INTRODUCTION
miRNAs (microRNAs) are a subset of small, typically 21-23 nt in length, non-coding RNAs evolutionarily conserved in many organisms as disparate as yeast, fruit flies, human and plants[1]. This growing family of small RNAs was first discovered in Caenorhabditis elegans in 1993[2] and newly honored as a milestone in the process of the gene concept[3]. Until now hundreds of miRNAs have been identified in many organisms by using experimental and bioinformatic prediction approaches. It is well known that, unlike its cousin signal interfering RNAs (siRNAs), miRNAs have the unique ability to negatively regulate gene expression involved in cell development, proliferation, apoptosis and the stress response[4]. Consequently, it is proposed that these biological properties of miRNAs could offer an access to many human diseases including cancers[5]. Recent findings have demonstrated that miRNAs play critical roles in human cancer, revealing that miRNAs could act as potential oncogenes and repressors[6,7]. Thus, this class of miRNAs are now dubbed ‘oncomirs’- miRNAs which is closely related to tumor[8].
Cancer is characterized by uncontrolled proliferation and the inappropriate survival of damaged cells. Although cells have evolutionarily developed several safeguards to prevent malignant transformation during development and adulthood, this normal process can be disrupted in cancer cells. Cancer cells can take advantage of their unique strategy to escape scrutiny during cell division. The oncogenesis conventionally refers to tumor suppressors and oncogenes, such as APC, κ-RAS, Myc, P53 and P21. Although these regulatory molecules do play critical roles in tumor development, recent intense interest is being attached to miRNAs.
We are just beginning to appreciate the novel involvement of miRNAs in human cancers but much more remain obscure. Few investigations to date have converged to support the concrete links between miRNAs and each cancer species. However, most cancer species share the same mechanisms that give rise to tumorigenesis even in different tissues, so the general mechanism may be applied extensively to each cancer species. The potential link between miRNAs and tumors discussed in this review will be limited to only a few of the elucidated cancers.
miRNA BIOGENESIS AND MECHANISMS OF GENE EXPRESSION CONTROL
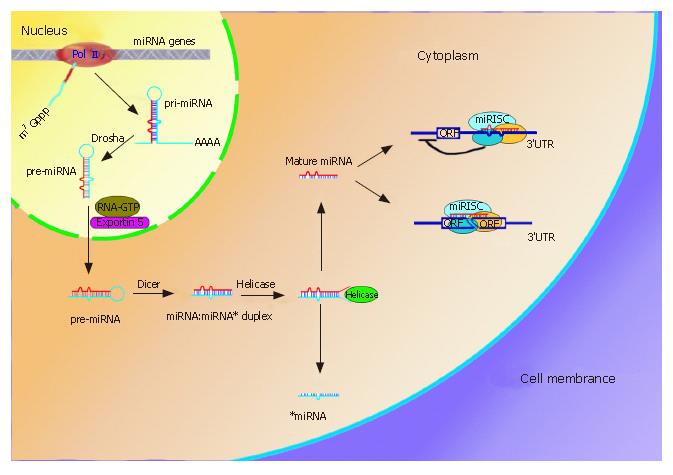
The biogenesis of miRNAs has recently been elucidated (Figure 1)[1]. RNA Pol II generally transcribes miRNA genes in the nucleus and gives rise to large primary miRNA (pri-miRNA) transcripts that, like mRNA, are capped at 5’ terminus and polyadenylated at 3’ terminus. The initial pri-miRNAs are then processed by RNase III, Drasha, to form 70-bp pre-miRNAs in the nucleus. The pre-miRNAs are exported into the cytoplasm by the RNA GTP-dependent transporter Exportin 5 and undergo an additional processing step to produce a miRNA:miRNA* duplex with the aid of another RNase III, Dicer. The RNA duplex is subsequently unwound by Helicase and the mature miRNA finally enter the RNA-induced silence complex (RISC)[9,10].
Figure 1 miRNAs biogenesis and two mechanisms involved in gene expression.
MiRNAs genes often cluster on the chromosome and are transcribed by RNA PolII to form pri-miRNAs in the nucleus. The pri-miRNAs then undergo procession by RNase III, Drasha, and are exported to cytoplasm by Exportin 5. Another RNase III, Dicer, further processes the pre-miRNA to generate a ~22nt miRNA:miRNA* duplex, where miRNA* is complementary to miRNA. Helicase can divide the duplex into two separate ones. Whereas miRNA* is degraded, mature miRNA can enter the the miRNA-induced silence complex (miRISC). The miRISC complex block protein synthesis by imperfectly binding to the 3’UTR of the mRNA (upper right), the other one is to endonucleolytically cleave the target mRNA by perfect or nearly perfect base pairing (lower right).
Two mechanisms of miRNA repression have been suggested depending on the degree of complementarities between the miRNAs and the target mRNAs (Figure 1)[4]. First, is interference with protein synthesis by binding to imperfect complementary sites within the 3’ untranslated region of the target mRNA. The 3’UTR of target mRNA usually contains multiple complementary sites for distinct miRNAs, but the precise mechanism is still poorly understood. Second, is endonucleolytic cleavage of the target mRNA by perfect base pairing. The latter was previously thought to function solely in flowering plants, but the paradigm that animal miRNAs do not affect th stability of imperfectly base-paired miRNA has been challenged recently. Lim et al[11] used microarray analysis to investigate changes in global mRNA level in HeLa cells in response to miRNAs that are normally undetectable in those malignant cells and observed the reduction of 100-200 genes at the mRNA level. This observed reduction in mRNA level was attributed to AU-rich elements (AREs) that induced mRNA turnover by degrading mRNAs in exosome[12]. AREs are often found in transcripts that encode cell proliferation factors (e.g., TNF-α, GM-CSF, c-Fos, IL-6 and IL-8), and therefore stability of these transcripts in the absence of specific miRNAs contributes to the cell proliferation that accelerate tumor formation and development[13]. Nevertheless, one recent finding indicated that the reverse step also exists in the latter mechanism, where the mRNA can be relieved from the miRNA-induced inhibition in cells subject to different stress conditions[14]. This repression and derepression of mRNA expression cooperatively contribute to the dynamic balance of mRNA in cells, but the detailed mechanism is still not clear.
miRNAs could provide a convenient and efficient pathway to manipulate gene expression at posttranscrip-tional level. Natural miRNAs exert their effects by base pairing with the target mRNAs in a much more compact and energy-efficient manner than protein encoding regulatory molecules like enzymes and hormones, which show a necessary adaptation to regulate gene expression in eukaryotes[15].
miRNA GENE CLUSTERS AND LOCI
The human genome contains up to 1000 miRNA genes, which constitute approximately 1-5% of the expressed genes[16]. miRNAs are endogenetically conserved with evolutionary plasticity in eukaryotic genomes and are often organized in tandem and closely clustered on the chromosome[17]. This arrangement can have particular significance in the control of gene expression. When clustered miRNAs have a similar sequence, miRNAs gene products may synchronize to regulate a set of mRNA targets. However, clusters can also contain miRNAs with different sequences that extensively deploy toward their specific targets. These closely related characteristics may allow miRNAs to function as pleiotropic regulators at the cellular level in many organisms.
Over half of miRNA genes (52.5%) are located in or near fragile sites or cancer-associated genomic regions[18]. These sites are preferential sites of sister chromatin exchange, translocation, deletion, amplification or integration of plasmid DNA and tumor-associated virus, which frequently cause the aberrant miRNA expression during pathogenesis. For instance, miR-15a and miR-16a genes, frequently deleted and/or underexpressed in patients with B cell chronic lymphocytic leukemia, map to 13q14 that is deleted in many cases[19]. This result highlights that aberrant miRNA expression is possibly geared towards the intrinsic defect that gives rise to tumors.
miRNA IN CANCER STEM CELLS
Most tissues contain rare cells that follow the norm of stem cell biology to tissue self-renewal and repair[20]. The unprecedented self-renewal rate of robust tissues (like intestine, skin, blood and breast) often parallels a high susceptibility to malignant transformation, because the molecular mechanisms that control homeostatic self-renewal and those underlie tumors are evidently symmetric[21]. Currently the emerging notion is that tumor might contain stem cell-like ‘cancer stem cells’-rare cells with indefinite proliferative potential that trigger tumor formation and growth, and with the presumed ability to transport new tumor seeds to distant sites[22]. Despite this controversial notion, one cancer (leukaemias) of the haematopoietic system provides the strong evidence that cancer cell proliferation is driven by cancer stem cells[23]. Given that cancer cells and normal stem cells share the similar potential to indefinite self-renew, it seems reasonable to propose that newly arising cancer cells appropriate the machinery for self-renewing cell division which is normally used in stem cells[24].
It is well known that miRNAs function as critical regulators of gene expression in the control of stem cells during development[5]. If miRNAs play a similar role in cancer stem cells then, in theory, it is possible support the hypothesis that several miRNAs appropriate the miRNA-mediated machinery for self-renewal in stem cells to develop tumors. Indeed, it has been validated that tumor tissues are constantly characterized with altered miRNA expressions.
A HALLMARK OF TUMOR
Currently the potential connection between miRNAs and cancers is just beginning to be appreciated. Cancer cells tend to undergo the distinct expression of miRNA, distinguishing them from the normal ones. Calin et al[25] first found that specific miRNA expression is abnormal in B-leukemia, suggesting that altered miRNA expression correlate with specific tumor development. In accordance with this, Michael et al[26] investigated possible changes at the miRNA level during tumorigenesis and showed the reduced accumulation of two specific miRNAs: miR-143 and miR-145, but consistent levels of the 70-bp precursor pre-miRNA in colorectal neoplasia as well as breast carcinoma, prostate carcinoma, chronic myelogenous leukemia and cervical carcinoma, implying that many tumors share aberrant specific miRNA expression. Recently Lu et al[27] found that cancer cell lines showed low miRNA expression profile when they used a new, bead-based flow cytometric miRNA expression profiling method to analyze a large-scale expression of 217 mammalian miRNA from 334 samples including multiple human cancers. Their study further demonstrated that tumors originating from tissue with a common embryonic source share the similar miRNA, but not mRNA, expression signature, and that distinct patterns of miRNA expression are consistent with the developmental history of human cancer, revealing that miRNAs could be used to classify different cancers and validate the developmental history of cancer. Volinia et al[7] further found that global miRNA expression signatures from solid tumors show a good separation between the different tissues, but not all miRNA expressions are underexpressed compared with their respective normal tissues. The studies support the hypothesis that the global change in miRNA expression is a hallmark of all human cancers, providing a hint that miRNAs correlate with various tumor development.
Moreover, impaired components of machinery mediating miRNA processing and miRNA-mediating gene repression give rise to tumorigenesis[28,29], demonstrating that altered specific miRNAs expression might play a causal role in the generation or/and maintenance of tumors. This description of cancers in molecular terms is likely to improve the way in which human cancers are diagnosed, classified, monitored, and (specially) treated, which will promise the emergency of the new era in cancer research in the future[30].
miRNA AND CELL CYCLE
Normal cells can tightly control cell proliferation and death by means of the cell cycle, thereby preventing malignant transformation during development and adulthood. The 3’UTR of mRNAs encoding many cell cycle-associated cytokines often contain binding sites to miRNAs, indicating that normal miRNAs are essential for cell cycle control. Hatfield et al[31] reported that Drosophila melanogaster germline stem cells subject to constitutively eliminated miRNA expression exhibited normal identity but were defective in cell cycle control, showing that miRNAs are essential in the control of cell cycle. Thus constitutive miRNAs are necessary to control cell cycle and maintain the balance of cell proliferation, differentiation and apoptosis, which play essential roles in preventing normal tissues from malignancy[32,33].
However, cancer cells are insensitive to cell division stop signals in an environment where most of the cells are quiescent. It is tempting to speculate that miRNAs could have a similar role in cancer cells and particularly rare cancer stem cells, where the disrupted miRNAs expression makes cells insensitive to environmental signals that normally stop the cell cycle. Brennecke et al[34] assigned a novel role to miRNA encoded by the bantam gene in control of cell proliferation and apoptosis during Drosophila development. Bantam miRNA could stimulate cell proliferation and simultaneously suppress apoptosis by manipulating the proapoptosis gene hid expression. It controls both cell growth and cycle progression in a coordinated manner, revealing that the putative vertebrate homologs of bantam miRNA genes may be oncogenes, whereas there are no homologues of bantam in human, other oncogenic miRNAs could play a similar role in control of cell proliferation and apoptosis. When the miRNA expression is impaired, normal tissues could have a high risk to develop tumor.
miRNA AND CANCER-ASSOCIATED SIGNALING PATHWAYS
The signaling transductions (Wnt, Notch, SHH and BMP) play essential roles in the processes of cell life at the molecular level, but these pathways controlling cell growth and differentiation in normal cell are almost invariably changed in cancer. Consistent with the altered miRNA expression in cancer cells, it is reasonable that the two can work together to control cell fate.
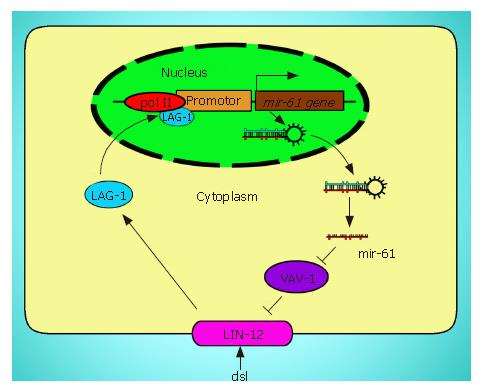
Several cancer-associated signaling pathways directly regulate expression of specific miRNA genes. Yoo et al[35] found that LIN-12/Notch signaling pathway directly binds to one miRNA gene, mir-61, and promote its expression in vulval precursor cells during C. elegans development. Stimulated expression of mir-61 gene subsequently represses the translation of Vav-1, the ortholog of the vav oncogene, who negatively regulates the lin-2 gene activity. These cyclic regulations form a positive feedback loop that helps maximize lin-12 activity and continually stimulate Notch signaling pathway (Figure 2). It has been known that Notch signaling pathway plays an important role in many cancer species. If the similar mechanisms exist in cancer cell, this positive loop could trigger and accelerate tumorigenesis.
Figure 2 The positive loop between mir-16 and LIN-12.
Exogenous signal activates LIN-12/Notch signaling pathway, which promotes the transcription of mir-61. Overexpression of mir-61 inhibits the activity of VAV-1 (purple) that reduces the activity of LIN-12 (pink), where LIN-12, mir-61, and VAV-1 form a feedback loop that helps maximize LIN-12 activity.
miRNAs can influence the signaling pathway by repressing several secreted signaling proteins. RAS is a signaling protein in many significant signaling pathways and its overexpression usually results in oncogenic transformation. The 3’UTL of the human RAS genes contains multiple let-7 complementary sites, allowing let-7 to regulate RAS expression. Johnson et al[36] found that let-7 expression is lower in lung tumors than in the normal lung tissue, providing a possible strategy to treat lung cancer by repairing mutated let-7 gene. It was reported that miR-143 and miR-145, lower in colorectal cancer than in normal tissues, are predicted to regulate several target mRNAs encoding components of signal transduction pathway (Raf, Rho, GTPase activating protein, G-protein γ, NF-κB and HGK)[26]. Hence, the direct and indirect interaction between miRNAs and secreted signaling protein can influence tumorigenesis.
Thus, it is supposed that miRNAs and signal as well as other regulatory molecules constitute a network where normal cells follow a rule to divide, differentiate, and die. When the regulatory network is impaired, cell cycle will be out of control, giving rise to tumor development. However, the networks of miRNAs and signals are largely elusive to date.
INTERACTION BETWEEN miRNA AND ONCOGENES
Due to mutation, many human proto-oncogenes can convert to oncogenes. These oncogenes often encode common regulatory molecules that can stimulate the tumor development in the body. For example, the proto-oncogene C-MYC encodes a helix-loop-helix leucine zipper transcriptional factor that regulates cell proliferation, growth and apoptosis. Recent findings revealed that c-Myc directly binds to the locus of a cluster of six miRNAs and stimulates their expression[37]. Overexpression of miR-17-5p and miR-20a, two miRNAs in this cluster, reduced the expression of E2F1 (one transcriptional factor). c-Myc and E2F1 are reciprocally induced in normal cell to form a putative positive feedback loop, like the one between mir-16 and LIN-12. So these two miRNAs provide a potent tool to dampen this reciprocal activation and tightly regulate c-Myc-mediated cellular proliferation in normal cells. When this cluster of miRNA genes is deleted or underexpressed, the cell cycle would be out of control and have a risk of tumorigenesis.
Notably, not all miRNAs function as tumor repressors in the body. For example, enforced expression of the mir-17-92 cluster positively cooperates with c-Myc expression to accelerate tumor development in a mouse B-cell lymphoma model, implicating that the mir-17-92 cluster as a potential oncogene[2]. The oncogenic miRNAs can be upregulated in cancer cells, which are consistent with the conclusion drawn by Volinia that not all miRNAs are underexpressed in cancer cells[7]. In fact, specific miRNA expression influencing cell fate is dependant on the milieu of miRNAs and their target mRNAs expressed in individual cell.
RULERS OF miRNA
As the pleiotropic regulators in cells, who regulate the expression of miRNA genes? As LIN-12/Notch signaling pathway directly regulates the expression of mir-16 gene discussed above[35], it seems that the regulation of miRNA expression follows the classic model widely used to control the mRNA expression. Taganov et al[38] recently reported that three putative NF-κB consensus binding sites locate upstream of the predicted miRNA-146 gene, so miRNA-146 is a NF-κB-dependent gene. Is this general model the common one or just an exceptional one with respect to diverse miRNAs and cytokines in cells? Our understanding of this knowledge awaits further investigations.
NOT THE END OF STORY
We have discussed the potential roles of miRNAs in cancers by means of cell cycle, signaling and oncogene, but they are just the tip of emerging iceberg, because exploding data show that miRNAs have a potentially much more widely influence over diverse developmental and physiological pathways than imagined. Recent evidence revealed that miRNAs could participate in genomic stability and epigenetic modification[2,39], in metabolic changes compatible with tumor formation, growth and metastasis[40], and in the immune response to virus-mediated infection[41-45]. Many more aspects of miRNAs are available for exploration, so this is not the whole story.
More intriguingly, the other cousin of miRNAs, piwi-interacting RNAs (piRNAs), have just been discovered in rat germline cells and are also shown to control gene expression involved in sperm development at posttranscriptional level[46]. The world of three small RNAs (siRNAs, miRNAs, and piRNAs) is undoubtedly yielding newly provocative insights and revolutionizing our thinking about genome control[47]. Although transcriptional control is the most prevalent form of gene expression control, it is by no means the only way in complex eukaryotes. For example, it is important for mature blood red cells to control the stability of expressed mRNAs accounting for no extra mRNA transcription any more or is for immune cells to make a rapid response to stress without mRNAs transcription initiation. Posttranscriptional control has been ignored, but many novel small RNAs are changing our thinking.
PROSPEROUS OUTLOOK IN miRNA
Evaluating the novel roles of miRNAs as repressors and oncogenes enriches our knowledge, which addresses the precise mechanism leading to tumorigenesis. The investigation will certainly bring about a potent tool to diagnose and treat human cancers, but a detailed, mechanistic understanding of miRNAs functions as oncogenes and tumor repressors is now retarded by lacking a valid and efficient biochemical technique to precisely identify miRNAs and their corresponding targets. It is estimated that many more miRNAs are still waiting to be discovered in the human genome and functions of most known miRNAs have not been elucidated. The other challenge is to accurately identify targets that are manipulated by miRNAs, because miRNAs can bind to their imperfect targets, even with no canonical complementarities that allow short stretch of mismatched base-pairs and G-U base pair. Other outstanding questions about miRNAs remain unresolved. What regulates the expression of miRNAs? Do distinct miRNAs have a direct function in cancer progression, or just simply differentially modulate in tumor? Who determines the opposite roles of miRNAs as both oncogenes and repressors? Do miRNAs act mainly to ‘fine-tune’ gene expression or more often as binary on/off switch? What factors affect the accessibility and efficacy of a miRNA at a 3’UTR? Another key one is how to apply this novel technique to cancer therapy. However, more sophisticated experimental approaches, in combination with computational prediction strategies, will shed light on these challenges.
It has been shown that miRNA expression profile is a more accurate signature than protein expression one, and several patients got better prognosis after repairing the abrogated miRNAs. We enthusiastically expect that traditional and bioinformatics technique will generate a tremendous amount of excitement and inspiration about miRNAs in future.
S- Editor Liu Y L- Editor Ma JY E- Editor Bai SH