Published online Nov 7, 2007. doi: 10.3748/wjg.v13.i41.5421
Revised: July 31, 2007
Accepted: August 14, 2007
Published online: November 7, 2007
The aetiology of primary sclerosing cholangitis (PSC) is not known. A more than 80-fold increased risk of PSC among first-degree relatives emphasizes the importance of genetic factors. Genetic associations within the human leukocyte antigen (HLA) complex on chromosome 6p21 were detected in PSC 25 years ago. Subsequent studies have substantiated beyond doubt that one or more genetic variants located within this genetic region are important. The true identities of these variants, however, remain to be identified. Several candidate genes at other chromosomal loci have also been investigated. However, according to strict criteria for what may be denominated a susceptibility gene in complex diseases, no such gene exists for PSC today. This review summarises present knowledge on the genetic susceptibility to PSC, as well as genetic associations with disease progression and clinical subsets of particular interest (inflammatory bowel disease and cholangiocarcinoma).
- Citation: Karlsen TH, Schrumpf E, Boberg KM. Genetic epidemiology of primary sclerosing cholangitis. World J Gastroenterol 2007; 13(41): 5421-5431
- URL: https://www.wjgnet.com/1007-9327/full/v13/i41/5421.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i41.5421
Primary sclerosing cholangitis (PSC) is a chronic inflammatory condition of unknown aetiology, characterised by progressive strictures of the intra- and extrahepatic bile ducts and eventually liver cirrhosis and liver failure[1,2]. No effective medical treatment is currently available[3,4], and PSC is the major indication for liver transplantation in the Scandinavian countries as well as the fifth leading indication for liver transplantation in the United States[5,6]. Population-based studies of disease frequency are available from Norway, Great Britain and The United States[7-9], and indicate comparable incidence (0.9-1.3 per 100000/year) and prevalence (8.5-14.2 per 100000) rates for these populations. The prevalence of PSC is probably lower in Southern European and Asian populations[10]. In contrast to the female predominance of many autoimmune diseases, approximately 2/3 of the PSC patients are male[11]. Affected individuals are young (less than 40 years at time of diagnosis), and median survival from time of diagnosis by cholangiography to death or liver transplantation is approximately 12 years[11].
Up to 80% of the PSC patients of Northern European origin have concurrent inflammatory bowel disease (IBD)[10]. The frequency in Southern Europe and Asia is lower (around 50% and 35%, respectively)[12-14]. According to standard criteria[15], the IBD phenotype in PSC has mainly been classified as ulcerative colitis (UC), although an association with colonic Crohn's disease also exists[16,17]. The increased frequency of a variety of other autoimmune diseases (e.g., type 1 diabetes) among patients with PSC does not seem related to the increase in IBD[18]. There is also an increased risk of cancer among the patients with PSC, not only cholangiocarcinoma of the biliary tract (approximately 13%-14% in Scandinavia)[19,20], but also other gastrointestinal malignancies (i.e., pancreatic and colorectal cancer)[19]. The diagnosis of cholangiocarcinoma is difficult because the cholangiographic changes may look similar to those found in PSC without cholangiocarcinoma[21]. As a result, the cancer is often recognised at an advanced stage when treatment by liver transplantation does not improve survival[22].
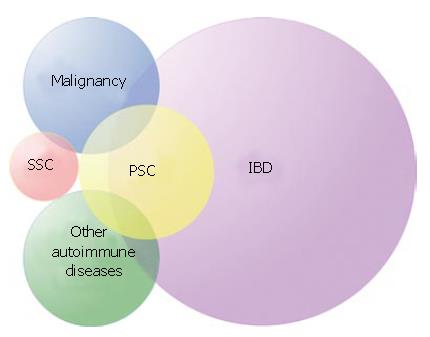
Smoking is the only environmental factor known to influence PSC susceptibility and is associated with a reduced risk of the disease[23]. Several genetic risk factors, however, have been repeatedly described throughout the 25 years since they were first detected[24,25]. The present editorial aims to summarise present knowledge on statistical associations between genetic variants and risk of PSC or particular characteristics of PSC. In genetic epidemiology, disease characteristics under study are called phenotypes. Etymologically, the pheno-prefix refers to "visible" or "evident". Phenotypes, also referred to as traits, may be dichotomous (e.g., PSC/healthy) or quantitative (e.g., the level of alkaline phosphatase in a blood sample from a PSC patient). The clinical definition of a disease is primarily made to decide whether a particular treatment or follow-up may be indicated for a patient or not. This practical aspect means that PSC as a clinical "diagnosis" does not necessarily equal the ideal "phenotype" for genetic association studies. The disease phenotype in such studies should be as homogeneous as possible, simply because the presence of irrelevant phenotypes in a study population will reduce the strength of effects to be identified. The clinical phenotype of PSC is compound (Figure 1).
In other diseases, susceptibility genes have been identified through genome-wide linkage scans followed by fine-mapping[26-28]. In PSC, the lack of families with affected sibling pairs has not allowed such studies to position susceptibility loci[28]. The search for PSC susceptibility genes has thus focused on plausible candidates with regard to function[25]. As a general basis for interpreting candidate gene association studies, an introduction to important concepts of such studies will be given, followed by a presentation and discussion of studies performed in PSC. We searched PubMed for relevant articles published up until the end of April 2007. We have also reviewed the reference lists of identified articles, as well as the reference lists of major immunogenetic- and hepatology conferences held over the last 2 years.
In genetic terms, PSC is considered a complex trait, meaning that polymorphisms in several genes along with environmental factors are required for disease development[27]. Heritability for a disease is measured by (a) concordance rates in monozygotic versus dizygotic twins and (b) relative risk in siblings of a patient (λs = prevalence among siblings divided by the general population prevalence). For monogenic disorders, λs ranges from several hundreds to several thousands, whereas values in complex traits are usually below 100. A strong genetic contribution to overall risk of PSC is supported by λs values of approximately 100[29], as compared with values of 15-35 for Crohn's disease and 6-9 for UC[30].
Polymorphisms are genetic variants that have arisen from mutational events in DNA[31]. Conventionally, to be denominated a polymorphism, a mutant variant should occur at a frequency of > 0.01 in the general population. A particular nucleotide (or nucleotide sequence) at a polymorphism is defined as an allele. The combination of alleles on the two chromosomes is termed the genotype of the individual at that position. A distinct combination of two or more alleles of polymorphisms that occur together on the same chromosome is defined as a haplotype.
When a mutation arises in a chromosomal region, it does so on a background of particular DNA variants that are already present in the population, i.e., the mutation is linked to these surrounding alleles by the integrity of the DNA molecule. Over time, recombination tends to separate a mutant allele from the alleles of the surrounding DNA. At the population level, the positive association that remains between particular alleles at linked polymorphisms is called linkage disequilibrium (LD), meaning that these alleles occur more frequently together than would be expected from their population frequencies. Recombination ultimately leads to loss of LD unless there is a selective advantage of particular allele combinations.
The relationship between disease phenotype and three of the genetic concepts described (polymorphisms, alleles and haplotypes), is the subject of genetic association studies. That is, the aim of genetic epidemiology is to identify alleles (or in diploid terms, genotypes) of polymorphisms that are associated with an increase or decrease in risk of disease or a particular characteristic of a disease. The advantage of LD is that all polymorphisms in a genetic region do not have to be genotyped to detect an association. This is because the causative variant will reside on the same haplotypes as other polymorphisms and can be indirectly detected by typing for these. The disadvantage of LD is that it may be almost impossible to determine which of a series of alleles in LD on a haplotype that is actually the causative variant. Most of the genetic variation (> 99%) in the human genome is believed to be without any phenotypic consequence[32].
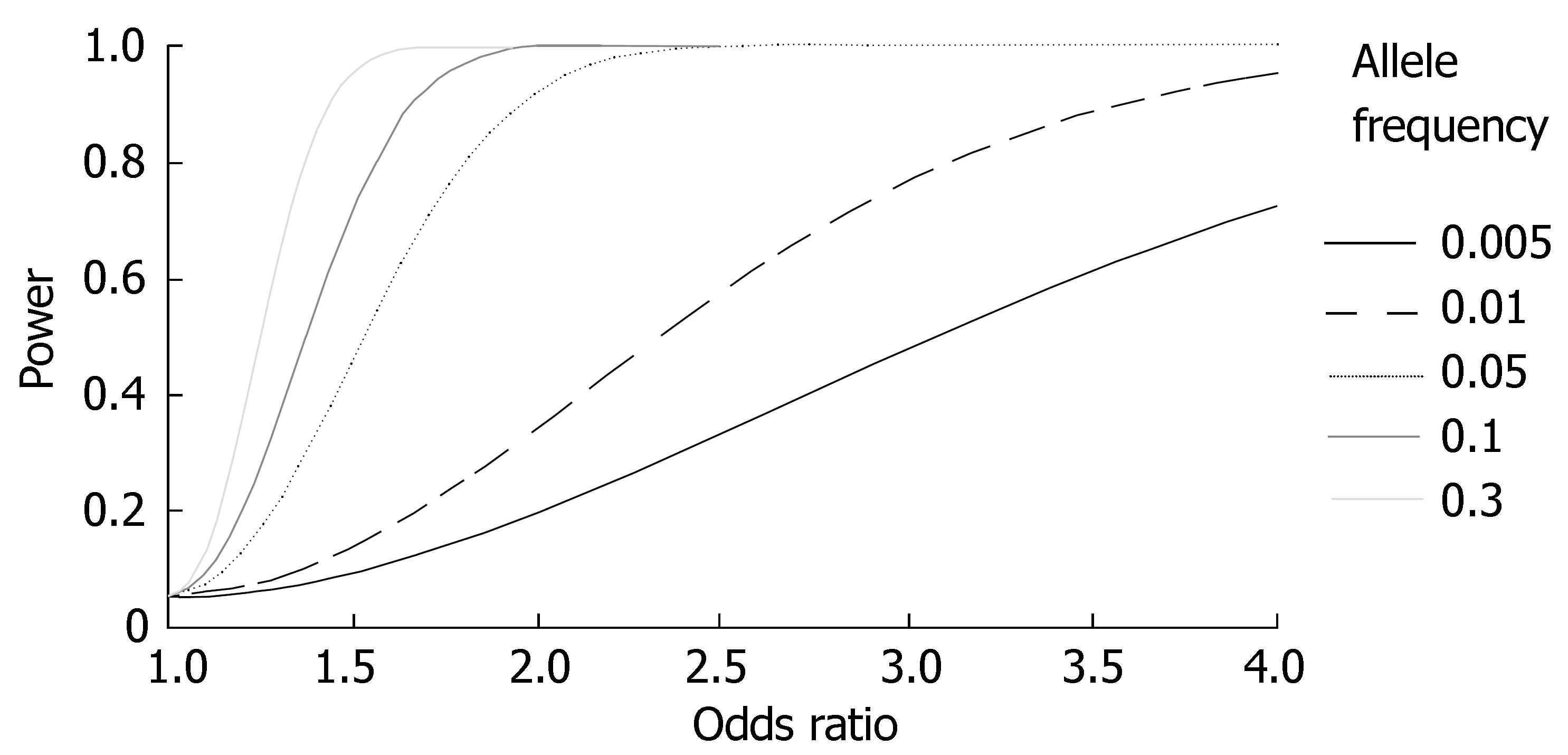
Because of the low prevalence, a major limiting factor for statistical power in studies of PSC susceptibility genes is sample size. Figure 2 illustrates the statistical power as a function of the effect size (odds ratio; OR) and allele frequency of a genetic variant for studies performed in the largest PSC population in which studies have been performed so far (n = 365)[33]. Two issues require mentioning. First, very weak effects (OR ≈ 1.0-1.3) are likely to be missed, even for populations of this size. Second, rare variants of importance for PSC susceptibility (allele frequency < 0.01) are likely to be missed unless the OR of the variant is very high (or low; ORs < 1 were not plotted for clarity).
An important controversy regarding the prospects of mapping the genetic predisposition to complex diseases is not related to statistical power, but the possible complexity of allelic variation at a susceptibility locus. Supporters of the "common-disease/common-variant" hypothesis argue that common diseases arise due to polymorphisms that are common (i.e., allele frequency > 0.10[34]) in the background population. Supporters of the "multiple rare variants" hypothesis point to the complexity observed at susceptibility loci in monogenic disorders, where multiple rare alleles define a similar phenotype (e.g., the hundreds of disease causing alleles at the cystic fibrosis transmembrane-conductance regulator locus)[35]. Possibly, susceptibility genes in complex diseases that are defined by multiple rare variants cannot be identified using regular LD based approaches[36]. Although PSC is relatively rare, the main HLA haplotypes that confer risk are relatively common (e.g., the frequency of the PSC associated ancestral HLA haplotype 8.1 is > 0.10 in Scandinavia[37]).
The abundance of false positive genetic association studies (i.e., typeIstatistical errors) represents a problem of legitimacy for this type of study design[38]. Simply using a P-value < 0.05 as "evidence" to distinguish between a "positive" and "negative" finding in these studies can be questioned[39]. The problem is partly related to the many statistical tests performed in these studies. The so-called Bonferroni correction (multiplying P-values with the number of comparisons that have been performed) is the most widely accepted strategy to account for this problem.
The Bonferroni approach has limitations. Due to the many tests that are theoretically possible throughout the genome, it can be argued that conservative significance levels of 10-5 or even 10-8 should be used for all tests[38,40]. Achieving such significance levels would require patient collections simply not available for rare diseases like PSC. The most recent proposal is that so-called permutation testing (in Latin, "permutare" means "change completely")within a dataset is the preferable strategy to take account of multiple testing[41]. In permutation tests, case/control assignment is shuffled randomly using a computer and tests are run over and over again to count how often the permuted dataset achieves the effect observed in the correctly ordered dataset. If the permuted dataset achieves an effect equal to or stronger than that observed in the original dataset in 500 out of 10000 analyses, this means that the probability of a typeIerror for a finding is 5%.
The problem of statistical significance in genetic association studies philosophically relates to the problem of causality for which criteria relevant to modern medicine were proposed by Sir Austen Bradford Hill in a classic essay in 1965[42]. These criteria point to factors in addition to the probability from statistical association tests (e.g., biological plausibility) that are required for a causal relationship to be established. This is also argued for in so-called Bayesian statistics, where the prior probability of a genetic variant to be associated (e.g., non-synonymous polymorphism in a gene which function is relevant to the disease phenotype), is accounted for when deciding on the posterior probability of whether or not a finding is valid[38]. In sum, circumstantial evidence (from functional studies or mouse models) is required to support findings if a genetic variant should be considered causative in terms of contributing to a disease phenotype[28], whatever the statistical evidence is available.
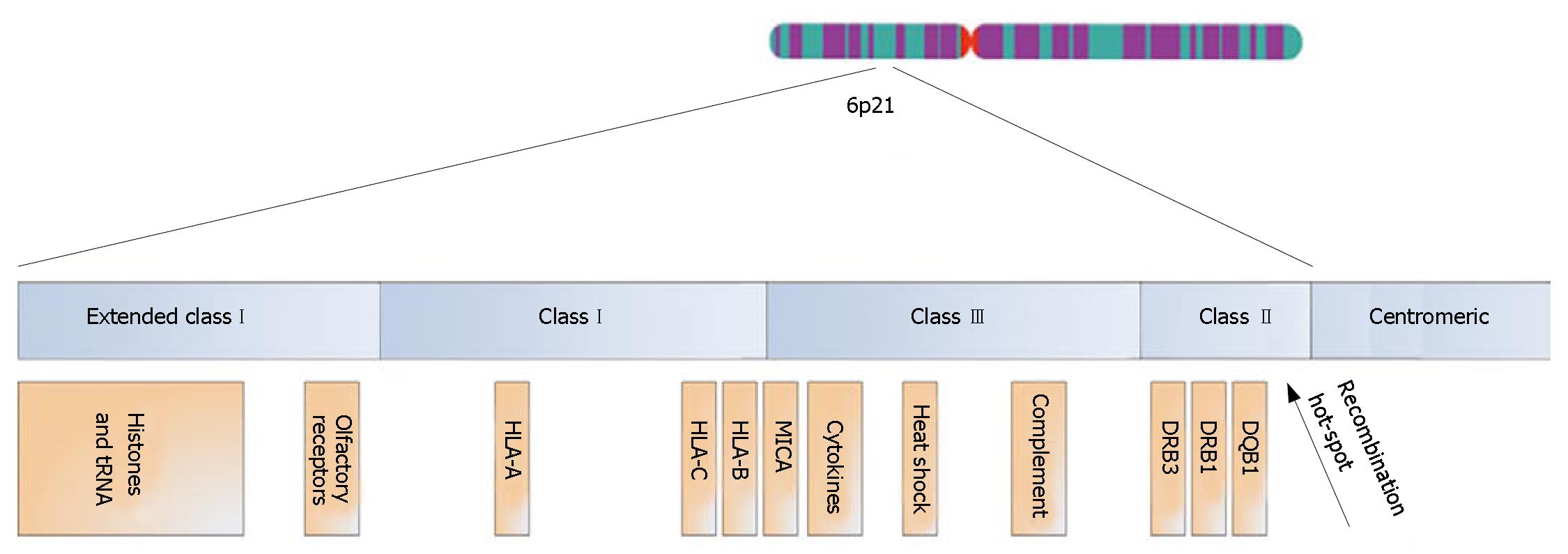
The HLA complex stretches across 7.6 million base pairs (bp) of DNA on the short arm of chromosome 6 and contains 252 expressed protein-coding genes, of which 28% are potentially related to immunological functions[43]. Throughout evolution of this genetic region[44], duplications have led to several gene clusters containing genes of similar function (Figure 3)[43]. HLA classImolecules (i.e., HLA-A, -B and -C) are expressed on all nucleated cells in the body and present intracellular/endogenous antigens to CD8+ T-lymphocytes. HLA classImolecules also serve as ligands for inhibitory killer immunoglobulin-like receptors (KIRs) on natural killer (NK) cells and γδ T-lymphocytes[45,46]. HLA class II molecules are expressed on antigen presenting cells (e.g., macrophages and dendritic cells) and present extracellular/exogenous antigens to CD4+ T-lymphocytes[45].
Sequence-based HLA-nomenclature was established in 1987[45]. The locus name is followed by an asterisk and two pairs of digits. The first pair of digits denominates the main type and is often similar to the serological type (e.g., DRB1*03 is the same as serological DR3, but DRB1*13 is only one of the DR6 alleles). The second pair of digits denominates the subtype (e.g., DRB1*0301 and DRB1*1301). Further definition is possible, since null alleles are suffixed by "N", and polymorphisms that do not alter the amino acid sequence of the peptide binding groove give rise to the fifth, sixth and seventh digits. In result, a complete sequence-based HLA allele name represents the haplotype of all alleles at all polymorphisms within the HLA gene at that chromosome.
LD between alleles at the HLA classIand II loci defines ancestral HLA haplotypes (AHs) and are named after which HLA-B allele they contain (e.g., the most common haplotype with HLA-B*08 is called AH8.1)[44]. Alleles of other genes are in LD with these ancestral haplotypes, and the co-occurrence of particular alleles across the entire HLA complex on one chromosome is called an extended HLA haplotype[47]. At the population level, the degree of conservation varies between different extended HLA haplotypes[48]. As examples of this phenomenon, an extended HLA haplotype with the HLA-B*08 and DRB1*0301 alleles (i.e., the AH8.1) is remarkably conserved in the Northern European population, whereas haplotypes carrying DRB1*04 alleles are considerably less conserved and may not even qualify for the denomination "extended haplotypes"[49].
A HLA association in PSC was first identified for HLA-B8 (i.e., HLA-B*0801) and DR3 (i.e., DRB1*0301)[24,50]. Later studies have verified that PSC associations exist also for the other alleles of the AH8.1 (the HLA-A1 allele[51], the HLA-C7 allele[52], the major histocompatibility complex classIchain-related A (MICA) *008/5.1 allele[53,54], and the tumour necrosis factor alpha (TNFα ) promoter -308 A allele[55,56]). This haplotype is associated with a wide range of autoimmune diseases[57,58]. A cross-European study (Norway, Sweden, Great Britain, Italy and Spain) concluded that a consistent, positive HLA class II association in PSC probably exists also for a haplotype that carries the DR6 (i.e., DRB1*1301) allele[37]. In individuals negative for DR3 and DR6, an association with haplotypes that carry the DR2 (i.e., DRB1*1501) allele can be found. Negative associations with HLA class II alleles have been reported for the DR4, DR7 and DR11 alleles[37,59,60], although primarily in populations of Northern European origin[56]. In Southern Europe, the picture is even more complex, since the DR4 allele seems to be consistent in LD with a predisposing variant in Italy[37,56], whereas a protective effect is noted in Spain[37].
Due to strong LD, an important question in HLA genetics is whether genetic associations are due to variation in the HLA classIor II genes (meaning that they arise because the patients are able to present particular antigens to the T-cell receptor)[61], or due to variation in neighbouring genes[62]. There is some degree of amino acid sequence similarity between several of the PSC associated HLA class II polypeptide variants[59,63]. However, no consistency has been found regarding these similarities[59]. The proposal of leucine at position 38 of the DRβ polypeptide as a critical determinant for PSC susceptibility relies heavily on the strong DRB3*0101 association in Northern European populations[63]. An early suggestion that a common denominator between haplotypes with the DRB1*0301 and DRB1*1301 alleles could be the DRB3*0101 allele (serologically DRw52a) was later withdrawn[64,65]. Another study found that the DRB1*1301-DRB3*0202 haplotype association is as strong as the DRB1*1301-DRB3*0101 association[37]. Taken together, the most interesting proposal of a single amino acid position in defining risk of PSC may rather relate to a protective effect in carriers of proline at position 55 of the DQβ polypeptide, which is common for DQ3 alleles known to be in LD with the protective DR4, DR7 and DR11 alleles[59]. However, no consistent risk allele is defined by this position[59], and to what extent the HLA class II molecules are of primary importance in the PSC pathogenesis should probably not be concluded based on present evidence.
The PSC-associated MICA*008/5.1 allele has been proposed as a common denominator between the PSC-associated A*01-C*07-B*08-DRB1*0301-DQB1*0201 and A*03-C*07-B*07-DRB1*1501-DQB1*0602 haplotypes[53,59]. MICA functions as a ligand for the activating NKG2D receptor on NK cells[66]. It was recently recognised that the two risk haplotypes in question share alleles not only at MICA, but also at the neighbouring HLA-B and -C loci, when these are defined according to the KIR binding properties of the HLA classImolecules[67]. The PSC-associated HLA-B and -C KIR ligand genotypes may result in decreased inhibition of NK cells and several subsets of T-lymphocytes that express KIRs[46,68]. Such combinations of KIR and HLA classIligand variants have been shown to increase susceptibility to other autoimmune diseases[46]. How the PSC-associated MICA*008/5.1 allele may cause disease is not known. This allele is also associated with an increased risk of other autoimmune conditions[69,70], and may thus also result in an increased activity of cells expressing the NKG2D receptor, acting in synergy with the loss of inhibition resulting from the PSC associated HLA classIligand genotypes. The fact that the MICA 5.1 allele was recently shown to confer protection against cholangiocarcinoma is in line with an activating effect[71]. Some studies report an increased frequency of NK cells in the portal infiltrate of patients with PSC when compared with other liver diseases[72,73], and also in the intestinal mucosa of patients with PSC without IBD compared with IBD patients without liver disease[74]. Taken together with the genetic findings in this region of the HLA complex (Figure 3), further studies on the role of these cells in PSC seem warranted.
In sum, the HLA association in PSC is likely to be complex. Multiple risk variants may exist[25], some of which may be associated not only with PSC, but autoimmunity in general.
Summarising the published genetic association studies in PSC, it seems proven beyond doubt that one or more genetic variants located within the HLA complex are important. The true identities of these variants, as discussed above, are not known. The situation is even less clear with regard to other susceptibility loci. Given the large number of protein coding genes in the human genome (25-35000)[32], selecting candidate genes for association studies is an extremely difficult task. According to strict criteria for what may be denominated a susceptibility gene in complex diseases (consistent statistical evidence, functional consequence of identified mutation, relevant tissue expression, etc.)[28], no such gene exists for PSC. A summary of studies performed is given in Table 1. So far, most attention has been given to genes known to be of importance in other autoimmune diseases. The association between PSC and IBD has also inspired some of the studies, as well as the observation of PSC-like changes in cystic fibrosis[75].
| Gene | Chromosome | N (PSC) | Primary finding | Reference | Replicationfinding | Reference |
| IL-1 | 2q | 40 | Negative | [76] | Negative | [77] |
| IL-10 | 1q | 96 | Negative | [77] | Negative | [55] |
| MMP1 | 11q | 165 | Negative | [78] | NA | - |
| MMP3 | 11q | 111 | Positive | [79] | Negative | [78] |
| CCR5 | 3p | 71 | Positive | [80] | Negative | [33] |
| ICAM-1 | 19p | 104 | Positive | [81] | Negative | [82] |
| CFTR | 7q | 29 | Negative | [83] | Negative | [84,85] |
| MDR3 | 7q | 37 | Negative | [86] | Negative | [87] |
| BSEP | 2q | 37 | Negative | [86] | NA | - |
| AIRE | 21q | 60 | Negative | [88] | NA | - |
| NRAMP1 | 2q | 40 | Negative | [89] | NA | - |
| CTLA4 | 2q | 144 | Negative | [90] | NA | - |
| FOXP3 | X | 195 | Negative | [91] | NA | - |
Two of the negative findings are of particular interest and will be discussed in greater detail. First, studies in limited populations (n < 50) have pointed to a non-significant increase of particular multidrug resistance gene 3 (MDR3) variants among PSC patients as compared with healthy controls[86,87]. Knock-out mice for this phospholipid transporter gene (called mdr2 in mice) spontaneously develop hepatic lesions resembling PSC[92], possibly due to loss of protection of the biliary epithelium from toxic bile acids. Second, it cannot be formally ruled out that the 32 bp deletion of the chemokine receptor 5 (CCR5) gene and the E/E genotype of the K469E SNP in the intercellular adhesion molecule 1 (ICAM-1) gene may confer population specific effects[80,81,93]. Both genes are plausible candidate genes in PSC. The CCR5 may be involved in the recruitment of intestinally activated lymphocytes via portal expression of CCR5 ligands (e.g., the macrophage inflammatory protein-1α and β), and ICAM-1 may play a similar role in recruiting leukocytes to an inflamed liver by interacting with the β2-integrin ligand. The negative findings in the replication series referred to in Table 1 state it unlikely that genetic variants of these receptors are of primary importance in the pathogenesis of PSC. The receptors may, however, still be involved in the disease process along with other CCRs and adhesion molecules [e.g., CCR9 and the mucosal addressin cell adhesion molecule 1 (MAdCAM-1)[94,95]].
The most prominent features of PSC along with the biliary changes are inflammatory bowel disease, cholangiocarcinoma and other autoimmune diseases (Figure 1).
The increased frequency of autoimmune diseases among patients with PSC is possibly due to the increased frequency of the AH8.1 among the patients[58,96]. Similarly, an increased frequency of IBD risk alleles among patients with PSC could contribute to the co-occurrence of these two phenotypes. Several IBD susceptibility genes have been identified during the last 6 years through the application of genome-wide linkage screens and subsequent fine-mapping approaches[26]. To determine if the high frequency of IBD among patients with PSC could be due to genetic risk factors shared with IBD in general, we recently genotyped key polymorphisms of known IBD susceptibility genes in a large cohort of Scandinavian PSC patients[97]. The following genes were studied: caspase activating recruitment domain 15 (CARD15), toll-like receptor 4 (TLR-4), caspase activating recruitment domain 4 (CARD4), solute carrier family 22, member 4 and 5 (SLC22A4 and SLC22A5), Drosophila discs large homolog 5 (DLG5) and multidrug resistance gene 1 (MDR1)[26,98]. No significant PSC associations were detected for any of the investigated polymorphisms[97]. These negative findings add to notions that the IBD phenotype in PSC may be a "third" IBD phenotype[99], possibly distinct from UC and Crohn's disease not only in clinical presentation, but also with regard to genetic susceptibility.
It is of interest to know whether genetic associations detected in PSC may be of particular importance for the IBD phenotype among the PSC patients or patients with IBD in general. In a recent study of HLA alleles in PSC and UC patients of the same ethnicity[100], the only parallel association detected was a protective effect of the DRB1*0404 allele, more pronounced among the PSC patients than among the patients with UC without liver disease. No association with any of the main PSC risk alleles (DRB1*0301, DRB1*1301 or DRB1*1501) was found among the regular UC patients. Interestingly, a non-significant trend towards a higher frequency of the DRB1*1501 allele was noted among the patients with PSC and concurrent IBD compared with PSC patients without IBD, and the possibility should be held open that this HLA haplotype may harbour genetic variants of particular importance for the IBD phenotype in PSC. A similar notion can be made with regard to the MMP3 5A allele association detected by Satsangi et al[79]. Although the replication study by Wiencke et al[78] failed to confirm an overall association with PSC susceptibility, a significant association was evident when PSC patients with UC were compared with UC patients without liver disease.
The study by Wiencke et al[78] also detected a possible association between cholangiocarcinoma and the MMP1 1G allele. Although the number of patients with cholangiocarcinoma in this series was too small for conclusive statistics to be performed (n = 15), the 100% occurrence of this allele among the cholangiocarcinoma patients warrants future replication attempts in other study populations. Recently, a highly significant association between polymorphisms in the NKG2D gene and cholangiocarcinoma in PSC was detected[71]. Previous studies have highlighted the importance of this activating NK cell receptor in protection against other cancer types[66]. Persistent exposure to effector molecules of inflammatory pathways (e.g., IL-6[101]), along with chronic cholestasis[102], is probably important for the malignant transformation of cholangiocytes. The study by Melum et al[33] points to the possible role of NK cell activity in protection against neoplastic cells. Polymorphisms of the NKG2D gene along with other parameters may also prove important in identifying PSC patients at a particular low risk of developing cholangiocarcinoma.
There is an increasing interest in so-called "modifier genes" in complex diseases (as compared with "susceptibility genes"),initiated by the recognition of the influence of such genes on disease expression (e.g., severity) in monogenic disorders like cystic fibrosis and haemochromatosis[103-105]. Modifier genes may point to biochemical and physiological systems of relevance to prognosis and are therefore of great clinical interest. Although PSC should be considered a progressive condition culminating in death or liver transplantation in most cases[106], the clinical course for each individual patient varies considerably[107,108]. In terms of disease course, indicators of PSC severity (e.g., portal hypertension and need for liver transplantation) are more likely to represent a particular disease stage than to serve as valid measures of disease progression. The most precise strategy for performing enquiries on effects from genotypes on disease course in PSC is thus to compare absolute survival time (defined as time from diagnosis until death or liver transplantation) using Kaplan-Meyer analyses, or calculating the relative risk for death and/or liver transplantation from Cox regressions[109,110].
We have recently observed that genetic variants of the steroid and xenobiotic receptor (SXR) are associated with a more aggressive disease course in PSC[110]. The SXR is a ligand-dependent transcription factor known to mediate protection against bile acid-induced liver injury in cholestatic animal models[111,112]. In this perspective, our data may suggest that the activity of bile acid detoxification systems could be of importance for disease progression in PSC. Interestingly, the SXR ligand rifampicin has been used in the treatment of cholestatic pruritus[113], and it has also been shown that ursodeoxycholic acid is able to activate SXR in human hepatocytes[114]. However, the SXR may also influence inflammatory pathways via the pro-inflammatory transcription factor nuclear factor kappa B (NF-κB)[115], as well as liver fibrogenesis and thus cirrhosis via direct effects on hepatic stellate cells and Kuppfer cells[116]. Further studies are needed to clarify the functional consequences of various polymorphisms of the SXR gene in patients with PSC.
The SXR variants associated with death or liver transplantation in our study were not associated with PSC susceptibility[110]. However, also for some of the disease- associated variants in the HLA complex, modifier effects have been observed. The first notion was made by Gow et al[117] who described an unusually aggressive disease progression in four patients carrying the DR4 allele. Later, Boberg et al[109] found that DR4 positive patients have an increased risk of cholangiocarcinoma, but do formally not experience an accelerated disease progression. In this study, an increased risk of death or liver transplantation was observed in patients heterozygous for the DR3-DQ2 haplotype. As long as the causative variants along the HLA haplotypes in question have not been identified, one can only hypothesize upon a biological explanation for these observations. Given the complexity of the HLA associations in PSC, it is even possible that other variants within this region may be important for disease progression than those primarily important for disease susceptibility. However, for the same reasons it has been difficult to pinpoint susceptibility genes in this region (strong LD, multiple genes of immunological relevance, etc.), such modifier genes may prove hard to identify conclusively.
Although several important findings have been made during the past 25 years since the first genetic association study in PSC was performed[24], PSC remains an enigmatic disease and future studies are warranted. With an ever increasing availability of methods for efficient genotyping of polymorphisms[118], a critical limitation for such studies in PSC is the availability of well-characterised patient materials. Collaborative efforts will be necessary to achieve patient collections required for detecting the modest effects (Figure 2), as well as for replicating results of uncertain validity[33]. Such collaborations are now being undertaken in other diseases[119], and have successfully aided in clarifying genetic associations found in PSC[37].
In terms of future research strategies, several proposals can be made. First, dissection of the widely replicated HLA-associated susceptibility to PSC should be considered a priority. Detailed maps of genetic markers in this region are now available[120]. It is anticipated that the systematic application of such marker maps in populations of an appropriate size may lead to the identification of true, disease causing variants in this difficult region[62].
Second, some biological pathways are pointed to by existing findings (e.g., the possible importance of bile acid homeostasis in influencing disease progression), and further candidate gene studies of critical components of these systems may identify additional risk factors. There is increasing awareness of the importance of interaction between polymorphisms in functionally related genes in complex diseases, i.e., epistasis[121,122]. In some cases, epistatic considerations have proven necessary for the detection of effects from genetic variation on a phenotype of interest[123,124]. These observations have implications for study design in future candidate gene studies in PSC. Polymorphisms not only in single genes, but in relevant panels of several genes encoding proteins with closely related functions, should be investigated.
Finally, two recent advances in the genetic research field now make genome-wide studies feasible also for case-control materials. First, the human haplotype map project (HAPMAP) was recently completed[125]. In the project, 3.9 million SNPs have been genotyped in families of three different ethnicities (at the time of writing). Results from the project enable researchers worldwide to efficiently select SNPs throughout the genome that are prone to cover genetic variation of interest to a project[126,127]. Second, although costs are high, genotyping technology now allows for the typing of 100000's of SNPs simultaneously in the same DNA sample[118]. Emerging reports provide proof-of-concept for genome-wide case-control studies[128,129]. However, there are still statistical problems to be solved regarding the many tests performed and risk of false positive results[130]. As evident from Figure 2, only strong effects may be detectable, and prospects may not yet justify the costs. However, sooner or later genome-wide studies seem warranted, also in PSC. Possibly, PSC susceptibility genes will be identified that would otherwise never have been included in hypothesis-driven candidate gene studies of the type performed so far[131].
| 1. | Schwartz si, Dale WA. Primary sclerosing cholangitis; review and report of six cases. AMA Arch Surg. 1958;77:439-451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 44] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 2. | Chapman RW, Arborgh BA, Rhodes JM, Summerfield JA, Dick R, Scheuer PJ, Sherlock S. Primary sclerosing cholangitis: a review of its clinical features, cholangiography, and hepatic histology. Gut. 1980;21:870-877. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 522] [Cited by in RCA: 516] [Article Influence: 11.2] [Reference Citation Analysis (1)] |
| 3. | Cullen SN, Chapman RW. Review article: current management of primary sclerosing cholangitis. Aliment Pharmacol Ther. 2005;21:933-948. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 51] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 4. | Olsson R, Boberg KM, de Muckadell OS, Lindgren S, Hultcrantz R, Folvik G, Bell H, Gangsøy-Kristiansen M, Matre J, Rydning A. High-dose ursodeoxycholic acid in primary sclerosing cholangitis: a 5-year multicenter, randomized, controlled study. Gastroenterology. 2005;129:1464-1472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 284] [Cited by in RCA: 266] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 5. | Brandsaeter B, Friman S, Broomé U, Isoniemi H, Olausson M, Bäckman L, Hansen B, Schrumpf E, Oksanen A, Ericzon BG. Outcome following liver transplantation for primary sclerosing cholangitis in the Nordic countries. Scand J Gastroenterol. 2003;38:1176-1183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 48] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 6. | Talwalkar JA, Lindor KD. Primary sclerosing cholangitis. Inflamm Bowel Dis. 2005;11:62-72. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 82] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 7. | Boberg KM, Aadland E, Jahnsen J, Raknerud N, Stiris M, Bell H. Incidence and prevalence of primary biliary cirrhosis, primary sclerosing cholangitis, and autoimmune hepatitis in a Norwegian population. Scand J Gastroenterol. 1998;33:99-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 362] [Cited by in RCA: 331] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 8. | Kingham JG, Kochar N, Gravenor MB. Incidence, clinical patterns, and outcomes of primary sclerosing cholangitis in South Wales, United Kingdom. Gastroenterology. 2004;126:1929-1930. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 91] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 9. | Bambha K, Kim WR, Talwalkar J, Torgerson H, Benson JT, Therneau TM, Loftus EV, Yawn BP, Dickson ER, Melton LJ. Incidence, clinical spectrum, and outcomes of primary sclerosing cholangitis in a United States community. Gastroenterology. 2003;125:1364-1369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 316] [Cited by in RCA: 303] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 10. | Schrumpf E, Boberg KM. Epidemiology of primary sclerosing cholangitis. Best Pract Res Clin Gastroenterol. 2001;15:553-562. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 51] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 11. | Broomé U, Olsson R, Lööf L, Bodemar G, Hultcrantz R, Danielsson A, Prytz H, Sandberg-Gertzén H, Wallerstedt S, Lindberg G. Natural history and prognostic factors in 305 Swedish patients with primary sclerosing cholangitis. Gut. 1996;38:610-615. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 596] [Cited by in RCA: 549] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 12. | Okolicsanyi L, Fabris L, Viaggi S, Carulli N, Podda M, Ricci G. Primary sclerosing cholangitis: clinical presentation, natural history and prognostic variables: an Italian multicentre study. The Italian PSC Study Group. Eur J Gastroenterol Hepatol. 1996;8:685-691. [PubMed] |
| 13. | Escorsell A, Parés A, Rodés J, Solís-Herruzo JA, Miras M, de la Morena E. Epidemiology of primary sclerosing cholangitis in Spain. Spanish Association for the Study of the Liver. J Hepatol. 1994;21:787-791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 103] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 14. | Takikawa H, Takamori Y, Tanaka A, Kurihara H, Nakanuma Y. Analysis of 388 cases of primary sclerosing cholangitis in Japan; Presence of a subgroup without pancreatic involvement in older patients. Hepatol Res. 2004;29:153-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 89] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 15. | Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347:417-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2693] [Cited by in RCA: 2772] [Article Influence: 115.5] [Reference Citation Analysis (3)] |
| 16. | Fausa O, Schrumpf E, Elgjo K. Relationship of inflammatory bowel disease and primary sclerosing cholangitis. Semin Liver Dis. 1991;11:31-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 216] [Cited by in RCA: 190] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 17. | Rasmussen HH, Fallingborg JF, Mortensen PB, Vyberg M, Tage-Jensen U, Rasmussen SN. Hepatobiliary dysfunction and primary sclerosing cholangitis in patients with Crohn's disease. Scand J Gastroenterol. 1997;32:604-610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 95] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 18. | Saarinen S, Olerup O, Broomé U. Increased frequency of autoimmune diseases in patients with primary sclerosing cholangitis. Am J Gastroenterol. 2000;95:3195-3199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 129] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 19. | Bergquist A, Ekbom A, Olsson R, Kornfeldt D, Lööf L, Danielsson A, Hultcrantz R, Lindgren S, Prytz H, Sandberg-Gertzén H. Hepatic and extrahepatic malignancies in primary sclerosing cholangitis. J Hepatol. 2002;36:321-327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 531] [Cited by in RCA: 479] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 20. | Schrumpf E, Abdelnoor M, Fausa O, Elgjo K, Jenssen E, Kolmannskog F. Risk factors in primary sclerosing cholangitis. J Hepatol. 1994;21:1061-1066. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 74] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 21. | Boberg KM, Jebsen P, Clausen OP, Foss A, Aabakken L, Schrumpf E. Diagnostic benefit of biliary brush cytology in cholangiocarcinoma in primary sclerosing cholangitis. J Hepatol. 2006;45:568-574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 112] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 22. | Lazaridis KN, Gores GJ. Primary sclerosing cholangitis and cholangiocarcinoma. Semin Liver Dis. 2006;26:42-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 95] [Article Influence: 4.8] [Reference Citation Analysis (1)] |
| 23. | Loftus EV, Sandborn WJ, Tremaine WJ, Mahoney DW, Zinsmeister AR, Offord KP, Melton LJ. Primary sclerosing cholangitis is associated with nonsmoking: a case-control study. Gastroenterology. 1996;110:1496-1502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 83] [Article Influence: 2.8] [Reference Citation Analysis (1)] |
| 24. | Schrumpf E, Fausa O, Førre O, Dobloug JH, Ritland S, Thorsby E. HLA antigens and immunoregulatory T cells in ulcerative colitis associated with hepatobiliary disease. Scand J Gastroenterol. 1982;17:187-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 135] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 25. | Donaldson PT, Norris S. Immunogenetics in PSC. Best Pract Res Clin Gastroenterol. 2001;15:611-627. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 40] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 26. | Gaya DR, Russell RK, Nimmo ER, Satsangi J. New genes in inflammatory bowel disease: lessons for complex diseases? Lancet. 2006;367:1271-1284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 173] [Article Influence: 8.7] [Reference Citation Analysis (1)] |
| 27. | Rioux JD, Abbas AK. Paths to understanding the genetic basis of autoimmune disease. Nature. 2005;435:584-589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 144] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 28. | Glazier AM, Nadeau JH, Aitman TJ. Finding genes that underlie complex traits. Science. 2002;298:2345-2349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 618] [Cited by in RCA: 562] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 29. | Bergquist A, Lindberg G, Saarinen S, Broomé U. Increased prevalence of primary sclerosing cholangitis among first-degree relatives. J Hepatol. 2005;42:252-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 82] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 30. | Mathew CG, Lewis CM. Genetics of inflammatory bowel disease: progress and prospects. Hum Mol Genet. 2004;13 Spec No 1:R161-R168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 83] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 31. | Strachan T, Read A. Human Molecular Genetics, 2nd ed. Oxford: BIOS Scientific Publishers Ltd 1999; . |
| 32. | Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigo R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X. The sequence of the human genome. Science. 2001;291:1304-1351. |
| 33. | Melum E, Karlsen TH, Broomé U, Thorsby E, Schrumpf E, Boberg KM, Lie BA. The 32-base pair deletion of the chemokine receptor 5 gene (CCR5-Delta32) is not associated with primary sclerosing cholangitis in 363 Scandinavian patients. Tissue Antigens. 2006;68:78-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 11] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 34. | Zondervan KT, Cardon LR. The complex interplay among factors that influence allelic association. Nat Rev Genet. 2004;5:89-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 373] [Cited by in RCA: 359] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 35. | Terwilliger JD, Weiss KM. Linkage disequilibrium mapping of complex disease: fantasy or reality? Curr Opin Biotechnol. 1998;9:578-594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 213] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 36. | Weiss KM, Terwilliger JD. How many diseases does it take to map a gene with SNPs? Nat Genet. 2000;26:151-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 329] [Cited by in RCA: 293] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 37. | Spurkland A, Saarinen S, Boberg KM, Mitchell S, Broome U, Caballeria L, Ciusani E, Chapman R, Ercilla G, Fausa O. HLA class II haplotypes in primary sclerosing cholangitis patients from five European populations. Tissue Antigens. 1999;53:459-469. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 105] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 38. | Colhoun HM, McKeigue PM, Davey Smith G. Problems of reporting genetic associations with complex outcomes. Lancet. 2003;361:865-872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 843] [Cited by in RCA: 831] [Article Influence: 36.1] [Reference Citation Analysis (0)] |
| 39. | Trikalinos TA, Ntzani EE, Contopoulos-Ioannidis DG, Ioannidis JP. Establishment of genetic associations for complex diseases is independent of early study findings. Eur J Hum Genet. 2004;12:762-769. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 64] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 40. | Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516-1517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3635] [Cited by in RCA: 3368] [Article Influence: 112.3] [Reference Citation Analysis (0)] |
| 41. | Cardon LR, Bell JI. Association study designs for complex diseases. Nat Rev Genet. 2001;2:91-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 946] [Cited by in RCA: 870] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 42. | Hill AB. The Environment and Disease: Association or Causation? Proc R Soc Med. 1965;58:295-300. [PubMed] |
| 43. | Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK, Lush MJ, Povey S, Talbot CC, Wright MW. Gene map of the extended human MHC. Nat Rev Genet. 2004;5:889-899. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 780] [Cited by in RCA: 853] [Article Influence: 38.8] [Reference Citation Analysis (0)] |
| 44. | Dawkins R, Leelayuwat C, Gaudieri S, Tay G, Hui J, Cattley S, Martinez P, Kulski J. Genomics of the major histocompatibility complex: haplotypes, duplication, retroviruses and disease. Immunol Rev. 1999;167:275-304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 240] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 45. | Marsh SGE, Parham P, Barber LD. The HLA factsbook. London, San Diego: Academic Press Inc 1999; . |
| 46. | Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5:201-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 873] [Cited by in RCA: 916] [Article Influence: 43.6] [Reference Citation Analysis (0)] |
| 47. | Dorak MT, Shao W, Machulla HK, Lobashevsky ES, Tang J, Park MH, Kaslow RA. Conserved extended haplotypes of the major histocompatibility complex: further characterization. Genes Immun. 2006;7:450-467. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 60] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 48. | Ahmad T, Neville M, Marshall SE, Armuzzi A, Mulcahy-Hawes K, Crawshaw J, Sato H, Ling KL, Barnardo M, Goldthorpe S. Haplotype-specific linkage disequilibrium patterns define the genetic topography of the human MHC. Hum Mol Genet. 2003;12:647-656. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 91] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 49. | Blomhoff A, Olsson M, Johansson S, Akselsen HE, Pociot F, Nerup J, Kockum I, Cambon-Thomsen A, Thorsby E, Undlien DE. Linkage disequilibrium and haplotype blocks in the MHC vary in an HLA haplotype specific manner assessed mainly by DRB1*03 and DRB1*04 haplotypes. Genes Immun. 2006;7:130-140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 37] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 50. | Chapman RW, Varghese Z, Gaul R, Patel G, Kokinon N, Sherlock S. Association of primary sclerosing cholangitis with HLA-B8. Gut. 1983;24:38-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 175] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 51. | Donaldson PT, Farrant JM, Wilkinson ML, Hayllar K, Portmann BC, Williams R. Dual association of HLA DR2 and DR3 with primary sclerosing cholangitis. Hepatology. 1991;13:129-133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 95] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 52. | Moloney MM, Thomson LJ, Strettell MJ, Williams R, Donaldson PT. Human leukocyte antigen-C genes and susceptibility to primary sclerosing cholangitis. Hepatology. 1998;28:660-662. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 53. | Norris S, Kondeatis E, Collins R, Satsangi J, Clare M, Chapman R, Stephens H, Harrison P, Vaughan R, Donaldson P. Mapping MHC-encoded susceptibility and resistance in primary sclerosing cholangitis: the role of MICA polymorphism. Gastroenterology. 2001;120:1475-1482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 60] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 54. | Wiencke K, Spurkland A, Schrumpf E, Boberg KM. Primary sclerosing cholangitis is associated to an extended B8-DR3 haplotype including particular MICA and MICB alleles. Hepatology. 2001;34:625-630. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 55] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 55. | Mitchell SA, Grove J, Spurkland A, Boberg KM, Fleming KA, Day CP, Schrumpf E, Chapman RW. Association of the tumour necrosis factor alpha -308 but not the interleukin 10 -627 promoter polymorphism with genetic susceptibility to primary sclerosing cholangitis. Gut. 2001;49:288-294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 68] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 56. | Neri TM, Cavestro GM, Seghini P, Zanelli PF, Zanetti A, Savi M, Podda M, Zuin M, Colombo M, Floreani A. Novel association of HLA-haplotypes with primary sclerosing cholangitis (PSC) in a southern European population. Dig Liver Dis. 2003;35:571-576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 19] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 57. | Candore G, Lio D, Colonna Romano G, Caruso C. Pathogenesis of autoimmune diseases associated with 8.1 ancestral haplotype: effect of multiple gene interactions. Autoimmun Rev. 2002;1:29-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 168] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 58. | Price P, Witt C, Allcock R, Sayer D, Garlepp M, Kok CC, French M, Mallal S, Christiansen F. The genetic basis for the association of the 8.1 ancestral haplotype (A1, B8, DR3) with multiple immunopathological diseases. Immunol Rev. 1999;167:257-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 394] [Cited by in RCA: 399] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 59. | Donaldson PT, Norris S. Evaluation of the role of MHC class II alleles, haplotypes and selected amino acid sequences in primary sclerosing cholangitis. Autoimmunity. 2002;35:555-564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 44] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 60. | Wiencke K, Karlsen TH, Boberg KM, Thorsby E, Schrumpf E, Lie BA, Spurkland A. Primary sclerosing cholangitis is associated with extended HLA-DR3 and HLA-DR6 haplotypes. Tissue Antigens. 2007;69:161-169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 36] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 61. | Sollid LM, Markussen G, Ek J, Gjerde H, Vartdal F, Thorsby E. Evidence for a primary association of celiac disease to a particular HLA-DQ alpha/beta heterodimer. J Exp Med. 1989;169:345-350. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 670] [Cited by in RCA: 682] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 62. | Valentonyte R, Hampe J, Huse K, Rosenstiel P, Albrecht M, Stenzel A, Nagy M, Gaede KI, Franke A, Haesler R. Sarcoidosis is associated with a truncating splice site mutation in BTNL2. Nat Genet. 2005;37:357-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 336] [Cited by in RCA: 317] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 63. | Farrant JM, Doherty DG, Donaldson PT, Vaughan RW, Hayllar KM, Welsh KI, Eddleston AL, Williams R. Amino acid substitutions at position 38 of the DR beta polypeptide confer susceptibility to and protection from primary sclerosing cholangitis. Hepatology. 1992;16:390-395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 86] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 64. | Prochazka EJ, Terasaki PI, Park MS, Goldstein LI, Busuttil RW. Association of primary sclerosing cholangitis with HLA-DRw52a. N Engl J Med. 1990;322:1842-1844. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 110] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 65. | Inability to attribute susceptibility to primary sclerosing cholangitis to specific amino acid positions of the HLA-DRw52a allele. N Engl J Med. 1991;325:1251-1252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 66. | Hayashi T, Imai K, Morishita Y, Hayashi I, Kusunoki Y, Nakachi K. Identification of the NKG2D haplotypes associated with natural cytotoxic activity of peripheral blood lymphocytes and cancer immunosurveillance. Cancer Res. 2006;66:563-570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 109] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 67. | Karlsen TH, Boberg KM, Olsson M, Sun JY, Senitzer D, Bergquist A, Schrumpf E, Thorsby E, Lie BA. Particular genetic variants of ligands for natural killer cell receptors may contribute to the HLA associated risk of primary sclerosing cholangitis. J Hepatol. 2007;46:899-906. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 67] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 68. | Nelson GW, Martin MP, Gladman D, Wade J, Trowsdale J, Carrington M. Cutting edge: heterozygote advantage in autoimmune disease: hierarchy of protection/susceptibility conferred by HLA and killer Ig-like receptor combinations in psoriatic arthritis. J Immunol. 2004;173:4273-4276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 207] [Cited by in RCA: 206] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 69. | Gambelunghe G, Ghaderi M, Tortoioli C, Falorni A, Santeusanio F, Brunetti P, Sanjeevi CB, Falorni A. Two distinct MICA gene markers discriminate major autoimmune diabetes types. J Clin Endocrinol Metab. 2001;86:3754-3760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 46] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 70. | Stastny P. Introduction: MICA/MICB in innate immunity, adaptive immunity, autoimmunity, cancer, and in the immune response to transplants. Hum Immunol. 2006;67:141-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 30] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 71. | Melum E, Karlsen TH, Boberg KM, Bergquist A, Thorsby E, Schrumpf E, Lie BA. Genetic variation in the receptor-ligand pair NKG2D-MICA is strongly associated with development of cholangiocarcinoma in patients with primary sclerosing cholangitis. J Hepatol. 2007;46:S49. |
| 72. | Hashimoto E, Lindor KD, Homburger HA, Dickson ER, Czaja AJ, Wiesner RH, Ludwig J. Immunohistochemical characterization of hepatic lymphocytes in primary biliary cirrhosis in comparison with primary sclerosing cholangitis and autoimmune chronic active hepatitis. Mayo Clin Proc. 1993;68:1049-1055. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 83] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 73. | Hata K, Van Thiel DH, Herberman RB, Whiteside TL. Phenotypic and functional characteristics of lymphocytes isolated from liver biopsy specimens from patients with active liver disease. Hepatology. 1992;15:816-823. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 39] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 74. | Silvain C, Zeevi A, Saidman S, Duquesnoy RJ, Van Thiel DH. Phenotypic and functional characteristics of colonic lymphocytes isolated from patients with primary sclerosing cholangitis and inflammatory bowel disease. Hepatogastroenterology. 1995;42:250-258. [PubMed] |
| 75. | Abdalian R, Heathcote EJ. Sclerosing cholangitis: a focus on secondary causes. Hepatology. 2006;44:1063-1074. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 135] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 76. | Stokkers PC, van Aken BE, Basoski N, Reitsma PH, Tytgat GN, van Deventer SJ. Five genetic markers in the interleukin 1 family in relation to inflammatory bowel disease. Gut. 1998;43:33-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 56] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 77. | Donaldson PT, Norris S, Constantini PK, Bernal W, Harrison P, Williams R. The interleukin-1 and interleukin-10 gene polymorphisms in primary sclerosing cholangitis: no associations with disease susceptibility/resistance. J Hepatol. 2000;32:882-886. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 18] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 78. | Wiencke K, Louka AS, Spurkland A, Vatn M, Schrumpf E, Boberg KM. Association of matrix metalloproteinase-1 and -3 promoter polymorphisms with clinical subsets of Norwegian primary sclerosing cholangitis patients. J Hepatol. 2004;41:209-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 29] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 79. | Satsangi J, Chapman RW, Haldar N, Donaldson P, Mitchell S, Simmons J, Norris S, Marshall SE, Bell JI, Jewell DP. A functional polymorphism of the stromelysin gene (MMP-3) influences susceptibility to primary sclerosing cholangitis. Gastroenterology. 2001;121:124-130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 61] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 80. | Eri R, Jonsson JR, Pandeya N, Purdie DM, Clouston AD, Martin N, Duffy D, Powell EE, Fawcett J, Florin TH. CCR5-Delta32 mutation is strongly associated with primary sclerosing cholangitis. Genes Immun. 2004;5:444-450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 40] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 81. | Yang X, Cullen SN, Li JH, Chapman RW, Jewell DP. Susceptibility to primary sclerosing cholangitis is associated with polymorphisms of intercellular adhesion molecule-1. J Hepatol. 2004;40:375-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 49] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 82. | Bowlus CL, Karlsen TH, Broomé U, Thorsby E, Vatn M, Schrumpf E, Lie BA, Boberg KM. Analysis of MAdCAM-1 and ICAM-1 polymorphisms in 365 Scandinavian patients with primary sclerosing cholangitis. J Hepatol. 2006;45:704-710. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 13] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 83. | Girodon E, Sternberg D, Chazouillères O, Cazeneuve C, Huot D, Calmus Y, Poupon R, Goossens M, Housset C. Cystic fibrosis transmembrane conductance regulator (CFTR) gene defects in patients with primary sclerosing cholangitis. J Hepatol. 2002;37:192-197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 34] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 84. | Sheth S, Shea JC, Bishop MD, Chopra S, Regan MM, Malmberg E, Walker C, Ricci R, Tsui LC, Durie PR. Increased prevalence of CFTR mutations and variants and decreased chloride secretion in primary sclerosing cholangitis. Hum Genet. 2003;113:286-292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 70] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 85. | Gallegos-Orozco JF, E Yurk C, Wang N, Rakela J, Charlton MR, Cutting GR, Balan V. Lack of association of common cystic fibrosis transmembrane conductance regulator gene mutations with primary sclerosing cholangitis. Am J Gastroenterol. 2005;100:874-878. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 29] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 86. | Pauli-Magnus C, Kerb R, Fattinger K, Lang T, Anwald B, Kullak-Ublick GA, Beuers U, Meier PJ. BSEP and MDR3 haplotype structure in healthy Caucasians, primary biliary cirrhosis and primary sclerosing cholangitis. Hepatology. 2004;39:779-791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 118] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 87. | Rosmorduc O, Hermelin B, Boelle PY, Poupon RE, Poupon R, Chazouillères O. ABCB4 gene mutations and primary sclerosing cholangitis. Gastroenterology. 2004;126:1220-1222; author reply 1222-1223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 23] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 88. | Vogel A, Liermann H, Harms A, Strassburg CP, Manns MP, Obermayer-Straub P. Autoimmune regulator AIRE: evidence for genetic differences between autoimmune hepatitis and hepatitis as part of the autoimmune polyglandular syndrome type 1. Hepatology. 2001;33:1047-1052. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 40] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 89. | Stokkers PC, de Heer K, Leegwater AC, Reitsma PH, Tytgat GN, van Deventer SJ. Inflammatory bowel disease and the genes for the natural resistance-associated macrophage protein-1 and the interferon-gamma receptor 1. Int J Colorectal Dis. 1999;14:13-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 90. | Wiencke K, Boberg KM, Donaldson P, Harbo H, Ling V, Schrumpf E, Spurkland A. No major effect of the CD28/CTLA4/ICOS gene region on susceptibility to primary sclerosing cholangitis. Scand J Gastroenterol. 2006;41:586-591. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 91. | Karlsen TH, Flåm S, Schrumpf E, Broomé U, Thorsby E, Vatn M, Boberg KM, Lie BA. Polymorphisms of the forkhead box P3 gene on chromosome X in primary sclerosing cholangitis and ulcerative colitis. Genes Immun. 2005;6:56 (abstract). |
| 92. | Popov Y, Patsenker E, Fickert P, Trauner M, Schuppan D. Mdr2 (Abcb4)-/- mice spontaneously develop severe biliary fibrosis via massive dysregulation of pro- and antifibrogenic genes. J Hepatol. 2005;43:1045-1054. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 201] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 93. | Henckaerts L, Fevery J, Van Steenbergen W, Verslype C, Yap P, Nevens F, Roskams T, Libbrecht L, Rutgeerts P, Vermeire S. CC-type chemokine receptor 5-Delta32 mutation protects against primary sclerosing cholangitis. Inflamm Bowel Dis. 2006;12:272-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 14] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 94. | Adams DH, Eksteen B. Aberrant homing of mucosal T cells and extra-intestinal manifestations of inflammatory bowel disease. Nat Rev Immunol. 2006;6:244-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 245] [Cited by in RCA: 223] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 95. | O'Mahony CA, Vierling JM. Etiopathogenesis of primary sclerosing cholangitis. Semin Liver Dis. 2006;26:3-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 94] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 96. | Cullen S, Chapman R. Primary sclerosing cholangitis. Autoimmun Rev. 2003;2:305-312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 29] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 97. | Karlsen TH, Hampe J, Wiencke K, Schrumpf E, Thorsby E, Lie BA, Broomé U, Schreiber S, Boberg KM. Genetic polymorphisms associated with inflammatory bowel disease do not confer risk for primary sclerosing cholangitis. Am J Gastroenterol. 2007;102:115-121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 35] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 98. | Ho GT, Soranzo N, Nimmo ER, Tenesa A, Goldstein DB, Satsangi J. ABCB1/MDR1 gene determines susceptibility and phenotype in ulcerative colitis: discrimination of critical variants using a gene-wide haplotype tagging approach. Hum Mol Genet. 2006;15:797-805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 93] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 99. | Loftus EV, Harewood GC, Loftus CG, Tremaine WJ, Harmsen WS, Zinsmeister AR, Jewell DA, Sandborn WJ. PSC-IBD: a unique form of inflammatory bowel disease associated with primary sclerosing cholangitis. Gut. 2005;54:91-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 515] [Cited by in RCA: 529] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 100. | Karlsen TH, Boberg KM, Vatn M, Bergquist A, Hampe J, Schrumpf E, Thorsby E, Schreiber S, Lie BA. Different HLA class II associations in ulcerative colitis patients with and without primary sclerosing cholangitis. Genes Immun. 2007;8:275-278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 44] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 101. | Kobayashi S, Werneburg NW, Bronk SF, Kaufmann SH, Gores GJ. Interleukin-6 contributes to Mcl-1 up-regulation and TRAIL resistance via an Akt-signaling pathway in cholangiocarcinoma cells. Gastroenterology. 2005;128:2054-2065. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 186] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 102. | Komichi D, Tazuma S, Nishioka T, Hyogo H, Chayama K. Glycochenodeoxycholate plays a carcinogenic role in immortalized mouse cholangiocytes via oxidative DNA damage. Free Radic Biol Med. 2005;39:1418-1427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 47] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 103. | Drumm ML, Konstan MW, Schluchter MD, Handler A, Pace R, Zou F, Zariwala M, Fargo D, Xu A, Dunn JM. Genetic modifiers of lung disease in cystic fibrosis. N Engl J Med. 2005;353:1443-1453. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 356] [Cited by in RCA: 345] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 104. | Jacolot S, Le Gac G, Scotet V, Quere I, Mura C, Ferec C. HAMP as a modifier gene that increases the phenotypic expression of the HFE pC282Y homozygous genotype. Blood. 2004;103:2835-2840. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 105] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 105. | Le Gac G, Scotet V, Ka C, Gourlaouen I, Bryckaert L, Jacolot S, Mura C, Férec C. The recently identified type 2A juvenile haemochromatosis gene (HJV), a second candidate modifier of the C282Y homozygous phenotype. Hum Mol Genet. 2004;13:1913-1918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 74] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 106. | Porayko MK, LaRusso NF, Wiesner RH. Primary sclerosing cholangitis: a progressive disease? Semin Liver Dis. 1991;11:18-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 27] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 107. | Schrumpf E, Fausa O, Elgjo K, Kolmannskog F. Hepatobiliary complications of inflammatory bowel disease. Semin Liver Dis. 1988;8:201-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 52] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 108. | Balasubramaniam K, Wiesner RH, LaRusso NF. Primary sclerosing cholangitis with normal serum alkaline phosphatase activity. Gastroenterology. 1988;95:1395-1398. [PubMed] |
| 109. | Boberg KM, Spurkland A, Rocca G, Egeland T, Saarinen S, Mitchell S, Broomé U, Chapman R, Olerup O, Pares A. The HLA-DR3,DQ2 heterozygous genotype is associated with an accelerated progression of primary sclerosing cholangitis. Scand J Gastroenterol. 2001;36:886-890. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 40] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 110. | Karlsen TH, Lie BA, Frey Frøslie K, Thorsby E, Broomé U, Schrumpf E, Boberg KM. Polymorphisms in the steroid and xenobiotic receptor gene influence survival in primary sclerosing cholangitis. Gastroenterology. 2006;131:781-787. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 44] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 111. | Xie W, Radominska-Pandya A, Shi Y, Simon CM, Nelson MC, Ong ES, Waxman DJ, Evans RM. An essential role for nuclear receptors SXR/PXR in detoxification of cholestatic bile acids. Proc Natl Acad Sci USA. 2001;98:3375-3380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 614] [Cited by in RCA: 579] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 112. | Stedman CA, Liddle C, Coulter SA, Sonoda J, Alvarez JG, Moore DD, Evans RM, Downes M. Nuclear receptors constitutive androstane receptor and pregnane X receptor ameliorate cholestatic liver injury. Proc Natl Acad Sci USA. 2005;102:2063-2068. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 171] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 113. | Khurana S, Singh P. Rifampin is safe for treatment of pruritus due to chronic cholestasis: a meta-analysis of prospective randomized-controlled trials. Liver Int. 2006;26:943-948. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 114] [Article Influence: 5.7] [Reference Citation Analysis (1)] |
| 114. | Schuetz EG, Strom S, Yasuda K, Lecureur V, Assem M, Brimer C, Lamba J, Kim RB, Ramachandran V, Komoroski BJ. Disrupted bile acid homeostasis reveals an unexpected interaction among nuclear hormone receptors, transporters, and cytochrome P450. J Biol Chem. 2001;276:39411-39418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 297] [Cited by in RCA: 283] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 115. | Zhou C, Tabb MM, Nelson EL, Grün F, Verma S, Sadatrafiei A, Lin M, Mallick S, Forman BM, Thummel KE. Mutual repression between steroid and xenobiotic receptor and NF-kappaB signaling pathways links xenobiotic metabolism and inflammation. J Clin Invest. 2006;116:2280-2289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 290] [Cited by in RCA: 325] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 116. | Wright MC. The impact of pregnane X receptor activation on liver fibrosis. Biochem Soc Trans. 2006;34:1119-1123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 30] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 117. | Gow PJ, Fleming KA, Chapman RW. Primary sclerosing cholangitis associated with rheumatoid arthritis and HLA DR4: is the association a marker of patients with progressive liver disease? J Hepatol. 2001;34:631-635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 16] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 118. | Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1893] [Cited by in RCA: 1794] [Article Influence: 85.4] [Reference Citation Analysis (0)] |
| 119. | Rich SS, Concannon P, Erlich H, Julier C, Morahan G, Nerup J, Pociot F, Todd JA. The Type 1 Diabetes Genetics Consortium. Ann N Y Acad Sci. 2006;1079:1-8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 98] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 120. | de Bakker PI, McVean G, Sabeti PC, Miretti MM, Green T, Marchini J, Ke X, Monsuur AJ, Whittaker P, Delgado M. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet. 2006;38:1166-1172. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 642] [Cited by in RCA: 622] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 121. | Gregersen JW, Kranc KR, Ke X, Svendsen P, Madsen LS, Thomsen AR, Cardon LR, Bell JI, Fugger L. Functional epistasis on a common MHC haplotype associated with multiple sclerosis. Nature. 2006;443:574-577. [PubMed] |
| 122. | Cordell HJ. Epistasis: what it means, what it doesn't mean, and statistical methods to detect it in humans. Hum Mol Genet. 2002;11:2463-2468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 757] [Cited by in RCA: 715] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 123. | Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, Cheng J, Goedert JJ, Vlahov D, Hilgartner M. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872-874. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 916] [Cited by in RCA: 930] [Article Influence: 42.3] [Reference Citation Analysis (0)] |
| 124. | Martin MP, Gao X, Lee JH, Nelson GW, Detels R, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, Trowsdale J. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31:429-434. [PubMed] |
| 125. | A haplotype map of the human genome. Nature. 2005;437:1299-1320. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4621] [Cited by in RCA: 4260] [Article Influence: 202.9] [Reference Citation Analysis (0)] |
| 126. | Conrad DF, Jakobsson M, Coop G, Wen X, Wall JD, Rosenberg NA, Pritchard JK. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat Genet. 2006;38:1251-1260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 374] [Cited by in RCA: 359] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 127. | de Bakker PI, Burtt NP, Graham RR, Guiducci C, Yelensky R, Drake JA, Bersaglieri T, Penney KL, Butler J, Young S. Transferability of tag SNPs in genetic association studies in multiple populations. Nat Genet. 2006;38:1298-1303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 172] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 128. | Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461-1463. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2428] [Cited by in RCA: 2345] [Article Influence: 117.3] [Reference Citation Analysis (1)] |
| 129. | Yang Z, Camp NJ, Sun H, Tong Z, Gibbs D, Cameron DJ, Chen H, Zhao Y, Pearson E, Li X. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science. 2006;314:992-993. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 543] [Cited by in RCA: 557] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 130. | Wang WY, Barratt BJ, Clayton DG, Todd JA. Genome-wide association studies: theoretical and practical concerns. Nat Rev Genet. 2005;6:109-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 831] [Cited by in RCA: 763] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 131. | Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1519] [Cited by in RCA: 1477] [Article Influence: 77.7] [Reference Citation Analysis (0)] |
| 132. | Jeffreys AJ, Ritchie A, Neumann R. High resolution analysis of haplotype diversity and meiotic crossover in the human TAP2 recombination hotspot. Hum Mol Genet. 2000;9:725-733. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 152] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
S- Editor Liu Y L- Editor Wang XL E- Editor Wang HF