically negative control revealed methylation of SFRP2. The sensitivity of SFRP2, HPP1 and MGMT assay for detecting CRC and precancerous lesions (including 10 advanced adenomas and 1 UC with dysplasia) was 90.5%, 71.4% and 46.0%, respectively. The specificity of SFRP2, HPP1 and MGMT for detecting CRC and precancerous lesions was 83.3%, 85.4% and 93.8%, respectively. These results indicated a 93.7% sensitivity and a 77.1% specificity of the combined test of the three genes for detecting CRC and precancerous lesions.
| Diagnosis | No | Gene locus | Total | ||
| SFRP2 | HPP1 | MGMT | |||
| Cancer | 52 | 49 (94.2) | 37 (71.2) | 25 (48.1) | 50 (96.2) |
| Adenomas | 21 | 11 (52.4) | 12 (57.1) | 6 (28.6) | 15 (71.4) |
| Advanced adenomas | 10 | 7 (70.0) | 7 (70.0) | 4 (40.0) | 8 (80.0) |
| Hyperplastic polyp | 8 | 3 (37.5) | 2 (25.0) | 1 (12.5) | 3 (37.5) |
| Ulcerative colitis | 6 | 1 (16.7) | 1 (16.7) | 0 (0) | 1 (16.7) |
| Normal | 24 | 1 (4.2) | 0 (0) | 0 (0) | 1 (4.2) |
To examine the coincidence of methylation status between fecal DNA and DNA in its original colon caner, 15 pairs of samples with matched tissue and fecal samples were tested for their methylation status. Methylated SFRP2, HPP1 and MGMT were found in 14, 11 and 8 of 15 CRC tissue samples, respectively. In contrast, none of the 8 samples from normal colonic epithelia revealed any detectable methylated DNA. The MSP results were highly consistent between paired fecal and tissue samples, except that two patients had detectable methylated HPP1 or MGMT only in tissue samples.
No correlation was observed between stool DNA hypermethylation and nodal status, Dukes stage, tumor size or tumor locus (data not shown).
In this study, we explored the feasibility of detecting methylated fecal DNA as a screening tool for CRC and precancerous lesions. The results showed that DNA methylation could be detected in feces of patients with CRC and precancerous lesions with a high sensitivity (93.7%) and specificity (77.1%).
We detected DNA methylation of SFRP2, HPP1 and MGMT in fecal samples from patients with colorectal tumor and non-tumor diseases. It has been shown that these three genes can be methylated in patients with CRC and precancerous lesions[6-8,12-16]. Of the genes chosen for this study, SFRP2 gene belongs to a new class of tumor suppressor genes (SFRPs), which are secreted glycoproteins working as inhibitory modulators of a putative tumorigenic pathway (the Wnt signaling pathway). Aberrant promoter methylation of SFRP genes has been described in CRC by several studies[7,15]. Müller et al[7] reported that SFRP2 is methylated differentially in stools of patients with or without CRC, and SFRP2 hypermethylation is proposed as a sensitive marker, detecting 77%-90% of CRCs. However, the sample size used in their study was small and the prevalence of methylated SFRP2 in patients with benign colorectal diseases and in other races or ethnic group was not addressed. In this study, we detected methylated SFRP2 in fecal samples from Chinese patients with CRC, benign colorectal diseases and in healthy individuals, simultaneously. Our results show that hypermethylated SFRP2 is detectable in stools of Chinese patients with CRC and precancerous lesions, suggesting that it is a sensitive marker for detecting CRC in healthy controls with 94.2% of sensitivity and 95.2% of specificity, higher than those in a previous report[8]. The different results may be due to the assay-related differences or a different patient population. In addition, methylation of this gene was found in one colonoscopically negative case, which may be most likely owing to the SFRP2 methylation occurring frequently in premaligant aberrant crypt foci (ACF) that is overlooked at colonoscopy[15].
HPP1 encodes a transmembrane protein containing follistatin and epidermal growth factor–like domains and contains a CpG island in its promoter region that can be aberrantly hypermethylated in CRC and its precursor lesions[13,14,16-18], indicating that HPP1 gene methylation is an early epigenetic alteration in colorectal carcinogenesis and may be a valuable molecular marker for early diagnosis of CRC. The results of our study indicate that HPP1 methylation could be frequently detected in most stool samples from patients with CRC (71.2%) and advanced adenoma (57.1%).
MGMT is an important DNA repair enzyme that protects cells from the carcinogenic effects of alkylating agents by removing adducts from the O6 position of guanine, thus preventing G:C to A:T transition. It was reported that MGMT gene inactivation caused by promoter methylation plays a key role in a great variety of human tumors[12]. Leung et al[6] showed that 45% hypermethylated MGMT are detected in stools of CRC patients in a small-scale feasibility study without healthy subjects as controls. The positive rate of MGMT hypermethylation in our study was comparable with their results.
Of the three genes, SFRP2 is the most informative marker, reaching a sensitivity of 90.5% for CRC and precancerous lesions. In our study, all the 32 samples with methylated MGMT revealed methylated SFRP2, suggesting that combined detecting of MGMT could not increase the detection rate of SFRP2 for CRC, while a combined test of SFRP2 and HPP1 increased the detection rate (93.7%) for CRC and precancerous lesions.
No significant correlation was found between the presence of methylation in fecal DNA and any clinicopathologic characteristics of cancer in our study, indicating that these methylation markers in later stage CRC are as sensitive as in early stage CRC, and may be equivalently sensitive to proximal or distal cancers. However, Ebert et al[16] reported that HPP1 methylation is linked to the location of the primary tumor in the colon and is more frequently methylated in colon cancer compared to rectal cancer. The reasons for the divergent results may include assay-related differences, different patient population and samples.
Adenomas are the precursor of most sporadic CRCs. Hyperplastic polyp-serrated adenoma to carcinoma sequence is characterized by recognizable histological changes. Colorectal lesions, starting with dysplastic ACF, hyperplastic polyp (HP) and benign tubular adenoma have the potential to progress to advanced adenomas, which have a significant potential to transform into CRC[17]. Although both ACF and HP are common lesions and the great majority would not transform into clinically significant polyps. HP occurring in the right colon and in the setting of hyperplastic polyposis has the greatest potential to transform into CRC[17,18]. It is unclear if these lesions with methylated DNA would develop into cancer. However, DNA methylation is more common in advanced adenomas. All these findings suggest that methylation may reflect the malignant potential of these lesions. UC-associated CRC is different from sporadic carcinoma. The molecular mechanisms underlying the progression of UC to dysplasia and carcinoma have not been fully understood. One of the UC patients with methylated SFRP2 and HPP1 in feces was also diagnosed with high-grade dysplasia, indicating that DNA methylation is not only a frequent event in colorectal tumor, but also detectable in fecal samples from patients with HP and UC. Although detection of early stage CRC represents an important screening goal, efforts should be directed toward detection of dysplastic cells present in patients with benign diseases, such as adenoma, HP and UC. Whether hypermethylation of these three genes can identify these non-malignant diseases, including self-limiting HPs, non-dysplastic ACF and UC, is still unclear. Recent studies indicate that methylation patterns can define a subset of HPs with a significant malignant potential[17,19]. Thus, patients with methylated fecal DNA may belong to high-risk individuals and need more accurate/specific clinical examinations and follow-up.
Although FOBT is a valuable noninvasive screening method that reduces the risk of CRC-related death, it has a limited sensitivity. Colonoscopy, with a high sensitivity and specificity for detection of CRC and large adenomas, requires a thorough bowel preparation, which causes discomfort and small but non-negligible risk of major complications in patients. In contrast, using aberrant gene methylation as a molecular marker seems to offer a potentially powerful approach to population-based screening for CRC and precancerous lesions. However, further prospective study is needed to validate the epigenetic markers. In addition, simplified, non-expensive and automatized assays may be important for population-based screening.
| 1. | Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ. Cancer statistics, 2006. CA Cancer J Clin. 2006;56:106-130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4370] [Cited by in RCA: 4261] [Article Influence: 213.1] [Reference Citation Analysis (0)] |
| 2. | Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1182] [Cited by in RCA: 1194] [Article Influence: 59.7] [Reference Citation Analysis (0)] |
| 3. | Kondo Y, Issa JP. Epigenetic changes in colorectal cancer. Cancer Metastasis Rev. 2004;23:29-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 224] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 4. | Chan AO, Broaddus RR, Houlihan PS, Issa JP, Hamilton SR, Rashid A. CpG island methylation in aberrant crypt foci of the colorectum. Am J Pathol. 2002;160:1823-1830. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 172] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 5. | Belshaw NJ, Elliott GO, Williams EA, Bradburn DM, Mills SJ, Mathers JC, Johnson IT. Use of DNA from human stools to detect aberrant CpG island methylation of genes implicated in colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2004;13:1495-1501. [PubMed] |
| 6. | Leung WK, To KF, Man EP, Chan MW, Bai AH, Hui AJ, Chan FK, Lee JF, Sung JJ. Detection of epigenetic changes in fecal DNA as a molecular screening test for colorectal cancer: a feasibility study. Clin Chem. 2004;50:2179-2182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 38] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 7. | Müller HM, Oberwalder M, Fiegl H, Morandell M, Goebel G, Zitt M, Mühlthaler M, Ofner D, Margreiter R, Widschwendter M. Methylation changes in faecal DNA: a marker for colorectal cancer screening? Lancet. 2004;363:1283-1285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 205] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 8. | Petko Z, Ghiassi M, Shuber A, Gorham J, Smalley W, Washington MK, Schultenover S, Gautam S, Markowitz SD, Grady WM. Aberrantly methylated CDKN2A, MGMT, and MLH1 in colon polyps and in fecal DNA from patients with colorectal polyps. Clin Cancer Res. 2005;11:1203-1209. [PubMed] |
| 9. | Lenhard K, Bommer GT, Asutay S, Schauer R, Brabletz T, Göke B, Lamerz R, Kolligs FT. Analysis of promoter methylation in stool: a novel method for the detection of colorectal cancer. Clin Gastroenterol Hepatol. 2005;3:142-149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 91] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 10. | Chen WD, Han ZJ, Skoletsky J, Olson J, Sah J, Myeroff L, Platzer P, Lu S, Dawson D, Willis J. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J Natl Cancer Inst. 2005;97:1124-1132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 269] [Cited by in RCA: 288] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 11. | Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821-9826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4183] [Cited by in RCA: 4262] [Article Influence: 142.1] [Reference Citation Analysis (12)] |
| 12. | Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793-797. [PubMed] |
| 13. | Young J, Biden KG, Simms LA, Huggard P, Karamatic R, Eyre HJ, Sutherland GR, Herath N, Barker M, Anderson GJ. HPP1: a transmembrane protein-encoding gene commonly methylated in colorectal polyps and cancers. Proc Natl Acad Sci USA. 2001;98:265-270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 72] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 14. | Sato F, Shibata D, Harpaz N, Xu Y, Yin J, Mori Y, Wang S, Olaru A, Deacu E, Selaru FM. Aberrant methylation of the HPP1 gene in ulcerative colitis-associated colorectal carcinoma. Cancer Res. 2002;62:6820-6822. [PubMed] |
| 15. | Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Chen WD, Pretlow TP, Yang B, Akiyama Y, Van Engeland M. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36:417-422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 821] [Cited by in RCA: 844] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 16. | Ebert MP, Mooney SH, Tonnes-Priddy L, Lograsso J, Hoffmann J, Chen J, Röcken C, Schulz HU, Malfertheiner P, Lofton-Day C. Hypermethylation of the TPEF/HPP1 gene in primary and metastatic colorectal cancers. Neoplasia. 2005;7:771-778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 53] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 17. | Wynter CV, Walsh MD, Higuchi T, Leggett BA, Young J, Jass JR. Methylation patterns define two types of hyperplastic polyp associated with colorectal cancer. Gut. 2004;53:573-580. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 107] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 18. | Goldstein NS, Bhanot P, Odish E, Hunter S. Hyperplastic-like colon polyps that preceded microsatellite-unstable adenocarcinomas. Am J Clin Pathol. 2003;119:778-796. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 166] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 19. | Wynter CV, Kambara T, Walsh MD, Leggett BA, Young J, Jass JR. DNA methylation patterns in adenomas from FAP, multiple adenoma and sporadic colorectal carcinoma patients. Int J Cancer. 2006;118:907-915. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 19] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
S- Editor Liu Y L- Editor Wang XL E- Editor Lu W