Published online Sep 14, 2007. doi: 10.3748/wjg.v13.i34.4593
Revised: June 13, 2007
Accepted: June 18, 2007
Published online: September 14, 2007
AIM: To evaluate the significance of BNIP3 in the pathogenesis of pancreatic cancer, we analyzed the relationship between the expression of BNIP3 and survival rate of the patients with pancreatic cancer, or chemosensitivities in pancreatic cancer cell lines, particularly for gemcitabine, the first-line anti-tumor drug for pancreatic cancer.
METHODS: To compare the expression level of BNIP3 with the resistance to gemcitabine, eight pancreatic cancer cell lines were subjected to gemcitabine treatment and the quantitative real-time RT-PCR method was used to evaluate BNIP3 expression. Immunohistochemical analysis was also performed using 22 pancreatic cancer specimens to study relationship between BNIP3 expression and survival rate.
RESULTS: Although no significantly positive association between BNIP3 mRNA level and gemcitabine chemosensitivity was observed, pancreatic cancer cell lines that were sensitive to gemcitabine treatment tended to show high levels of BNIP3 expression. The converse, an absence of BNIP3 expression, was not correlated with gemcitabine resistance. We further compared the BNIP3 expression profiles of resected primary pancreatic cancer specimens with the prognosis of the patients, and found a tendency of favorable prognosis and low BNIP3 expression.
CONCLUSION: High levels of BNIP3 expression cannot be used as one of the predicting factors for gemcitabine chemosensitivity, and some yet to be known factors will have to fill the gap for the accurate prediction of pancreatic cancer chemosensitivity to gemcitabine. However, BNIP3 expression may have an impact on prediction of prognosis of patients with pancreatic cancer.
- Citation: Ishida M, Sunamura M, Furukawa T, Akada M, Fujimura H, Shibuya E, Egawa S, Unno M, Horii A. Elucidation of the relationship of BNIP3 expression to gemcitabine chemosensitivity and prognosis. World J Gastroenterol 2007; 13(34): 4593-4597
- URL: https://www.wjgnet.com/1007-9327/full/v13/i34/4593.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i34.4593
Pancreatic adenocarcinoma is a common cancer with an extremely poor prognosis around the world because of its aggressive invasive capacity, early metastasis, resistance to existing chemotherapeutic agents and radiation therapy, and lack of specific symptoms that help in finding patients at early stages for curative operation. To improve the horrible prognosis, we need to find novel approaches to both diagnosis and treatment that are more efficient than currently available techniques.
BNIP3, the hypoxia-inducible proapoptotic gene belonging to the BCL2 family, was originally identified as the gene encoding a protein that interacts with adenovirus E1B 19-kDa protein[1]. The expression of BNIP3 is increased under hypoxic conditions by a transcription factor, hypoxia-inducible factor 1α (HIF1α)[2,3], and leads to cell death by two different pathways; (1) heterodimerization with the anti-apoptotic protein BCL2, and (2) opening the mitochondrial permeability transition pores by direct contact with its outer membrane. Thus, BNIP3 is considered to be a key regulator of hypoxia-induced cell death[4,5,6].
Hypoxia is a common phenomenon in solid tumors and has been proven to occur in pancreatic cancer, but BNIP3 expression was shown to be decreased in pancreatic cancer compared with normal pancreas due to the hypermethylation of its promoter[7]. On the other hand, high expression of BNIP3 in pancreatic cancer cell lines has been reported to be associated with the chemosensitivity to gemcitabine 2', 2'-difluorodeoxycitidine. (Gemzar, Eli-Lilly, Indianapolis, IN), a novel pyrimidine nucleoside analogue[8]. It was expected that downregulation of proapoptotic BNIP3 might contribute to chemo-resistance, survival and progression of pancreatic cancer in a hypoxic environment. In the present study, we analyzed the expression of BNIP3 in several pancreatic cancer cell lines to explore the association with chemosensitivity in pancreatic cancer.
Two human pancreatic cancer cell lines (CFPAC1 and SUIT2) were obtained from Cancer Research UK Research Services (London, UK). PK-1, PK-8, PK-9, PK-45, and PK-59 were established at Tohoku University from patients with pancreatic cancer[9,10]. PANC-1 was obtained from Cell Resource Center for Biomedical Research, Institute for Development, Aging, and Cancer, Tohoku University (Sendai, Japan). All cells were maintained in RPMI 1640 containing 10% fetal bovine serum under an atmosphere of 5% CO2 with humidity at 37°C.
To assess cell proliferation, the MTT test was used as described previously[11]. Cells were resuspended in fresh medium and seeded in 96-well plates at concentration of 5 × 103 cells/well. Cells were incubated for 24 h at 37°C, and then gemcitabine was added to each well at various concentrations. Each plate was incubated at 37°C for 72 h. An aliquot of 10 μL of 3-(4, 5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (2.5 mg/mL) was added to each well, and the plate was incubated at 37°C for 2 h. The absorbance was measured at 560 nm using a microplate reader.
Total RNAs were extracted from the harvested cells using an RNeasy mini kit (QIAGEN, Tokyo, Japan) according to the supplier’s instructions. Each purified RNA was dissolved in RNase-free water, and its concentration was measured by optical absorbance at an aliquot of 10 μg total RNA and Super Script II Reverse Transcriptase (Invitrogen, Carlsbad, CA) by the methods described previously[12]. The synthesized cDNA was used for a quantitative real-time PCR analysis using an ABI PRISM 7000 Sequence Detection System (Applied Biosystems, Foster City, CA) following the manufacturer’s instructions. Specific primers and common probe were designed by using the Primer Express software (Applied Biosystems), and their nucleotide sequences are: 5'-tggacggagtagctccaagagc-3' (forward primer), 5'-agaagccctgttggtatcttgtg-3' (reverse primer), 5'-tctcactgtgacagtccacctcgc-3' (TaqMan probe). These primers were purchased from Nihon Gene Research Laboratories (Sendai, Japan). Expression of the β2-microglobulin (B2M) gene was monitored as an internal control, and the nucleotide sequences for the primers and the probe were described previously[13]. Amplifications were carried out in the reaction mixture in 25 μL containing 5 μL of cDNA samples and 12.5 μL of 2 × ABsolute QPCR ROX Mix (ABgene, Epsom, UK), and the final concentration of 0.2 μmol/L of each primer pair and 0.4 μmol/L of the probe were added in a program comprised of 2 min at 50°C, 15 min at 95°C, followed by 40 cycles consisting of 15 s at 95°C and 1 min at 60°C. The expression ratio of BNIP3/B2M was calculated and used. Each experimental reaction was performed in triplicate.
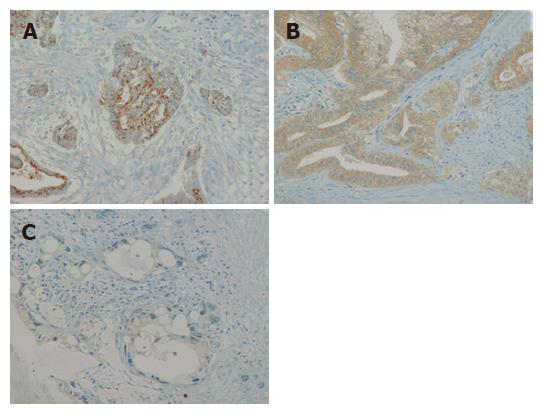
Pancreatic cancer specimens were obtained from 22 patients between 1995 and 2004. All the patients had undergone surgery at Tohoku University Hospital. Stages were defined according to the general rules of the Japan Pancreas Society[14]. These patients underwent either pancreaticoduodenectomy or distal pancreatectomy with nodal dissection followed by similar chemotherapy including gemcitabine treatment. All of the patients’ prognoses until 2005 were also determined. Twelve patients were male and 10 patients were female, and the mean age of operation was 64.0 years old. Specimens for immunohistochemistry were fixed in 10% formalin and embedded in paraffin for histological analysis. Research protocols for this study were approved by the Ethics Committee of the Tohoku University School of Medicine. Antigen retrieval was performed by boiling the slides in 10 mmol/L citrate buffer twice for 10 min. Peroxidase was quenched with a 3% H2O2 solution in 30% methanol. After an overnight incubation with mouse anti-BNIP3 antibody clone Ana 40 (Sigma-Aldrich, St. Louis, MO) at 4°C, slides were washed with PBS supplemented with 0.05% Tween-20 and exposed to the HRPO-linked anti-mouse secondary antibody for 45 min at room temperature. Color reaction was carried out by incubation for 4 min with liquid DAB+ substrate and counterstaining by Mayer’s hematoxylin solution. Staining patterns were divided into three groups following the report by Erkan et al[15]; punctate perinuclear staining, diffuse cytoplasmic staining, and negative/faint staining.
All experiments were done in duplicate or triplicate. A two-tailed Student’s t-test was applied for statistical analysis of comparative data. Overall survival curves were generated according to the Kaplan-Meier method. P < 0.05 was considered statistically significant.
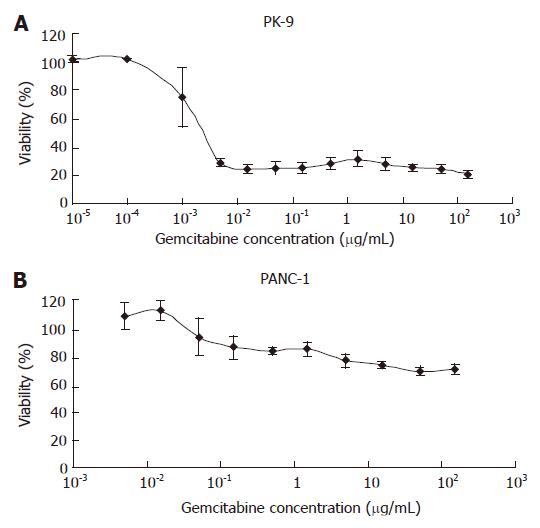
The response of eight pancreatic cancer cell lines to gemcitabine treatment was investigated using the MTT assay. Representative dose-response curves are shown in Figure 1; PANC-1 is a representative resistant cell line, whereas PK-9 is a representative sensitive cell line. The pancreatic cancer cell lines were divided into three groups according to their IC50 values as shown in Table 1. Judging from the IC50 value, two cell lines (PK-9 and SUIT2) showed high sensitivities to gemcitabine; less than 30% of the cells survived in the presence of 0.1 μg/mL of gemcitabine for 72 h (Figure 1A). In contrast, PANC-1 and PK-59 showed very low sensitivity; more than 50% of the cells survived even in the presence of more than 100 μg/mL of the drug for 72 h (Figure 1B). The remaining four cell lines (CFPAC1, PK-1, PK-8 and PK-45) showed moderate sensitivity (IC50 values 0.1-100 μg/mL) and were classified as intermediately sensitive cell lines.
| IC50 (μg/mL) | Cell lines | |
| Sensitive group | < 0.1 | PK-9, SUIT2 |
| Intermediate group | 0.1-100 | CFPAC1, PK-1, PK-8, PK-45 |
| Resistant group | > 100 | PK-59, PANC-1 |
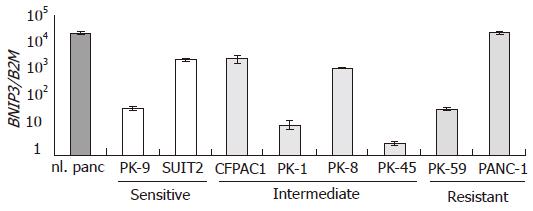
The expression of BNIP3 mRNA was examined in all the pancreatic cancer cell lines by quantitative real-time RT-PCR. Among the cell lines analyzed, BNIP3 expression levels varied by 104 fold, as shown in Figure 2. Cell lines were arranged by their IC50 values from left to right. PANC-1 showed the level of BNIP3 expression comparable to normal pancreas, but PK-45 and PK-1 showed highly suppressed levels of expression. No significant correlation was observed between the expression level of BNIP3 and the IC50 for gemcitabine.
Next we examined the expression level of BNIP3 before and after gemcitabine treatment to see if gemcitabine affects the BNIP3 expression. After administration of 1 μg/mL of gemcitabine to PANC-1, PK-45, PK-9, and SUIT2, the cells were harvested on d 0, 1, 2, and 3. No significant alterations in BNIP3 expression were observed in these cell lines (data not shown).
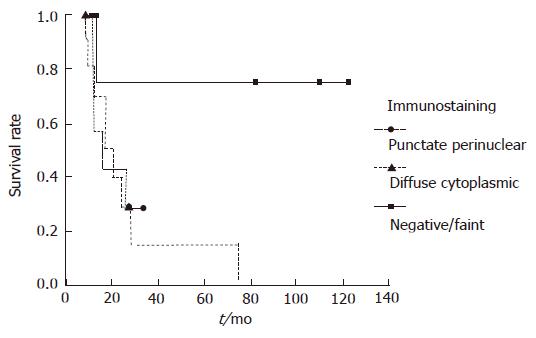
To explore the question of whether the expression level of BNIP3 affects the prognosis, we performed immunohistochemical analyses of resected tumor tissues. The staining patterns of BNIP3 were divided into three groups[15]. Representative examples are shown in Figure 3. Among the 22 evaluated specimens, four patients were classified as negative, 11 patients displayed diffuse cytoplasmic staining and 7 showed the punctate perinuclear pattern. The groups were comparable in terms of tumor morphology, stage, and year of treatment including usage of gemcitabine. Although significant differences were not calculated, a tendency between long survival of the patients and negative/faint BNIP3 expression was observed (Figure 4).
Recent studies have reported that BNIP3 expression is silenced in pancreatic cancer by hypermethylation of its promoter and that loss of BNIP3 expression contributes to chemoresistance and worsened prognosis[7,8,15]. Furthermore, siRNA-mediated knockdown of BNIP3 caused chemoresistance to gemcitabine[8]. The information was supposed to give us some clues to finding some efficient novel methods for treatment of pancreatic cancer patients. As the first step for invention of such an efficient method for treatment, we tried to confirm these reported results by comparing BNIP3 expression levels and gemcitabine chemosensitivity in several pancreatic cancer cell lines. However, our results did not support the previous report by Akada et al[8]. The expression levels of BNIP3 in our series of pancreatic cancer cell lines, which were basically the same as used in the previous study, did not reproduce the previous results; the chemosensitivities of pancreatic cancer cell lines to gemcitabine were quite different. Representative gemcitabine sensitive cell lines (CFPAC1 and SUIT2) and the representative resistant cell line (PK-59) were used in both studies, but PK-9, which was previously scored as moderately sensitive to gemcitabine, was actually classified as the most sensitive cell line in our study. PANC-1, which was reported also as “moderate” was the most resistant cell line. In addition, PK-8, which was not tested in the previous study, showed resistance to gemcitabine even with high expression of BNIP3. The cause of these discrepancies of the gemcitabine sensitivity between the two studies is unclear; the difference of the methods used in these studies, different culture media, and, probably, cell density, may have caused the discrepancy to some extent. Our immunohistochemical studies also showed results opposite to those reported by Erkan et al[15]; loss of BNIP3 expression seemed to correlate with better survival of patients.
In our present study, we could not detect any clear relationship between BNIP3 expression and chemosensitivity that might associate with a favorable patient survival rate. BNIP3 is a proapoptotic protein that was considered to have some effect on the chemosensitivity and growth of the pancreatic cancer cell in itself, so it is not difficult to think that overexpression of BNIP3 leads to cell death and that inhibition of BNIP3 leads cells survive longer. Our present results by immunostaining supported this idea, albeit no significant difference was obtained. As the number of cases increases, a significant difference will probably be observed. However, pancreatic cancer involves very complicated molecular changes, and those BNIP3-positive cancer cells that appear to be weak to the cytotoxic effect and less aggressive may have other genetic and epigenetic changes which can compensate for the proapoptotic effect of BNIP3 and eliminate the differences between BNIP3-positive and –negative cancer cells with regard to chemosensitivity and survival. Furthermore, chemosensitivities can be caused by factors other than those we have discussed so far, such as upregulation of the transportation system to eliminate the drugs from the cells or upregulation of metabolism. Any association between chemosensitivity and BNIP3 expression may yet give us a clue to find a way to invent efficient methods for treatment; further studies are necessary to explain the discrepancy.
We are grateful to Dr. BLS Pierce (the University of Maryland University College) for editorial work in the preparation of this manuscript.
| 1. | Boyd JM, Malstrom S, Subramanian T, Venkatesh LK, Schaeper U, Elangovan B, D'Sa-Eipper C, Chinnadurai G. Adenovirus E1B 19 kDa and Bcl-2 proteins interact with a common set of cellular proteins. Cell. 1994;79:341-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 373] [Cited by in RCA: 372] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 2. | Bruick RK. Expression of the gene encoding the proapoptotic Nip3 protein is induced by hypoxia. Proc Natl Acad Sci USA. 2000;97:9082-9087. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 579] [Cited by in RCA: 626] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 3. | Sowter HM, Ratcliffe PJ, Watson P, Greenberg AH, Harris AL. HIF-1-dependent regulation of hypoxic induction of the cell death factors BNIP3 and NIX in human tumors. Cancer Res. 2001;61:6669-6673. [PubMed] |
| 4. | Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 2007;26:1324-1337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1641] [Cited by in RCA: 1618] [Article Influence: 85.2] [Reference Citation Analysis (0)] |
| 5. | Chen G, Cizeau J, Vande Velde C, Park JH, Bozek G, Bolton J, Shi L, Dubik D, Greenberg A. Nix and Nip3 form a subfamily of pro-apoptotic mitochondrial proteins. J Biol Chem. 1999;274:7-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 220] [Cited by in RCA: 237] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 6. | Yasuda M, Theodorakis P, Subramanian T, Chinnadurai G. Adenovirus E1B-19K/BCL-2 interacting protein BNIP3 contains a BH3 domain and a mitochondrial targeting sequence. J Biol Chem. 1998;273:12415-12421. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 189] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 7. | Okami J, Simeone DM, Logsdon CD. Silencing of the hypoxia-inducible cell death protein BNIP3 in pancreatic cancer. Cancer Res. 2004;64:5338-5346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 178] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 8. | Akada M, Crnogorac-Jurcevic T, Lattimore S, Mahon P, Lopes R, Sunamura M, Matsuno S, Lemoine NR. Intrinsic chemoresistance to gemcitabine is associated with decreased expression of BNIP3 in pancreatic cancer. Clin Cancer Res. 2005;11:3094-3101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 116] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 9. | Fukushige S, Waldman FM, Kimura M, Abe T, Furukawa T, Sunamura M, Kobari M, Horii A. Frequent gain of copy number on the long arm of chromosome 20 in human pancreatic adenocarcinoma. Genes Chromosomes Cancer. 1997;19:161-169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 10. | Kobari M, Hisano H, Matsuno S, Sato T, Kan M, Tachibana T. Establishment of six human pancreatic cancer cell lines and their sensitivities to anti-tumor drugs. Tohoku J Exp Med. 1986;150:231-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 34] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 11. | Hata T, Furukawa T, Sunamura M, Egawa S, Motoi F, Ohmura N, Marumoto T, Saya H, Horii A. RNA interference targeting aurora kinase a suppresses tumor growth and enhances the taxane chemosensitivity in human pancreatic cancer cells. Cancer Res. 2005;65:2899-2905. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 179] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 12. | Mori Y, Shiwaku H, Fukushige S, Wakatsuki S, Sato M, Nukiwa T, Horii A. Alternative splicing of hMSH2 in normal human tissues. Hum Genet. 1997;99:590-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 13. | Xu S, Furukawa T, Kanai N, Sunamura M, Horii A. Abrogation of DUSP6 by hypermethylation in human pancreatic cancer. J Hum Genet. 2005;50:159-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 103] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 14. | Japan pancreatic society. Classification of Pancreatic Carcinoma, Second English Edition. Tokyo, Kanehara & Co., Ltd. 2003;23. |
| 15. | Erkan M, Kleeff J, Esposito I, Giese T, Ketterer K, Büchler MW, Giese NA, Friess H. Loss of BNIP3 expression is a late event in pancreatic cancer contributing to chemoresistance and worsened prognosis. Oncogene. 2005;24:4421-4432. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 172] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
Supported in part by Grants-in-Aid and the 21st Century COE Program Special Research Grant from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and by a Grant-in-Aid for Cancer Research from the Ministry of Health, Labour and Welfare of Japan
S- Editor Liu Y L- Editor Alpini GD E- Editor Yin DH